Abstract
Purpose
Causal mechanisms underlying systemic inflammation in spinal & widespread pain remain an intractable experimental challenge. Here we examined whether: (i) associations between blood C-reactive protein (CRP) and chronic back, neck/shoulder & widespread pain can be explained by shared underlying genetic variants; and (ii) higher CRP levels causally contribute to these conditions.
Methods
Using genome-wide association studies (GWAS) of chronic back, neck/shoulder & widespread pain (N = 6063–79,089 cases; N = 239,125 controls) and GWAS summary statistics for blood CRP (Pan-UK Biobank N = 400,094 & PAGE consortium N = 28,520), we employed cross-trait bivariate linkage disequilibrium score regression to determine genetic correlations (rG) between these chronic pain phenotypes and CRP levels (FDR < 5%). Latent causal variable (LCV) and generalised summary data-based Mendelian randomisation (GSMR) analyses examined putative causal associations between chronic pain & CRP (FDR < 5%).
Results
Higher CRP levels were genetically correlated with chronic back, neck/shoulder & widespread pain (rG range 0.26–0.36; P ≤ 8.07E-9; 3/6 trait pairs). Although genetic causal proportions (GCP) did not explain this finding (GCP range − 0.32–0.08; P ≥ 0.02), GSMR demonstrated putative causal effects of higher CRP levels contributing to each pain type (beta range 0.027–0.166; P ≤ 9.82E-03; 3 trait pairs) as well as neck/shoulder pain effects on CRP levels (beta [S.E.] 0.030 [0.021]; P = 6.97E-04).
Conclusion
This genetic evidence for higher CRP levels in chronic spinal (back, neck/shoulder) & widespread pain warrants further large-scale multimodal & prospective longitudinal studies to accelerate the identification of novel translational targets and more effective therapeutic strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Accumulating evidence points to inflammatory mechanisms driving the development & maintenance of chronic spinal pain [1]. Systematic reviews report higher levels of blood inflammatory markers (e.g., C-reactive protein [CRP]) in back pain [2], neck pain [2, 3] and fibromyalgia [4]. Higher CRP is associated with worse clinical outcomes in these patients, including increased pain, disability & hyperalgesia [5, 6]. Furthermore, CRP and interleukin (IL)-6 were shown to be systemically elevated in a large cohort of individuals with early-acute back pain [5], with the levels resolving at three (IL-6) and six (CRP) months later in those that recovered best over 12 months [7, 8]. Similarly, patients with acute whiplash-associated disorder [6] show raised CRP which is normalised by 3 months in those that recover, but persists in those who do not. Despite these links between systemic inflammatory responses and clinical outcomes in spinal & widespread pain conditions, the underlying specific pathways & mechanisms remain unclear.
Large-scale human genome-wide association studies (GWAS) have delivered novel insights into the mechanisms underpinning chronic pain [9, 10]. Particularly promising has been the discovery of specific shared genetic variants (i.e., genetic correlations) between chronic back & neck pain, widespread pain and a range of traits such as depression, obesity & poor sleep [10]—which helps to explain these common chronic disease comorbidities. Only recently have GWAS data been used to explore the relationship between systemic inflammation & pain conditions. Kasher et al. [11] reported genetic correlations between higher circulating CRP and an increased risk of chronic back pain. Using Mendelian randomisation analyses, they also suggested that CRP had a causal influence on back pain. However, it is not known whether CRP and other spinal pain phenotypes (e.g., neck pain) and/or widespread pain have a shared genetic basis, or whether CRP has a causal effect on these conditions.
Characterising the genetic contributions to systemic inflammation in chronic spinal & widespread pain remains a significant hurdle in better understanding their pathoetiology and developing new effective treatments. Demonstrating a causal role of such systemic factors is a critical step towards improving the management & prevention chronic spinal & widespread pain. Here, we leveraged large-scale genetic data to: (i) determine whether associations between blood CRP and chronic back, neck/shoulder & widespread pain can be explained by shared underlying genetic variants (genetic correlations); and (ii) assess whether higher CRP has a causal role in these conditions.
Methods
Study design & sample
An overview of the analysis pipeline is shown in Fig. 1. We performed GWAS of chronic back pain, neck/shoulder pain & widespread pain using the UK Biobank dataset (application number 25331) as described in prior studies [9, 10]. Briefly, the UK Biobank is a large-scale dataset comprising > 500,000 people aged 40–69 years in the United Kingdom. Data collected spans medical & psychosocial factors, blood markers, neuroimaging traits and genetics. UK Biobank holds research ethics approval from the North West Multi-centre Research Ethics Committee (Manchester, United Kingdom). All participants provided written informed consent.
GWAS dataset for chronic spinal & widespread pain phenotypes
Chronic pain phenotypes
We selected chronic back pain & neck/shoulder pain as spinal pain phenotypes, in addition to chronic widespread pain, which also encompasses pain at the back & neck (i.e., pain all over the body). In UK Biobank, neck pain & shoulder pain are pooled into one phenotype. We defined each chronic pain phenotype using self-report answers to the following questions: Have you had [back pains/neck or shoulder pains/pain all over the body] for more than 3 months? (Questionnaire field IDs: 3571/3404/2956). These questions could be answered with Yes, No, Don’t know, or Prefer not to answer. Participants who responded Yes to a question were defined as cases for that pain phenotype. We defined controls as participants who denied experiencing pain at any body site for more than three months (N = 239,125). Participants who preferred not to answer were excluded from the study. Number of cases for each GWAS were as follows: back pain (N = 79,089); neck/shoulder pain (N = 72,216); widespread pain (N = 6063) (Table 1).
GWAS analysis
GWAS analyses were undertaken using REGENIE (v1.0.6.2) [12] to assess associations between the chronic pain phenotypes & genetic variants. This method uses a logistic mixed model that accounts for cryptic relatedness by modelling a genetic relationship between individuals as a random effect. We used age, sex, genotyping array and the top 10 principal components derived from genetic data as co-variates for the GWAS analyses. Quality control procedures consisted of exclusion of variants with: (i) minor allele frequency < 0.005; (ii) imputation quality < 0.6; and (iii) deviating from Hardy–Weinberg equilibrium (P < 1E-05). We excluded individuals’ data if genotype-derived principal components 1 and 2 were > 6 standard deviations from those of the 1000 Genomes European population (i.e., data were restricted to individuals with European heritage).
Genetic correlation analyses (pain vs CRP)
We employed cross-trait bivariate linkage disequilibrium (LD) score regression to estimate genetic correlations between the three chronic pain phenotypes & blood CRP concentration. LD-score regression estimates genetic correlations across traits through leveraging the relationship between LD & GWAS summary statistics, accounting for potential sample overlap across studies [13]. These analyses were performed with the Complex Traits Genetics Virtual Lab (CTG-VL—http://genoma.io) [14] using previously published GWAS summary statistics for CRP concentration [15, 16]. CTG-VL is a publicly available web platform compiled with > 1600 GWAS summary statistics that is used for running analyses across a comprehensive range of complex traits & disorders (mostly from UK Biobank releases).
A genetic correlation (rG) ranges from − 1 to 1 and quantifies the genome-wide genetic concordance between a pair of traits. Genetic correlation estimates approaching 1 or − 1 indicate (respectively) that a proportion of genetic variants have concordant or divergent effects on both traits. Conversely, a genetic correlation that approaches 0 indicates there is little genetic concordance between the traits or that the genetic predictors of the traits are largely independent. rG estimates were calculated using two CRP traits from: (i) UK Biobank (Pan-UK Biobank—N = 400,094 [15]); and (ii) the Population Architecture using Genomics & Epidemiology (PAGE) consortium (N = 28,520 [16]). We corrected for multiple comparisons using the Benjamini–Hochberg false discovery rate (FDR < 5%) procedure, which was applied across genetic correlations for each chronic pain type GWAS.
Latent causal variable analyses (pain vs CRP)
In instances of non-zero rG estimates (FDR < 5%), latent causal variable (LCV) analyses were employed to quantify the genetic causal proportion (GCP) [17, 18], which enabled determination of whether a genetic correlation may reflect horizontal or vertical pleiotropy. That is, whether chronic pain and CRP concentrations are independently affected by the same set of genetic variants (horizontal pleiotropy) or by causal pathways (vertical pleiotropy) [19]. LCV analyses assume a latent ‘causal’ variable mediates the genetic correlation between two traits. This method quantifies the degree to which one trait is correlated with the latent ‘causal’ variable, using mixed fourth moments of the bivariate effect size distribution across all SNPs in both GWAS to estimate the posterior mean for GCP. It accounts for overlapping samples (i.e., GWAS data for chronic pain & CRP levels were both from UK Biobank). A GCP estimate ranges from − 1 to 1, whereby GCP > 0.6 indicates greater partial genetic causality for Trait A (chronic pain) on Trait B (CRP) (i.e., chronic pain has a causal effect on CRP level). A GCP < − 0.6 indicates greater partial genetic causality for Trait B on Trait A (i.e., CRP level has a causal effect on chronic pain). We applied a Benjamini–Hochberg false discovery rate (FDR < 5%) across GCPs for each chronic pain phenotype.
Generalised summary-data-based Mendelian randomisation (pain vs CRP)
As LCV analyses are unable to detect bidirectional causal relationships, we performed Mendelian randomisation analyses with the Generalised Summary-data-based Mendelian Randomisation (GSMR) method [20] implemented in CTG-VL using GWAS summary statistics of chronic pain (back, neck/shoulder, widespread pain) and that of CRP (UK Biobank & PAGE studies). GSMR was performed in a bidirectional way (i.e., to test whether chronic pain affects CRP or vice versa). For each analysis, we selected SNPs with P < 5E-08 and minor allele frequency > 1% as instrumental variables.
Results
Genetic correlation analyses (pain vs CRP)
Cross-trait bivariate LD-score regression revealed 3 out of 6 significant genetic correlations (Table 2; Figs. 2 and 3). These were between each chronic pain type and CRP from the UK Biobank. The significant rG estimates were all positive (range: 0.26–0.36; P ≤ 8.07E-09), suggesting that genetic variants associated with higher CRP concentrations were also associated with an increased risk of the respective chronic pain phenotype. No significant rG estimates were found with traits from the PAGE consortium (range: 0.00–0.05; P ≥ 0.55).
Heatmap of genetic correlations (rG) between chronic back, neck/shoulder & widespread pain phenotypes and blood C-reactive protein (CRP) levels. The direction of correlation is represented by colours (i.e., red for positive rG & blue for negative rG), with the darker shading indicating stronger associations. The asterisk denotes significant rG at 5% false discovery rate. Significant positive genetic correlations were observed between UK Biobank CRP levels and each chronic pain type, which suggests that genetic variants associated with higher CRP were also associated with an increased risk of the respective chronic pain conditions
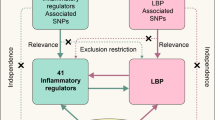
Schematic summary of key results. Significant positive genetic correlations (rG) indicated genetic variants associated with higher C-reactive protein (CRP) were also associated with chronic neck/shoulder, back & widespread pain. Genetic causal proportion (GCP) estimates were not consistent with causal relationships between CRP and the chronic pain conditions explaining these genetic correlations. Mendelian randomisation (MR) analyses suggested a bidirectional causal association between CRP & chronic neck/shoulder pain, and potential causal effects of CRP on chronic back & widespread pain. These findings replicate and expand upon the study by Kasher et al. [11], which reported a genetic correlation and putative causal effect between CRP & chronic back pain
Latent causal variable analyses (pain vs CRP)
GCP values for the three trait pairs with significant genetic correlations are presented in Table 2. For chronic back pain (GCP [S.E.] − 0.32 [0.44]; P = 0.46) and widespread pain (− 0.08 [0.76]; P = 0.92), GCP was non-significant. For chronic neck/shoulder pain (− 0.15 [0.06]; P = 0.02), GCP was significant but |GCP|< 0.6. These results suggest there were no causal relationships between CRP and chronic back, neck/shoulder or widespread pain (Fig. 3).
Generalised summary-data-based Mendelian randomisation (pain vs CRP)
Table 3 and Fig. 3 present the Mendelian randomisation results. CRP from UK Biobank showed a putative causal effect on chronic back pain (beta [S.E.] 0.077 [0.010], P = 1.32E-13), neck/shoulder pain (0.027 [0.011], P = 9.82E-03) and widespread pain (0.166 [0.032], P = 1.64E-07). Chronic neck/shoulder pain also had a putative causal effect on CRP (UK Biobank) (0.071 [0.021], P = 6.97E-04). The number of SNPs used in each analysis is shown in Table 3. There were insufficient SNPs to analyse chronic widespread pain versus CRP.
Discussion
Higher levels of blood CRP and chronic spinal (back, neck/shoulder) & widespread pain were found to have a common genetic basis, which suggests genetic variants are shared by individuals who are at risk of elevated CRP and having these pain conditions. In addition, there were genetic casual effects of CRP on back, neck/shoulder & widespread pain, and of neck/shoulder pain on CRP. These findings provide initial evidence supporting genetically driven inflammation in the development of chronic spinal & widespread pain conditions.
This study has demonstrated a genetic contribution of systemic inflammation in chronic spinal & widespread pain. It also extends upon prior studies reporting genetic correlations between CRP and back pain [11], osteoarthritis [21] & rheumatoid arthritis [21], by finding a genetic basis to higher CRP across a wider range of chronic pain phenotypes. Moreover, we have provided corroborating evidence for clinical studies showing higher CRP levels and other inflammatory markers in chronic back, neck & widespread pain [2,3,4]. Chronic musculoskeletal disorders (including spinal & widespread pain) are often co-morbid with many medical conditions that are also associated with higher CRP (e.g., obesity, cardiovascular disease) [22]. As such, the genetic correlations between chronic spinal/widespread pain & higher CRP could also reflect an underlying core basis that is shared across many chronic disease syndromes. For example, genetic correlations have been shown between CRP and obesity [21], cardiovascular conditions [21] & depression [23], while chronic spinal & widespread pain are also genetically correlated with these disorders [10].
The Mendelian randomisation results suggest a putative bidirectional causal relationship between chronic neck/shoulder pain & higher CRP, as well as a causal role of CRP in chronic back & widespread pain. These findings are consistent with a recent Mendelian randomisation study reporting a causal effect of CRP on back pain [11], while also extending upon that work by finding a causal effect on neck/shoulder & widespread pain. Together these studies provide genetic evidence for a causal pathway between CRP and chronic spinal & widespread pain. Other converging support comes from clinical & benchtop evidence of pathophysiological interactions between the immune & nervous systems in response to acute & chronic musculoskeletal conditions (reviewed in [1] & [24]). Higher levels of systemic inflammatory markers, including CRP, are often associated with worse clinical outcomes (e.g., higher pain intensity, hyperalgesia [5, 6]), which may reflect immune-mediated peripheral nociceptor sensitisation & central amplification of nociceptive signalling [24, 25]. While the clinical practice implications of this work require further investigation, within the context of increasing convergent evidence for immune contributions to the development & maintenance of chronic pain, our study has shed new light on the genetic causal factors in chronic spinal & widespread pain. Large-scale genetic analyses [10, 11, 21] also enable the identification of genetically supported risk & protective factors to enhance the design of translationally targeted diagnostic & therapeutic trials.
In the current study, GWAS analyses of chronic pain were limited to people of European ancestry, so it is unclear whether the findings can be extrapolated to non-European populations. The LCV analyses provided estimates of the likelihood of observed genetic correlations being attributable to a genetic causal effect of one trait (pain) upon another (CRP) or vice versa [18]. A drawback of this method is it cannot identify bidirectional relationships (i.e., reciprocal relationships from pain→CRP & CRP→pain). While Mendelian randomisation analyses have the capacity to identify bidirectional relationships, caution is warranted in interpreting those findings as both the chronic pain & CRP GWAS data were from the UK Biobank (overlapping samples), which can bias estimates towards the confounded associations. To mitigate this issue, we also included CRP GWAS data from the PAGE consortium, although it was a comparatively much smaller sample (N = 28,520 [16]), which likely accounted for the non-significant results with this dataset. In addition, the PAGE consortium dataset comprised people with non-European ethnicities, which may have influenced the results given the European ancestry of the chronic spinal & widespread pain GWAS data.
To further advance our understanding of systemic inflammation in chronic spinal & widespread pain, major concerted investigations are needed such as: (i) large-scale GWAS of pain-specific clinical measures (e.g., pain intensity, disability) with a wider spectrum of inflammatory markers (e.g., cytokines) in both European & non-European populations; (ii) prospective longitudinal pharmacogenomics studies examining biomarkers of treatment response (e.g., anti-inflammatory drugs); and (iii) expanding the application of Mendelian randomisation to other chronic pain diagnoses (e.g., whiplash-associated disorder, non-traumatic neck pain) and putative mechanistic subtypes (e.g., neuropathic, nociplastic) along with the use of multivariable approaches.
Conclusions
The application of statistical genetics methods to population-level data has enabled the elucidation of otherwise-intractable causal factors & directions in common chronic disorders, particularly over recent years. The genetic evidence demonstrated here for higher CRP in chronic pain conditions represents a promising foundation for further large-scale & translationally focussed studies to accelerate the development of novel therapeutic technologies & strategies.
Data availability statement
Summary statistics for the GWAS used in this study are publicly available & can be accessed in CTG-VL (https://vl.genoma.io) and CTG-VIEW (https://view.genoma.io). Results from downstream analyses for the chronic pain phenotypes reported in this study are available at CTG-VIEW.
References
Klyne DM, Barbe MF, James G, Hodges PW (2021) Does the interaction between local and systemic inflammation provide a link from psychology and lifestyle to tissue health in musculoskeletal conditions? Int J Mol Sc 22:7299
Canlı K, Billens A, Van Oosterwijck J, Meeus M, De Meulemeester K (2022) Systemic cytokine level differences in patients with chronic musculoskeletal spinal pain compared to healthy controls and its association with pain severity: a systematic review. Pain Med. https://doi.org/10.1093/pm/pnac091
Farrell SF, de Zoete RMJ, Cabot PJ, Sterling M (2020) Systemic inflammatory markers in neck pain: a systematic review with meta-analysis. Eur J Pain 24:1666–1686
O’Mahony LF, Srivastava A, Mehta P, Ciurtin C (2021) Is fibromyalgia associated with a unique cytokine profile? A systematic review and meta-analysis. Rheumatology 60:2602–2614
Klyne DM, Barbe MF, Hodges PW (2017) Systemic inflammatory profiles and their relationships with demographic, behavioural and clinical features in acute low back pain. Brain Behav Immun 60:84–92
Sterling M, Elliott JM, Cabot PJ (2013) The course of serum inflammatory biomarkers following whiplash injury and their relationship to sensory and muscle measures: a longitudinal cohort study. PLoS ONE 8:e77903
Klyne DM, Barbe MF, Hodges PW (2021) Relationship between systemic inflammation and recovery over 12 months after an acute episode of low back pain. Spine J. https://doi.org/10.1016/j.spinee.2021.09.006
Klyne DM, Barbe MF, van den Hoorn W, Hodges PW (2018) ISSLS PRIZE IN CLINICAL SCIENCE 2018: longitudinal analysis of inflammatory, psychological, and sleep-related factors following an acute low back pain episode-the good, the bad, and the ugly. Eur Spine J 27:763–777
Farrell SF, Campos AI, Kho PF, de Zoete RMJ, Sterling M, Rentería ME, Ngo TT, Cuéllar-Partida G (2021) Genetic basis to structural grey matter associations with chronic pain. Brain 144:3611–3622
Farrell SF, Kho PF, Lundberg M, Campos AI, Rentería ME, de Zoete RMJ, Sterling M, Ngo TT, Cuellar-Partida G (2022) A shared genetic signature for common chronic pain conditions and its impact on biopsychosocial traits. J Pain. https://doi.org/10.1016/j.jpain.2022.10.005
Kasher M, Williams FMK, Freidin MB, Cherny SS, Malkin I, Livshits G (2022) Insights into the pleiotropic relationships between chronic back pain and inflammation-related musculoskeletal conditions: rheumatoid arthritis and osteoporotic abnormalities. Pain. https://doi.org/10.1097/j.pain.0000000000002728
Mbatchou J, Barnard L, Backman J, Marcketta A, Kosmicki JA, Ziyatdinov A, Benner C, O’Dushlaine C, Barber M, Boutkov B, Habegger L, Ferreira M, Baras A, Reid J, Abecasis G, Maxwell E, Marchini J (2021) Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet 53:1097–1103
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM (2015) An atlas of genetic correlations across human diseases and traits. Nat Genet 47:1236–1241
Cuéllar-Partida G, Lundberg M, Kho PF, D’Urso S, Gutiérrez-Mondragón LF, Ngo TT, Hwang L-D (2019) Complex-traits genetics virtual lab: a community-driven web platform for post-GWAS analyses. bioRxiv. https://doi.org/10.1101/518027518027
Pan-UKB Team. Pan-UK Biobank (2020) Pan-ancestry genetic analysis of the UK Biobank 2020 Available from: https://pan.ukbb.broadinstitute.org
Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, Highland HM, Patel YM, Sorokin EP, Avery CL, Belbin GM, Bien SA, Cheng I, Cullina S, Hodonsky CJ, Hu Y, Huckins LM, Jeff J, Justice AE, Kocarnik JM, Lim U, Lin BM, Lu Y, Nelson SC, Park S-SL, Poisner H, Preuss MH, Richard MA, Schurmann C, Setiawan VW, Sockell A, Vahi K, Verbanck M, Vishnu A, Walker RW, Young KL, Zubair N, Acuña-Alonso V, Ambite JL, Barnes KC, Boerwinkle E, Bottinger EP, Bustamante CD, Caberto C, Canizales-Quinteros S, Conomos MP, Deelman E, Do R, Doheny K, Fernández-Rhodes L, Fornage M, Hailu B, Heiss G, Henn BM, Hindorff LA, Jackson RD, Laurie CA, Laurie CC, Li Y, Lin D-Y, Moreno-Estrada A, Nadkarni G, Norman PJ, Pooler LC, Reiner AP, Romm J, Sabatti C, Sandoval K, Sheng X, Stahl EA, Stram DO, Thornton TA, Wassel CL, Wilkens LR, Winkler CA, Yoneyama S, Buyske S, Haiman CA, Kooperberg C, Le Marchand L, Loos RJF, Matise TC, North KE, Peters U, Kenny EE, Carlson CS (2019) Genetic analyses of diverse populations improves discovery for complex traits. Nature 570:514–518
Haworth S, Kho PF, Holgerson PL, Hwang L-D, Timpson NJ, Rentería ME, Johansson I, Cuellar-Partida G (2021) Assessment and visualization of phenome-wide causal relationships using genetic data: an application to dental caries and periodontitis. Eur J Hum Genet 29:300–308
O’Connor LJ, Price AL (2018) Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet 50:1728–1734
Jordan DM, Verbanck M, Do R (2019) HOPS: a quantitative score reveals pervasive horizontal pleiotropy in human genetic variation is driven by extreme polygenicity of human traits and diseases. Genome Biol 20:222
Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, Robinson MR, McGrath JJ, Visscher PM, Wray NR, Yang J (2018) Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 9:224
Si S, Li J, Tewara MA, Xue F (2021) Genetically determined chronic low-grade inflammation and hundreds of health outcomes in the UK biobank and the Finngen population: a phenome-wide Mendelian randomization study. Front Immunol 12:720876
Fayaz A, Ayis S, Panesar SS, Langford RM, Donaldson LJ (2016) Assessing the relationship between chronic pain and cardiovascular disease: a systematic review and meta-analysis. Scand J Pain 13:76–90
Kappelmann N, Arloth J, Georgakis MK, Czamara D, Rost N, Ligthart S, Khandaker GM, Binder EB (2021) Dissecting the association between inflammation, metabolic dysregulation, and specific depressive symptoms: a genetic correlation and 2-sample Mendelian randomization study. JAMA Psychiat 78:161–170
Baral P, Udit S, Chiu IM (2019) Pain and immunity: implications for host defence. Nat Rev Immunol 19:433–447
Mustafa S, Bajic JE, Barry B, Evans S, Siemens KR, Hutchinson MR, Grace PM (2023) One immune system plays many parts: the dynamic role of the immune system in chronic pain and opioid pharmacology. Neuropharmacology 228:109459
Acknowledgements
AIC & ML were supported by a UQ Research Training Scholarship from The University of Queensland. ML thanks the support of the Commonwealth Scientific & Industrial Research Organisation through a Postgraduate Top-Up Scholarship. PFK was supported by an Australian Government Research Training Program Scholarship from Queensland University of Technology (QUT). DMK is supported by the Assistant Secretary of Defense for Health Affairs, endorsed by the U.S. Department of Defense through the FY19 Chronic Pain Management Research Program (Award No. W81XWH2010909). RECOVER Injury Research Centre (MS, SFF) receives unrestricted grant funding from the Motor Accident Insurance Commission (Queensland). The funders had no role in the design or interpretation of this study.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
SFF, MS, TTN & GC-P conceived the study. AIC & MER performed the GWAS. SFF & GC-P performed downstream analyses. All authors contributed to interpretation of results and manuscript preparation. TTN & GC-P co-supervised the study as part of a broader Precision Pain Medicine R&D program (led by TTN).
Corresponding author
Ethics declarations
Conflict of interest
GC-P contributed to this study while employed at The University of Queensland and is now an employee of Gilead Sciences. The other authors have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Farrell, S.F., Sterling, M., Klyne, D.M. et al. Genetic impact of blood C-reactive protein levels on chronic spinal & widespread pain. Eur Spine J 32, 2078–2085 (2023). https://doi.org/10.1007/s00586-023-07711-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00586-023-07711-7