Abstract
Purpose
This single-center study aimed to develop a convolutional neural network to segment multiple consecutive axial magnetic resonance imaging (MRI) slices of the lumbar spinal muscles of patients with lower back pain and automatically classify fatty muscle degeneration.
Methods
We developed a fully connected deep convolutional neural network (CNN) with a pre-trained U-Net model trained on a dataset of 3,650 axial T2-weighted MRI images from 100 patients with lower back pain. We included all qualities of MRI; the exclusion criteria were fractures, tumors, infection, or spine implants. The training was performed using k-fold cross-validation (k = 10), and performance was evaluated using the dice similarity coefficient (DSC) and cross-sectional area error (CSA error). For clinical correlation, we used a simplified Goutallier classification (SGC) system with three classes.
Results
The mean DSC was high for overall muscle (0.91) and muscle tissue segmentation (0.83) but showed deficiencies in fatty tissue segmentation (0.51). The CSA error was small for the overall muscle area of 8.42%, and fatty tissue segmentation showed a high mean CSA error of 40.74%. The SGC classification was correctly predicted in 75% of the patients.
Conclusion
Our fully connected CNN segmented overall muscle and muscle tissue with high precision and recall, as well as good DSC values. The mean predicted SGC values of all available patient axial slices showed promising results. With an overall Error of 25%, further development is needed for clinical implementation. Larger datasets and training of other model architectures are required to segment fatty tissue more accurately.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Low back pain (LBP) generally correlates with the grade of fatty infiltration of the lumbar multifidus muscle [1], independent of the weight and activity of the patient. Fatty degeneration is also present in lumbar spine segments with different modic changes, ranging from type I to III, due to disc degeneration [2]. In summary, degeneration of the lumbar paravertebral musculature seems to influence the outcomes of patients with degenerative spinal degeneration [3,4,5]. As described in more detail below, we questioned whether an automated method could reliably detect fatty infiltration in the paravertebral musculature. In recent years, artificial intelligence (AI) has been increasingly used for image analysis. In the field of medical image processing, computational models based on convolutional neural networks (CNNs) show promising results in pattern recognition and image segmentation. For this work, we specifically chose a model that uses a U-NET architecture, established as a fast and secure method for the automated semantic segmentation of biomedical images. This model uses a fully convolutional network and replaces upsampling operators with pooling operations and is specifically designed for image segmentation [6].
The implementation of such a model for semantic segmentation tasks in CT imaging precisely and reliably predicts the segmentation of subcutaneous and paraspinal muscles [7]. Approaches that implemented CNNs for MRI image processing showed that the segmentation of the sagittal spine can characterize pathologies in the intervertebral discs as well as automatically segment the spinal cord in defined single MRI slices [8]. The widespread adoption of U-net models is mainly attributed to their high utility and overall good segmentation, even with a limited amount of training data [9]. As outlined in Bardis et al., the performance of such a U-Net model directly scales with the number of available training data and may only plateau around a database of over 160 datasets, depending on the segmentation task. In response, our focus was to define reliable ground-truth data [10]. This study aims to develop a CNN to segment multiple consecutive axial magnetic resonance imaging (MRI) slices of the lumbar spinal muscles of patients with lower back pain and automatically classify fatty muscle degeneration.
Materials and methods
Study subjects
This retrospective study was approved by the local ethics committee (Approval Number: 298/20-ek) and was performed in accordance with the Declaration of Helsinki. During this process, the need for informed consent was waived. We included MRI examinations of patients with non-traumatic back pain of the lower spine. We recruited patients from 2015–2020 who underwent MRI diagnostics and were treated conservatively at our university hospital. Patients with traumatic injuries, age below 17 years, preexisting spine implants, detectable tumors, or infection were excluded. For inclusion, consecutive axial images of the lumbar spine of more than two lumbar segments from each patient were required. A total of 100 patients with 3,650 axial T2-weighed turbo spin echo (tse) sequence MRI images of the lumbar spine were included in our database.
MRI Imaging
Three different MRI scanners were used during the patient inclusion: 1.5 Tesla MRI (Aera, Siemens, Erlangen, Germany) or 3.0 Tesla MRI (Siemens Trio, Siemens or Philips Ingenia, Best, The Netherlands). All T2-weighted tse sequence of the lower spine were considered. There was no selection process concerning the quality of imaging; patients with movement artifacts or overall poor MRI quality were still included. Even though no quality metrics were collected, all included MRIs fulfilled following criteria: Good delineation of the extensor spinae muscle compartment, absence of strong motion artifacts, usable for clinical diagnosis without the need of repeating the examination.
Inclusion criteria and patient demographics
Overall, we included 100 patients selected from 133 patients with T2-weighted tse sequence MRI diagnostics in the years 2015–2020. Exclusion criteria were detection of fractures, tumors, spine implants, or infection in the examined patients at the time of MRI diagnostics. The exclusion was due to detected fractures (n = 14) and pre-existing spine implants (n = 19). Of the patient population, 55% were female (female = 55, male = 45), with a mean age of 68.1 ± 14.77 (range: 19–92). We selected patients with axial images of L1-S1 superior endplate, a minimum of 3 intervertebral segments was set as pre-condition for inclusion. Overall, we included 3650 images from 100 patients, with an average of 365 images per patient. Highest and lowest image count per patient were 19 and 140 images respectively.
Generation of ground truth labels and csv files
From the included patients, every axial T2-weighed tse sequence MRI image of the lumbar spine was extracted as a single axial slice and saved in digital imaging and communications in medicine (DICOM) format after being anonymized. Ground truth masks were created by a spine surgery specialist with ten years of clinical and radiological experience using segmentation software (Materialize-Mimics Version 22.0.0.524 Löwen, Belgium). We segmented the original images into labels of the erector spinae and multifidus muscles as a combined mask. The muscle segmentation area was defined as all tissues included by the fascia of the erector spinae and multifidus muscles. We decided to include the multifidus muscle given its high clinical relevance for lumbar spine stability [11] and the boundary between the multifidus and erector spinae is frequently indistinguishable. After muscle segmentation, individual threshold adaptation was applied to define the ground truth for the fatty tissue within the muscle group. For grading of fatty muscle degeneration of the lumbar erector spinae and multifidus muscles, a simplified Goutallier (SGC) classification system was used to differentiate between normal/mild (< 10% fat content), slight/moderate (< 50%), and severe (> 50%) fatty degeneration [12]. The classification was applied as the ground truth and the predicted fatty tissue divided by the overall muscle area. We calculated the data needed for classification from the segmented areas assessed by two independent researchers.
Labels were saved for both muscle sections, as well as separate labels for the right and left muscle sections. The same procedure was applied to fatty tissue segmentation labels. All labels were saved in three 3-channel.jpg files at 320 × 320-pixel resolution (Fig. 1c). All segmentations were controlled by a second reviewer (certified spine surgeon). Original images and labels were fitted using custom software.csv files for the direct implementation in a Python 3.6 environment to prepare the data for further processing.
Work steps for the development of the semantic segmentation model. (a) examples of MRI and ground truth segmentation masks before (original) and after applying augmentation functions (transformed), (b) Overview of the U-Net architecture, (c) Example for automatically segmented muscle tissue (left) and the respective ground data (right), (d) Visual concept for the dice similarity coefficient (DSC) metric with regards to ground truth and prediction masks
U-Net Model Training and Optimization
As our semantic segmentation model, we used a fully connected deep neural network based on the U-Net architecture (Fig. 1b) [13].
The established database of 100 patients resulted in 3650 images that were used to train segmentations models in a k-fold cross-validation study with k = 10. For each individual fold ten patients were randomly selected, and their respective images used as the test dataset. The remaining images of the database were then used as training data for the segmentation model of that particular fold. The training data was then further divided into training and validation datasets in a ratio of 80/20. Data augmentation was performed using shift, scale, and rotation operations, as well as elastic deformation and optical distortion on both images and masks (Fig. 1a) [14]. The model training was performed over 25–30 epochs. The selected hyperparameters were tuned manually prior to the study. As the objective function, we used a combination of dice and focal loss to include both a region-based model penalty for incorrect segmentation boundaries as well as a distribution-based penalty to prevent over-fitting behaviour owing to the relatively small number of training images [15]. All data processing and deep learning model computations were performed using the Pytorch V 1.7.0 framework. The development, training, and validation were performed using Windows 10 Pro, Intel I7-9700CPU, NVIDIA GTX 2060 8 GB RAM, 16 GB 3200 MHz System RAM, Visual Studio code V 1.55.1, and Python v3.6.
Statistical analysis
The segmentation results generated by the CNN-based network were compared with the ground truth labels. To validate the segmentation results, we calculated the dice similarity coefficient (DSC). We also calculated the cross-sectional area (CSA) and error of the segmented CSA for each image. Additionally, precision and recall for muscle and fatty tissues were included to further investigate the model performance. For each slice, the metrics were calculated and averaged for each patient. Additionally, the mean values for each metric were calculated across all slices (Graph Pad Prism software 7, La Jolla, USA).
Results
In total, 100 MRI of the lumbar spine with 3650 slices were used for automatic image segmentation. Descriptive data are shown in Table 1. Our U-Net-based network was able to segment 3,629 MRI slices for overall muscle area (99.45%), 3,633 slices for muscle tissue area (99.56%), and 3,507 slices for fatty tissue area (96.11%). The mean time for segmentation per image was 15 ms.
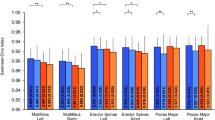
The mean segmentation area of all included MRI slices (n = 3,649) for muscle, muscle tissue, and fatty tissue did not differ significantly between ground truth and model segmentation for overall muscle but did for muscle tissue and fatty tissue (3,153.63 vs. 3,199.77 mm2, p < 0.30; 2,780.87 vs. 2,656.11 mm2, p < 0.0001; and 500.18 vs. 557.69, p < 0.0001, respectively). The DSC for overall muscle, muscle tissue, and fatty tissue was 0.91 ± 0.13 (95% CI Interval 0.90–0.91), 0.83 ± 0.14 (95% CI Interval 0.83–0.84), and 0.51 ± 0.25 (95% CI Interval 0.50–0.52), respectively (Fig. 2). Precision and recall were high for overall muscle segmentation and muscle tissue segmentation (0.92 and 0.90, and 0.82 and 0.86, respectively). Predicted segmentation of fatty tissue showed low values for precision and recall concordant with a low DSC (0.54 and 0.53, respectively). The mean CSA error resulted in similar results with low errors for overall muscle and muscle tissue segmentation (8.42 and 14.00 mm2), with larger errors for fatty tissue segmentation (40.74 mm2; Fig. 2). The mean results for all MRI slices of the datasets for overall muscle, muscle tissue, and fatty tissue segmentation prediction are shown in Table 2.
Scatterplots for dice similarity coefficient (DSC), CSA error, Violin plots for dice similarity coefficient (DSC), and cross-sectional area (CSA) error. (a) Scatterplots showing the dice similarity coefficient (DSC) and cross-sectional area (CSA) mean indicated as a line for muscle and fatty tissue; (b) Violine Plots showing the dice similarity coefficient (DSC) and cross-sectional area (CSA) median represented as a line for fatty tissue and muscle tissue, respectively
Results for the SGC showed that 75% of the patients were classified correctly by our network. None of the 100 patients in the ground truth or predicted groups were classified as SGC III. For ground truth data, 47 patients were classified as SGC I showing below 10 percent fatty tissue inside the muscle area and 53 patients as SGC II showing 10–50% fatty tissue respectively. Our network classified 54 patients as SDC I and 46 patients as SGC II, effectively underestimating segmented fatty tissue overall. Boxplots of ground truth and prediction show greater heterogeneity for SGC II supporting the thesis of bigger errors when more fatty tissue is present in the respective MRI images (Fig. 3).
Discussion
We successfully implemented a U-Net-based segmentation model to accurately identify the muscle area of the erector spinae and multifidus muscles in consecutive axial lumbar MRI slices of patients with lower back pain. To our knowledge, no studies have combined segmentation of the erector spinae and multifidus muscles with simultaneously implemented segmentation of intramuscular fatty tissue of the paravertebral lumbar muscles.
Recently, other publications showed good results using CNNs for muscle segmentation in CT imaging, with a DSC of 0.94 (standard deviation 0.04) for the erector spinae muscle group [16]. Comparatively speaking, the lower locational resolution and contrast dependencies (dependent on the weight of the patient, humidity, and others) are factors that make standardized segmentation on MRI imaging challenging [17]. For muscle fatty infiltrations in MRI imaging of the cervical spine, Weber et al. showed promising results to segment the deep cervical extensors, with a high DSC value of up to 0.887 for segmentation of the muscle area of the deep cervical spine extensors [18]. With a DSC of 0.91, our network can keep pace for segmentation of the extensor spinae compartment of the lower spine. A mean CSA error of 8% and high precision and recall underline this statement. This trend continued for the segmentation of the muscle tissue with a DSC of 0.83 and high precision and recall. Segmentation predictions of fatty tissue inside the muscle were subpar, with low DSC (0.51), precision and recall values, and the highest non-segmented slice (142 MRI slices) count without any fatty tissue identification. A reason for non-identification was the comparatively small fatty tissue area (mean, 500.18 mm2) that was more frequently present for slices in the lower lumbar area and smaller muscle areas. In segmented slices, a high mean CSA error of 40.74%, with a standard deviation of ± 106.72, showed a high variance in the segmented fatty tissue area.
We hypothesize that this effect was partly due to fatty streaks, representing most of the fatty tissue inside the muscle, which was strongly under- or over-segmented by the network. Caudal images showed lower muscle and fatty tissue areas, which could explain the poor quality of fat segmentation, especially in the lumbosacral spine (Fig. 4, lower rows).
Exemplary segmentation results of good and insufficient quality. First row: exemplary good results for automated muscle segmentation vs. ground truth label; second row: exemplary good result for fatty tissue segmentation; third row: insufficient muscle tissue segmentation vs. ground truth; fourth row: insufficient fat segmentation vs. ground truth
Comparing the overall score for the three-class SGC, 25 of 100 patients were labeled incorrectly compared to the ground truth. The high total error, mostly attributable to the low DSC in fatty tissue segmentation, opposes the direct clinical implementation of the system in this state.
We also evaluated threshold image processing for automated segmentation of fatty tissue after segmentation of the whole muscle area. Since we see great potential in the direct segmentation of fine grained areas like the fatty tissue in our use case, we decided against redevelopment. Other segmentation tasks can profit from trained networks abilitiy to segment these tissues with high accuracy and we see further potential in CNN based segmentation processes.
This study had several limitations. Since we conducted this study as a single-center study, we can only affirm results for three different MRI scanners, which limits the generalization of our results significantly. Images with low quality or noise were not excluded, which generally led to a larger potential segmentation prediction error.
Considering these limitations, segmentation prediction for overall muscle area and muscle tissue of the erector spinae and multifidus muscles were good and predicting the SGC from the mean values of all available axial slices per patient showed promising but not clinically relevant automated results.
Other architectures, especially generative adversarial networks (GANs), could be a way to improve model accuracy when segmenting more streaky areas, such as the segmentation of fatty tissue. Furthermore, the clinical correlation with age, weight, BMI, and height of the examined section of the spine would use this automated process, with more far-reaching implications for clinicians and stakeholders. Since lower back pain is the most prevalent symptom in orthopedics, we plan to expand our dataset and correlate clinical parameters with the results of fatty degeneration to gain insight into the connection between MRI-representative pathologies and patient symptoms.
Availability of data and material
The detailed datasets used during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- MRI:
-
Magnetic resonance imaging
- CNN:
-
Convolutional neural network
- AI:
-
Artificial intelligence
- ML:
-
Machine learning
- SD:
-
Standard deviation
- DSC:
-
Dice similarity coefficient
- CSA:
-
Cross-sectional area
- SGC:
-
Simplified Goutallier classification
- PACS:
-
Picture archiving and communication system
References
Kjaer P, Bendix T, Sorensen JS, Korsholm L, Leboeuf-Yde C (2007) Are MRI-defined fat infiltrations in the multifidus muscles associated with low back pain? BMC Med 5:2. https://doi.org/10.1186/1741-7015-5-2
Faur C, Patrascu JM, Haragus H, Anglitoiu B (2019) Correlation between multifidus fatty atrophy and lumbar disc degeneration in low back pain. BMC Musculoskelet Disord 20(1):414. https://doi.org/10.1186/s12891-019-2786-7
Miki T, Naoki F, Takashima H, Takebayashi T (2020) Associations between paraspinal muscle morphology, disc degeneration, and clinical features in patients with lumbar spinal stenosis. Prog rehabil med 5:20200015. https://doi.org/10.2490/prm.20200015
Duan P-G, Mummaneni PV, Guinn JMV, Rivera J, Berven SH, Chou D (2020) Is the Goutallier grade of multifidus fat infiltration associated with adjacent-segment degeneration after lumbar spinal fusion? Journal Neurosurg Spine 30:1–6. https://doi.org/10.3171/2020.6.SPINE20238
Stanuszek A, Jędrzejek A, Gancarczyk-Urlik E, Kołodziej I, Pisarska-Adamczyk M, Milczarek O et al (2021) Preoperative paraspinal and psoas major muscle atrophy and paraspinal muscle fatty degeneration as factors influencing the results of surgical treatment of lumbar disc disease. Arch Orthop Trauma Surg. https://doi.org/10.1007/s00402-021-03754-x
Kart T, Fischer M, Küstner T, Hepp T, Bamberg F, Winzeck S et al (2021) Deep learning-based automated abdominal organ segmentation in the UK Biobank and German national cohort magnetic resonance imaging studies. Invest Radiol 56(6):401–408. https://doi.org/10.1097/RLI.0000000000000755
Park HJ, Shin Y, Park J, Kim H, Lee IS, Seo DW et al (2020) Development and validation of a deep learning system for segmentation of abdominal muscle and fat on computed tomography. Korean J Radiol 21(1):88–100. https://doi.org/10.3348/kjr.2019.0470
Kim S, Bae WC, Masuda K, Chung CB, Hwang D (2018) Fine-Grain segmentation of the intervertebral discs from MR spine images using deep convolutional neural networks: BSU-net. Appl sci. 8(9):1656. https://doi.org/10.3390/app8091656
Siddique N, Sidike P, Elkin C, Devabhaktuni V (2021) U-Net and its variants for medical image segmentation: theory and applications. IEEE Access 9:82031–82057. https://doi.org/10.1109/ACCESS.2021.3086020
Bardis M, Houshyar R, Chantaduly C, Ushinsky A, Glavis-Bloom J, Shaver M (2020) Deep learning with limited data: organ segmentation performance by U net. In: Electronics 9(8):1199. https://doi.org/10.3390/electronics9081199
Ward SR, Kim CW, Eng CM, Gottschalk LJ, Tomiya A, Garfin SR, Lieber RL (2009) Architectural analysis and intraoperative measurements demonstrate the unique design of the multifidus muscle for lumbar spine stability. The J. bone and joint surg American Vol. 91(1):176–185
Slabaugh MA, Friel NA, Karas V, Romeo AA, Verma NN, Cole BJ (2012) Interobserver and intraobserver reliability of the Goutallier classification using magnetic resonance imaging: proposal of a simplified classification system to increase reliability. Am J Sports Med 40(8):1728–1734. https://doi.org/10.1177/0363546512452714
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional networks for biomedical image segmentation. Accessed on 6/19/2021 at http://arxiv.org/pdf/1505.04597v1
Buslaev A, Iglovikov VI, Khvedchenya E, Parinov A, Druzhinin M, Kalinin AA (2020) Albumentations: fast and flexible image augmentations. Information 11(2):125. https://doi.org/10.3390/info11020125
Wang P, Chung, ACS (2018) Focal Dice Loss and Image Dilation for Brain Tumor Segmentation. In: Stoyanov D, Taylor Z, Carneiro G, Syeda-Mahmood T, Martel A, Maier-Hein L et al. (Hg.): Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Bd. 11045. Cham: Springer International Publishing (Lecture Notes in Computer Science), p 119–127
Barnard R, Tan J, Roller B, Chiles C, Weaver AA, Boutin RD et al (2019) Machine learning for automatic paraspinous muscle area and attenuation measures on low-dose chest CT scans. Acad Radiol 26(12):1686–1694. https://doi.org/10.1016/j.acra.2019.06.017
Wong AKO, Manske SL (2020) A comparison of peripheral imaging technologies for bone and muscle quantification: a review of segmentation techniques. J Clin Densitom 3(1):92–107. https://doi.org/10.1016/j.jocd.2018.04.001
Weber K, Smith A, Wasielewski M, Eghtesad K, Upadhyayula P, Wintermark M (2019) Deep learning convolutional neural networks for the automatic quantification of muscle fat infiltration following whiplash injury. Scientific rep 9(1):7973. https://doi.org/10.1038/s41598-019-44416-8
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was carried out as part of the “AIQNET” Project—AI for clinical studies. It was funded by the “DLR” Project Management Agency for the Federal Ministry for Economic Affairs and Energy.
Author information
Authors and Affiliations
Contributions
DB, RB, and AV wrote the main manuscript text. CEH and AV performed the pre-segmentation of the back muscle; RB, JB, and DB wrote the code; JN evaluated measurements; JH evaluated radiological measurement; TN and CEH supervised the manuscript writing.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Ethics approval
This retrospective study protocol was approved by the ethics committee of the Medical Faculty at the University of Leipzig, Germany (Ethics Committee; 025/21-ek) and in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written consent was obtained from the participants. All relevant data generated or analyzed during the current study have been presented in the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
David Baur and Richard Bieck: Shared first authorship.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baur, D., Bieck, R., Berger, J. et al. Analysis of the paraspinal muscle morphology of the lumbar spine using a convolutional neural network (CNN). Eur Spine J 31, 774–782 (2022). https://doi.org/10.1007/s00586-021-07073-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00586-021-07073-y