Abstract
The main objective of this study was to shed light on the previously unknown arbuscular mycorrhizal fungal (AMF) communities in Southern Arabia. We explored AMF communities in two date palm (Phoenix dactylifera) plantations and the natural vegetation of their surrounding arid habitats. The plantations were managed traditionally in an oasis and according to conventional guidelines at an experimental station. Based on spore morphotyping, the AMF communities under the date palms appeared to be quite diverse at both plantations and more similar to each other than to the communities under the ruderal plant, Polygala erioptera, growing at the experimental station on the dry strip between the palm trees, and to the communities uncovered under the native vegetation (Zygophyllum hamiense, Salvadora persica, Prosopis cineraria, inter-plant area) of adjacent undisturbed arid habitat. AMF spore abundance and species richness were higher under date palms than under the ruderal and native plants. Sampling in a remote sand dune area under Heliotropium kotschyi yielded only two AMF morphospecies and only after trap culturing. Overall, 25 AMF morphospecies were detected encompassing all study habitats. Eighteen belonged to the genus Glomus including four undescribed species. Glomus sinuosum, a species typically found in undisturbed habitats, was the most frequently occurring morphospecies under the date palms. Using molecular tools, it was also found as a phylogenetic taxon associated with date palm roots. These roots were associated with nine phylogenetic taxa, among them eight from Glomus group A, but the majority could not be assigned to known morphospecies or to environmental sequences in public databases. Some phylogenetic taxa seemed to be site specific. Despite the use of group-specific primers and efficient trapping systems with a bait plant consortium, surprisingly, two of the globally most frequently found species, Glomus intraradices and Glomus mosseae, were not detected neither as phylogenetic taxa in the date palm roots nor as spores under the date palms, the intermediate ruderal plant, or the surrounding natural vegetation. The results highlight the uniqueness of AMF communities inhabiting these diverse habitats exposed to the harsh climatic conditions of Southern Arabia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Geological history and the influence of past climates of Southern Arabia (Preusser et al. 2002) have played a major role in shaping its unique environmental settings. Today, most of the area is hyper-arid (Fisher and Membery 1998), and half of the region has maximum summer temperatures exceeding 48°C (Glennie and Singhvi 2002). Agricultural ecosystems inherited infertile and light structured soils, soil salinity, drought (Ministry of Agriculture and Fisheries 1989, 1991), and high phosphorus (P) fixation (Cookson 1996). In the harsh environmental conditions of Southern Arabia, it can be assumed that the mycorrhizal symbiosis between plants and arbuscular mycorrhizal fungi (AMF) is a key factor for plant tolerance to such conditions. Moreover, native AMF communities can be expected to have distinctive physiologies adapted to withstand these conditions. As a multifunctional symbiosis (Newsham et al. 1995), the mycorrhiza is well known to play a crucial role in the P acquisition by plants (Smith and Gianinazzi-Pearson 1988). Through biochemical, biological, and biophysical mechanisms, AMF enhances soil aggregate stability (Rillig and Mummey 2006), a feature of particular relevance for the sandy soils prone to erosion. Under arid conditions, mycorrhizal plants were found also to maintain higher drought tolerance (Augé 2001) and to have better access to P than non-mycorrhizal ones (Neumann and George 2004).
The main crop in Southern Arabia is date palm (Phoenix dactylifera) (FAO 2009), the possibly most ancient crop in the world (Zaid and de Wet 1999). In addition to its high cultural and economical value, date palm still represents a survival crop (Al-Shahib and Marshall 2003) for the region and also for millions of people in many desert regions of the world. The decline of the world date production has reached an alarming level, and more research and global cooperation were called (FAO 2004). Date palms have already been shown to form AM symbiosis (Sabet 1940; Dreyer et al. 2006). Under greenhouse conditions, AMF promoted the growth of date palm seedlings, especially on nutrient-poor soils (Al-Whaibi and Khaliel 1994). AMF were also proposed as a potential biocontrol agent for bayoud disease (Jaiti et al. 2007; Jaiti et al. 2008), one of the most aggressive diseases threatening date palm (Zaid et al. 1999). It seems highly likely, therefore, that this symbiosis represents a major factor that needs to be considered in the effort to sustain productivity and fitness of date palms. However, a fundamental prerequisite for any management or promotion of this symbiosis is a basic understanding of the occurrence and diversity of AMF in the target ecosystems, because AMF diversity has significant ecological consequences: different members of the AMF community differ in their potential to promote plant fitness and to respond to environmental biotic and abiotic cues (Fitter 2005).
Such information on diversity of AMF associated with date palm in Southern Arabia or other parts of the world is scarce. To our knowledge, the only study on this topic was conducted in North Africa (Bouamri et al. 2006) and was based exclusively on the classical identification method by spore morphology. In contrast, molecular methods have the potential to detect and identify AMF directly in colonized roots. In particular, by comparing sequences from public data bases, taxa detected in one environment can be compared with those found in other ecosystems (Öpik et al. 2006).
AMF communities associated with date palms must have been shaped through a complex interaction between two main factors: firstly, the original local environmental settings, including the indigenous AMF community associated with the native plants prior to the introduction of date palms and, second, the impact of agricultural practices including the introduction of a new plant, physical soil disturbance, irrigation, and addition of chemical and organic fertilizers. It also includes the probability of accidental introduction of exogenous AMF through the agricultural inputs and machinery. AMF communities in agro-ecosystems were often found to differ from the communities in the surrounding natural habitats. While some agro-ecosystems in a hot and arid area were found to have higher AMF species richness and spore abundance (Li et al. 2007), others in temperate climate zones were found to have lower species richness and infectivity (Helgason et al. 1998; Oehl et al. 2003) compared to surrounding more natural habitats.
Our goal was to analyze AMF communities associated with date palm in Southern Arabia and to compare them with the communities associated with the local natural vegetation. Using spore morphotyping, we studied two differently managed date palm plantations at an oasis and an experimental station, a disturbed and dry area where a ruderal plant was growing at the experimental station, undisturbed habitat adjacent to the experimental station, and a distant natural habitat hosting a desert plant, located in Al-Sharqiya Sands. Additionally, molecular identification methods were applied to investigate the AMF communities colonizing the date palm roots from the two plantations.
Considering the characteristic environmental settings in the harsh climate of Southern Arabia, we hypothesized that date palms, ruderal, and natural vegetation are associated with unique AMF communities.
Materials and methods
Study habitats and vegetation
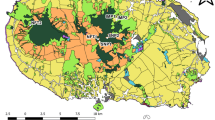
All study habitats (Fig. 1) are located within the territory of the Sultanate of Oman. These habitats are as follows:
Simplified map of Southern Arabia showing main geographical features of the area (modified from Preusser et al. 2002) including the sampling sites. The four sites are an oasis (filled triangle), an experimental station (empty triangle), an undisturbed habitat (filled circle), and sand dunes (empty circle)
Date palm plantations
The two date palm plantations were established in a sandy plain. The first was a farm located on the margin of the oasis of Al-Kamel (22°12′56″N, 59°12′9″E). It was cultivated mainly with date palms belonging to different cultivars planted randomly, among them are Khalas, Barmi, Madluki, Helali, Naghal, and Khunaizi. Most of these cultivars originated from the same oasis, but some were brought from areas in the interior of Oman around 300 km northwest. Soil traces from the original land usually are attached to the roots of the imported seedlings. Management followed local traditional farming practices since the establishment of the farm in 1992. The only source of fertilization was the manure produced on the same farm. Irrigation was performed by the traditional flooding system with water flowing from a natural source a few kilometers away through narrow channels called Aflaj (Al-Marshudi 2001; UNESCO 2006). The area between the palm trees was annually tilled using a tractor and then cultivated with Sorghum bicolor. To our knowledge, no synthetic pesticides or chemical fertilizers were used on this farm.
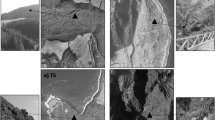
The second plantation was an experimental station (22°14′13″N, 59°11′5″E) belonging to the Ministry of Agriculture of Oman. It is situated 3 km northwest of the oasis, and it was cultivated mainly with date palms (Fig. 2a). The date palms, all of the same cultivar Khalas, were originally produced by the tissue culture (micropropagation) technique in the tissue culture laboratory of the Ministry of Agriculture of Oman located about 300 km northwest of the study site. Upon transfer to an unsterilized mixture of vermiculite and peat moss, the seedlings passed a period of weaning in a shadow house at the same laboratory. Then they were transferred to the plantation. The management followed conventional guidelines since the establishment of the station in 1990. Synthetic pesticides were used rarely, and chemical fertilizers (mainly NPK) were added according to recommended ecologically sound agricultural practices. A bubbler irrigation system was applied. All the agricultural inputs and managements, including irrigation, were directed to the root zone of the palm trees exclusively, leaving uncultivated and dry inter-cropping strip in these plantations which had lost its original native vegetation, apparently because of the mechanical disturbance but was recolonized by a ruderal plant, Polygala erioptera (Fig. 2b).
Undisturbed habitat
The third sampled site was an undisturbed habitat adjacent to the date palm plantation at the experimental station (22°14′11″N, 59°10′53″E). The natural vegetation of this habitat was also representative for the area surrounding the date palm plantation at the oasis. The vegetation consisted mainly of three perennial plant species, namely Zygophyllum hamiense (Fig. 2c), Salvadora persica (Fig. 2d), and Prosopis cineraria (Fig. 2e). The inter-plant area had traces of desiccated grass which seems to grow after rainfall and dry up during the dry season. According to the indigenous knowledge, the areas of both date palm plantations previously looked like this undisturbed natural habitat before agricultural use started.
Sand dunes
Lastly, a different natural habitat, about 50 km southwest of the three main study habitats, was chosen (21°52′39″N, 58°52′44″E). The location was at the bottom of an extinct Holocene lake (Radies et al. 2005) in the current sand sea (Preusser et al. 2002; Radies et al. 2004) that was previously called Wahiba Sands and presently Al-Sharqiya Sands. A native plant, Heliotropium kotschyi, occurring in this habitat (Fig. 2f) was included in our study.
Soil and root sampling
Soil samples were collected in August 2006 from each of the four study habitats. Four replicate plots of ca. 200 m2 were randomly chosen at each habitat. From each plot, four plants of each species were selected for obtaining a pooled sample per plant species. In the case of the date palms at the two plantations and the three perennial plant species in the undisturbed habitat, the rhizosphere soil was sampled by first digging a pit of 30-cm diameter and 30-cm depth, close to the plant stems to have access to the roots. Soil was then collected from the border of the pits in order to obtain a sample representing a vertical cross section across the root zones of these plants. Moreover, soil samples were taken below the ruderal plant P. erioptera at the experimental station, at the inter-plant area in the undisturbed habitat, and below H. kotschyi in the sand dunes by sampling the sandy soil around their root systems. This sampling procedure was chosen because the shallow root system did not allow making the soil cross-section. A total of 32 pooled soil samples were obtained as a result of 128 original samples.
For molecular analysis of AMF communities, roots of date palms were collected immediately from the sampled soil. They were washed with tap water and kept in centrifuge tubes in a cooling box for transportation. Four pooled samples were obtained from each date palm plantation. DNA was extracted from the roots as soon as they arrived to the laboratory.
In a preliminary experiment, root samples from five different date palm trees were collected in January 2004 at the experimental station.
Properties of soils from the study habitats are presented in Table 1. Analyzed soils represent pooled samples from the root zones of all plants in each habitat in exception of the experimental station where it represents only the date palms root zones.
Trap culturing the AMF
For each soil sample, two trap culture pots (0.85 L) were established. Pots were filled with 0.45 kg of an autoclaved substrate consisting of Terragreen (American aluminum oxide, oil dry US special, type III R, <0.125 mm; Lobbe Umwelttechnik, Iserlohn, Germany). The mycorrhizal inocula (50 g of soil sample) were spread as a layer on the surface of the substrate in the pots and covered with a thin layer of the substrate. Two trap culture methods were used. The first three trap plants (Plantago lanceolata, Hieracium pilosella, and Allium porrum) were randomly planted in every pot. Using more than one trap plant species was expected to increase the recovery of AMF species from the field soils, since different plants might trap different AMF. In the other trap culture system, S. bicolor was used as trap plant. Five 2-week-old AMF-free plantlets of S. bicolor were randomly planted in every pot. The trap cultures were kept in a greenhouse (temperature, 19–23°C; relative humidity, 50%) for 8 months and irrigated with an automatic irrigation system (Tropf-Blumat; Weninger GmbH, Telfs, Austria). The roots were used to assess the AMF colonization levels after 7 months of plant growth as an indicator for the infection potential.
Evaluation of AMF infection potential
The mycorrhizal colonization on S. bicolor was assessed on 30 root pieces (1-cm segments) from each replication. The root segments were incubated overnight at room temperature in 0.1% cotton blue (w/v) in lactic acid, destained in glycerol, and observed under a light microscope for quantification. Infection potential was defined as the percentage of root length colonized by AMF according to Trouvelot et al. (1986).
AMF spore isolation and identification
AMF spores occurring in each field soil sample and those propagated in the trap cultures were extracted by wet sieving and sucrose density gradient centrifugation (Daniels and Skipper 1982). Fifteen grams of air-dried field soil or 30 cm3 of harvested trap culture substrate were passed through 1000-, 500-, 125-, and 32-µm sieves. The 500-µm sieve was checked for large spores, spore clusters, and sporocarps whereas the contents of the 125- and 32-µm sieves were layered onto a water–sucrose solution (70% w/v) gradient and centrifuged at 900×g for 2 min. The resulting supernatant was washed with tap water for 2 min in a 32-µm sieve and transferred to petri dishes. Spores, spore clusters, and sporocarps obtained from all sieves were mounted on slides with polyvinyl-lactic acid-glycerol (Koske and Tessier 1983) or polyvinyl-lactic acid-glycerol mixed 1:1 (v/v) with Melzer’s reagent (Brundrett et al. 1994) and examined under a light microscope (Zeiss; Axioplan) at a magnification of up to ×400. The presence/absence of spores from all AMF species was determined for each sample. Spore abundance was assessed by measuring the total number of AMF spores extracted from the field soil samples while species richness was derived from the total number of AMF species found in the samples of both the field soils and the trap cultures.
Identification was based on original and recent species descriptions and identification manuals (Schenck and Perez 1990; the International Culture Collection of Arbuscular and Vesicular-Arbuscular Endomycorrhizal Fungi, INVAM: http://invam.caf.wvu.edu; Arbuscular Mycorrhizal Fungi (Glomeromycota), Endogone and Complexipes species deposited in the Department of Plant Pathology, University of Agriculture in Szczecin, Poland: http://www.agro.ar.szczecin.pl/~jblaszkowski/).
DNA extraction and polymerase chain reaction
Five centimeters of each date palm root sample were first ground in liquid nitrogen within a 1.5-ml microcentrifuge tube using a pellet pestle. A DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used according to the manufacturer’s instructions to extract the total DNA. A volume of 30 µl of elution buffer was used to elute the DNA. DNA extracts were used as template for the first polymerase chain reaction (PCR) reaction. PCR was performed in a nested procedure as previously described (Redecker 2000; Redecker et al. 2003) using Taq polymerase from Amersham (Basel, Switzerland), 2 mM MgCl2, 0.5 µM primers, and 0.13 mM of each desoxynucleotide. The PCR was performed using the universal eukaryote primers NS5 and ITS4 (White et al. 1990) and carried out as follows: 3 min at 94°C, followed by 30 cycles (45 s at 94°C, 50 s at 51°C, and 1 min 30 s at 72°C) and a final extension phase of 10 min at 72°C. For the second PCR, dilutions of 1:100 of the PCR products in TE buffer were used as a template, and a hot start at 61°C was performed to minimize the possibility of nonspecific amplification. Primer sequences are given in Redecker (2000) and Appoloni et al. (2008). Targeted AMF clades were Glomus group A, Glomus group B, Glomus group C, Acaulosporaceae, Archaeospora/Paraglomus, Gigasporaceae, and Paraglomus. The cycling parameters of the second PCR step were the same as those of the first one except that the annealing temperature was raised to 61°C. The second PCR step products were then run on agarose gels (2%:1% NuSieve/SeaKem, Cambrex Bio Science, Rockland, ME, USA) in Tris/Acetate buffer at 120 V for 25 min. When no second PCR product was observed, the amplification process was repeated using combinations of different dilutions of the template DNA and different dilutions of first PCR step products.
Cloning, RFLP analysis, and sequencing
PCR products were purified using the High Pure Kit from Hoffman LaRoche (Basel, Switzerland) and cloned using the pGEM-T vector (Promega/Catalys, Wallisellen, Switzerland). Thirty-four clones of each PCR product were chosen randomly, and the inserts were re-amplified. Digestion reactions of 3 h were performed using the restriction enzymes HinfI and MboI. Products of the reaction were revealed by agarose gels as described above. Representative clones of restriction types were then re-amplified and purified using the High Pure Kit and sequenced in both directions. The BigDye Terminator Cycle Sequencing Kit (ABI, Foster City, CA, USA) was used for labeling. Samples were run on an ABI 310 capillary sequencer.
Sequence analyses
The glomeromycotan origin of the sequences was initially tested by BLAST search. Using PAUP*4b10 (Swofford 2001), sequences from the present study were aligned to sequences in GenBank which showed high similarity to them, to other previously published sequences in addition to yet unpublished sequences from our own database. Phylogenetic trees were obtained by distance analysis (using the neighbor-joining algorithm) in PAUP*4b10 using the Kimura two-parameter model and a gamma shape parameter = 0.5. In the resulting phylogenetic trees, sequence phylotypes were defined in a conservative manner as consistently separated monophyletic groups. The sequence phylotypes were named based on the numerical system used to define groups detected by the group of Basel.
Statistical analysis
Significance of differences in AMF spore abundance, species richness, and infection potential between the samples coming from and within different habitats was tested using Fisher's least significant difference at P < 0.05 after one-way analysis of variance. The correlation between the AMF spore abundance and species richness and infection potential was calculated through a simple regression test. A dendrogram was obtained by cluster analysis displaying the similarity of AMF species composition across the host plants in all habitats. The group average clustering method and the squared Euclidean distance metric were used. All the statistical tests were performed using Statgraphics (version 3.1).
Results
AMF morphospecies composition
Altogether, 25 AMF taxa were detected by spore morphotyping in the 32 soil samples originating from the two date palm plantations, the disturbed inter-cropping area in the experimental station, the undisturbed habitat, and the sand dunes (Table 2). Out of the detected morphospecies, 18 species (72%) belonged to the genus Glomus, 2 species to each Scutellospora (8%) and Racocetra (8%), and only 1 species (4%) to each of the following genera: Acaulospora, Paraglomus, and Ambispora. Four of the Glomus species (Glomus sp. OMA3, Glomus sp. OMA5, Glomus sp. OMA8, and Glomus sp. OMA9) are undescribed. The most frequent species in the field samples was putatively assigned to Glomus aggregatum, with 26 occurrences in the 32 samples while the least frequent were Glomus microaggregatum, Racocetra gregaria, Acaulospora spinosa, Ambispora gerdemannii, Glomus sp. OMA7, and Glomus sp. OMA8, each with one rare occurrence. However, the most frequent species in the trap cultures was Glomus eburneum with 29 occurrences in the 32 trap culture pots. Most of the species detected in the field samples did not sporulate in the trap cultures.
As shown in Table 2, four AMF species (Glomus sinuosum, Glomus sp. OMA6, Scutellospora calospora, and Scutellospora sp. OMA10) were exclusively found in both date palm plantations. Other seven AMF species (G. microaggregatum, Glomus sp. OMA9, Racocetra fulgida, R. gregaria, A. spinosa, Glomus etunicatum, and Glomus sp. OMA13) were exclusively found in only one of the date palm plantations. G. aggregatum and G. eburneum were the only two species that were associated with all plants in the four studied habitats including the disturbed area in the experimental station. G. aggregatum, Glomus sp. OMA3, G. eburneum, and Glomus microcarpum were common to the date palm plantations and the undisturbed habitat. In this last location, A. gerdemannii, Glomus sp. OMA7, Glomus sp. OMA8, and Glomus sp. OMA 11 were detected as exclusive taxa. As a consequence of the exclusive presence of different AMF species in different habitats, two main different clusters were formed by the cluster analysis (Fig. 3) suggesting that the AMF communities under the date palms in the two plantations differ from those under the plants in the undisturbed habitat. The AMF community associated with the ruderal plant, P. erioptera, had AMF community composition resembling more to the undisturbed habitat than to that under the date palms. H. kotschyi growing in the sand dunes seems to be closer in its AMF community composition to the vegetation in the undisturbed habitat and to P. erioptera in the disturbed inter-crop area at the experimental station than to the date palms.
Dendrogram of cluster analysis based on the similarity of AMF morphospecies community composition across the host plants in all studied habitats. Two main clades were formed suggesting that AMF communities associated with date palm in the two plantations have different composition than that of vegetation in the undisturbed habitat (inter-plant area, S. persica, P. cineraria, and Z. hamiense), the disturbed area of the experimental station (P. erioptera), and the sand dunes (H. kotschyi). The group average clustering method and the squared Euclidean distance metric were used
AMF spore abundance, species richness, and infection potential
The mean spore abundance under the date palms in both plantations (58.8 and 82.3 spores at the oasis and the experimental station, respectively) was by far higher than that under the ruderal plant, P. erioptera (eight spores) and under natural vegetation in the undisturbed habitat (9.8, 7, 21.5, and 25.8 under Z. hamiense, S. persica, and P. cineraria and at the inter-plant area at the natural site, respectively; Fig. 4a). No AMF spores were found in the field soil coming directly from the root zone of H. kotschyi growing in the sand dunes. The spore abundance was higher under the date palms in the experimental station than under those in the oasis, and it varied among the different host plants in the undisturbed habitat. At this last habitat, the highest mean of spore abundance was found in the inter-plant area. This abundance was significantly higher than that found under Z. hamiense and S. persica but not that under P. cineraria.
Comparison of the effect of plant species in different study habitats on a AMF spore abundance in the field, b AMF species richness in the field and the trap cultures, and c the infection potential. Values are reported as means (±SD). Non-significant differences between the means are indicated by similar letters above their error bars
The AMF species richness shown (Table 2) represents the total number of species found in both the field and trap culture samples. Similar to spore abundance, date palms in both plantations were found to have significantly higher mean species richness (10.3 and 9.3 species at the oasis and the experimental stations, respectively) than the ruderal plant, P. erioptera (4.3 species) and the vegetation on the undisturbed habitat (4.5, 4, 6, and 5 for Z. hamiense, S. persica, P. cineraria, and inter-plant area, respectively) (Fig. 4b). The lowest mean of species richness (one species) was found for H. kotschyi growing in the sand dunes. Unlike in the case of the spore abundance, the date palms in the two plantations did not differ significantly in species richness. In the undisturbed habitat, the richness also did not vary greatly among different host plants. The highest richness at this habitat was found under P. cineraria where it was significantly higher than under both Z. hamiense and S. persica but not than at the undisturbed inter-plant area. Trap culture revealed two species associated with H. kotschyi growing in the sand dunes, G. aggregatum and G. eburneum (Table 2).
The AMF infection potential was higher in the samples from date palms of the oasis (62.6%) than from the ruderal plant, P. erioptera, of the experimental station (35.7%) and the vegetation in the undisturbed habitat (30.9%, 19%, 20.6%, 21.2% for Z. hamiense, S. persica, P. cineraria, and inter-plant area, respectively; Fig. 4c). However, there was no significant difference in the AMF infection potential between the samples from date palms (45.4%) and the ruderal plant, P. erioptera, in the experimental station and from Z. hamiense in the undisturbed habitat. The date palm samples from the two plantations had no statistically different infection potential, and no significant difference was recorded among samples from the different host plants in the undisturbed habitat. Similar to spore abundance and species richness, the lowest infection potential was found with samples from H. kotschyi growing in the sand dunes (2.7%). It was significantly lower than the infection potential of all the other plants in the other habitats except for that of S. persica growing in the undisturbed habitat.
The results of the simple regression analysis revealed a significant positive correlation between the AMF spore abundance in the field and the AMF infection potential (r = 0.676, P = 0.0001) (Fig. 5a). AMF species richness also had a positive correlation with the AMF infection potential (r = 0.729, P = 0.0001) (Fig. 5b).
Phylotypes detected in roots of date palms
The primer combinations GLOM1310-GLOM5.8R and GLOM1310-ITS4i were used successfully. These primers were designed to target Glomus group A (Schwarzott et al. 2001). In addition, the primer combination GIGA1313-GIGA5.8R, designed to target members of the Gigasporaceae, was successful in one of the samples originating from the date palms in the oasis. None of the primer combinations targeting the remaining groups of Glomeromycota yielded PCR products. Clones were selected for sequencing on the basis of their restriction fragment length polymorphism (RFLP) patterns. At least one clone representing each RFLP pattern found in each root sample was sequenced. In the samples of 2006, 263 clones were classified into 19 different RFLP patterns, and 33 representatives were sequenced. In the samples of 2004, 43 clones were classified into ten different RFLP patterns, and 16 samples were sequenced. The phylogenetic tree obtained from the sequences from the date palms at the two plantations in 2006 and from the experimental station only in 2004 is shown in Fig. 6. Based on this phylogeny, nine phylogenetic taxa colonizing the roots of the date palm were discerned. Eight of these nine taxa belong to the Glomus group A. Defining phylotypes in the Glomeromycota often causes problems because there is no molecular species concept and nuclear-encoded ribosomal RNA genes are polymorphic even within single spores (Sanders et al. 1995; Corradi et al. 2007).
Phylogenetic tree of sequences obtained in this study from the date palms in the oasis plantation (OA) in 2006 and the experimental station (ES) in 2004 and 2006 and sequences from public databases. Sequences obtained in the present study are shown in boldface and are labeled with the database accession number (e.g., GQ406077), the plantation (e.g., oasis OA), the root sample replicate (e.g., 1), RFLP pattern (e.g., A), and the sampling year. The tree was obtained by neighbor-joining analysis of 308 characters from the 18S rDNA. The numbers above the branches are neighbor-joining bootstrap values from 1,000 replications. The tree was rooted using Paraglomus occultum. Note: S. castanea, S. gregaria, and S. fulgida were all recently renamed R. castanea, R. gregaria, and R. fulgida, respectively (Oehl et al. 2008); Kuklospora colombiana is the former Entrophospora colombiana (Sieverding and Oehl 2006); Ambispora appendicula is the former Archaeospora leptoticha (Spain et al. 2006; Walker 2008)
Sequence types clustering in the GLOM-A32 group are associated with both a G. sinuosum and a Glomus clarum sequence at the same time. Some other sequences from semi-arid areas of Namibia also cluster in this group. GLOM-A33, GLOM-A38, GLOM-A39, and the Racocetra group (RACO1) are distantly related to AMF sequences. GLOM-A34, GLOM-A35, GLOM-A36, and GLOM-A37 are not closely related neither to sequences in GenBank nor in our own database. No members from Diversisporaceae, Archaeosporaceae, Acaulosporaceae, or Paraglomeraceae were detected in this study.
The GLOM A-32 phylotype was found exclusively in three out of the four root samples obtained from the date palms in the experimental station, the GLOM A37 phylotype was found in three of the four root samples from the date palms in the oasis, whereas the GLOM A-39 and RACO 1 groups each were present only in one root sample exclusively in the oasis. GLOM A-33 was found in both plantations but only in one root sample from each plantation.
Discussion
To our knowledge, this is the first study investigating AMF communities in Southern Arabia and uncovering AMF diversity associated with date palms using morphological and molecular approaches. The detection of 25 morphospecies from a limited number of habitats is remarkable, given that only two morphospecies were known so far from the whole Arabian Peninsula (Khaliel 1989).
Uncovering the AMF community associated with date palms in plantations and with the vegetation of neighboring undisturbed habitat aimed at elucidating the midterm effect of date palm introduction on the native AMF community. The underlying assumption is that the same AMF community was present in all the three studied habitats before the introduction of the date palms, and therefore, any shift in the AMF community associated with date palms would be due to the land-use change.
Date palms at both plantations had a different AMF community composition, higher spore abundance, and species richness than the vegetation at the undisturbed habitat. This suggests that there was a high impact of the introduction of date palms on the native AMF community. In spite of the mechanical disturbance of the uncultivated inter-crop area between the date palm trees in the experimental station, the AMF community under the dominant ruderal plant, P. erioptera, was closer in all the measured parameters to that of the plants in the natural vegetation of the adjacent undisturbed habitat. This is an indication that the propagules of the native AMF in the root zone of this ruderal plant were not affected by the mechanical disturbance, and therefore, the shift in the AMF communities observed in the root zone of date palms is most likely due to the agricultural inputs including irrigation.
Such an unusual trend of increasing AMF abundance and richness upon initiation of agricultural land use might be attributable to the particularity of the studied ecosystems (Ghazanfar and Fisher 1998; Glennie and Singhvi 2002). The natural undisturbed habitats are directly exposed to the harsh climate in Southern Arabia. Drought (Stahl and Christensen 1982; Cui and Nobel 1992) and heat (Koske 1987; Bendavid-Val et al. 1997) are both known for their differential effects on the biology of AMF and consequently on their community composition. Introducing agriculture with irrigation in such a habitat represents a drastic change of the environmental conditions, and thus, a dramatic shift in the AMF community is expected.
It has been reported that agricultural practices (Boddington and Dodd 2000), including soil fertilization (Johnson 1993; Toljander et al. 2008) can affect the species composition of AMF communities. Such practices can lead to a lower AMF species diversity as compared to the corresponding natural sites (Helgason et al. 1998; Oehl et al. 2003). Indeed, in the present study, some of the AMF species occurring in the undisturbed natural habitat were not detected under the date palms in the adjacent experimental station nor recovered in the respective trap cultures (Glomus sp. OMA5, Glomus macrocarpum, A. gerdemannii, Glomus sp. OMA7, Glomus sp. OMA8, Glomus sp. OMA11, and Glomus sp. OMA12). These species might have been sensitive to the habitat alterations imposed by the plantations. However, the overall species richness was higher under date palms than under the plants growing in the undisturbed habitat. A moderate P fertilization can enhance mycorrhiza formation (Bolan et al. 1984; Koide and Li 1990; Xavier and Germida 1997). Therefore, the sodium bicarbonate-extractable P levels of 71 and 93 ppm in the date palm plantations of the oasis and the experimental station, respectively, might fall within the range that promotes the mycorrhizal association (Schubert and Hayman 1986; Amijee et al. 1989). In an arid and hot ecosystem, similar observations were reported, namely that a low-input agricultural site had higher AMF diversity than an adjacent natural site (Li et al. 2007).
Some AMF species (G. sinuosum, Glomus sp. OMA6, S. calospora, and Scutellospora sp. OMA10, G. microaggregatum, Glomus sp. OMA9, R. fulgida, R. gregaria, A. spinosa, G. etunicatum, and Glomus sp. OMA13) were found exclusively under date palms, and thus, agricultural inputs might have introduced these species. Soil traces from the tree nurseries is usually attached to the roots of the imported seedlings. Thus, these AMF species might be exotic and might be able to use the new habitat with its low indigenous AMF diversity as an open ecological niche that can be easily colonized, leading to an increase in the AMF species richness.
The lowest AMF spore abundance and species diversity were found in the root zone of H. kotschyi growing in the middle of the sand sea, with only two species recovered in the trap cultures. This apparent rarity might be related to the physical instability of the sandy soil at this habitat. Uncovering a rare presence of AMF in this habitat contributes to the efforts (Dutton 1988) to understand and sustain the ecology of this sand sea ecosystem.
The degree of AMF root colonization of S. bicolor was used as an indirect estimation of the vitality and activity of the AMF community present in the studied habitats. A different response of S. bicolor to the AMF inocula obtained from under the date palms and the vegetation in the undisturbed habitat could indirectly indicate a change on the AMF community structure induced by the land-use change. A higher infection potential was found with the AMF inocula originating from the soil under the date palms than from under plants of the undisturbed habitat (Fig. 4c). This is probably due to the difference of both spore abundance (Fig. 4a) and species richness (Fig. 4b) observed at these habitats. Indeed, a positive correlation was found between AMF spore abundance and species richness and the AMF infection potential (Fig. 5a, b).
Sequences from a G. sinuosum isolate from Maryland, USA (AJ133706) and from spores obtained from field sites in a semi-dry region of Namibia and identified as G. clarum (AY285854) grouped within the GLOM A-32 clade. Another sequence from a G. clarum isolate in the database (AJ276084) was not closely related to G. sinuosum. Considering that G. sinuosum and G. clarum are easy to distinguish morphologically, this is surprising. If this discrepancy is not due to an error in assigning AY285854 to G. clarum, the most likely explanation would be that a species fitting the morphological description of G. clarum cannot be distinguished from G. sinuosum using 18S DNA sequences. We also found sporocarps of G. sinuosum in the same field but no G. clarum-like spores. Considering this fact and that sequences from cultivated isolates should be more reliable than field-collected spores, all available evidence supports assigning GLOM-A32 to G. sinuosum.
G. sinuosum was the only named species in our study that could be detected as a morphospecies in the root zone and as a phylospecies in the date palm roots. Although it was the dominant morphospecies in all the samples from date palms of both plantations, it was detected as phylospecies only in the roots of date palms growing in the experimental station. This species seems to be present in various undisturbed ecosystems. G. sinuosum-associated sequences obtained from different regions of the rDNA of AMF were found in unmanaged grassland in Denmark (Rosendahl and Stukenbrock 2004) and in a mountain meadow in Germany (Börstler et al. 2006). Its sporocarps were reported from semi-humid (Tchabi et al. 2008; Tchabi et al. 2009), semi-arid (Muthukumar and Udaiyan 2002), and arid (Shi et al. 2007) undisturbed soils and from low-input agricultural soils (Li et al. 2007). The present study adds date palm with its distinct habitat as a host for G. sinuosum.
The other morphospecies detected in the date palm rhizosphere might be potential colonizers whose life cycles are controlled by various factors (Azcón-Aguilar et al. 1999) including seasonal fluctuations of their activity (Hijri et al. 2006).
Four taxa were only distantly related to other environmental glomeromycotan phylotypes. GLOM-A33 was related to a sequence obtained from a sandy, alkaline soil (Kovács et al. 2007), two characteristics shared with the soil in the present study. The closest relative of GLOM-A38 was an AMF sequence obtained from a Swiss meadow (Sýkorová et al. 2007). GLOM-A39 was another group—in addition to GLOM-A32—that was associated with sequences obtained from roots and root zones of different plants growing in a semi-dry region of Namibia (Uhlmann et al. 2004). The closest relative to RACO 1 group was Racocetra castanea, the former Scutellospora castanea (Oehl et al. 2008).
Four other taxa (GLOM-A34, GLOM-A35, GLOM-A36, GLOM-A37) presently have no closely related sequences in the GenBank and, hence, can neither be related to taxonomically described AMF nor to environmental sequences. We hypothesize that these may be specialists for the ecosystems in Southern Arabia. This hypothesis is in agreement with the observation that there are groups which are specific to certain habitats (Öpik et al. 2006). Recently, an unusual composition of AMF community has been revealed from the distinct ecosystem of the geothermal Yellowstone National Park (USA) (Appoloni et al. 2008).
The exclusive occurrence of GLOM A-32 in the experimental station and GLOM A-37 in the oasis, both in three out of the four root samples, might indicate that these two groups are site-specialist AMF. Some studies showed a community differentiation between different sites, both in temperate (Börstler et al. 2006; Hijri et al. 2006) and in dry ecosystems (Wubet et al. 2006). This differentiation might be due to the differences in the agricultural practices that led to different soil chemical properties in the two plantations (Table 1). The two plantations differed also in date palm cultivar diversity, but whether there are cultivar-dependent differences in the association with AMF is not known. The occurrence of what seems to be site-specific taxa in these two plantations might imply that in other different agricultural systems, date palm might be associated with distinct different taxa, but to test this, a larger-scale survey is needed, involving multiple sites of each management type.
In addition to the observed unusual trend of increasing AMF abundance and richness under date palms supposedly due to agricultural land use, there are two noteworthy points to be mentioned about the composition of the revealed AMF communities. Firstly, neither any of the sequences found nor any morphospecies detected were related to the ubiquitous generalists Glomus intraradices and Glomus mosseae, the two globally most frequently found AMF (Öpik et al. 2006). We cannot completely rule out that these species might be rare in our study habitats and could potentially be revealed through a more intensive sampling effort and successive trap culturing techniques (Stutz and Morton 1996; Bever et al. 2001) but even such an unusual rarity would be an intriguing finding. Second, we found four undescribed AMF morphospecies and a considerable degree of molecular diversity that either is only distantly related to other known sequences or appears to be new to science. This community composition suggests a striking level of uniqueness most probably resulting from the characteristic environmental settings of Southern Arabia.
Uncovering AMF communities associated with vegetation in Southern Arabia and recognizing their distinctness are important steps toward a better understanding and appreciation of AMF regional and global diversity and ecology.
References
Al-Marshudi AS (2001) Traditional irrigated agriculture in Oman: operation and management of the Aflaj system. Water Int 26:259–264
Al-Shahib W, Marshall RJ (2003) The fruit of the date palm: its possible use as the best food for the future? Int J Food Sci Nutr 54:247–259
Al-Whaibi MH, Khaliel AS (1994) The effect of Mg on Ca, K and P content of date palm seedlings under mycorrhizal and non-mycorrhizal conditions. Mycoscience 35:213–217
Amijee F, Tinker PB, Stribley DP (1989) The development of endomycorrhizal root systems.VII. A detailed study of effects of soil phosphorus on colonization. New Phytol 111:435–446
Appoloni S, Lekberg Y, Tercek MT, Zabinski CA, Redecker D (2008) Molecular community analysis of arbuscular mycorrhizal fungi in roots of geothermal soils in Yellowstone National Park (USA). Microb Ecol 56:649–659
Augé RM (2001) Water relations, drought and vesicular-arbuscular mycorrhizal symbiosis. Mycorrhiza 11:3–42
Azcón-Aguilar C, Bago B, Barea JM (1999) Saprophytic growth of AMF. In: Varma A, Hock B (eds) Mycorrhiza: structure, function, molecular biology and biotechnology, 2nd edn. Springer, Berlin, pp 391–407
Bashan Y, Khaosaad T, Salazar BG, Ocampo JA, Wiemken A, Oehl F, Vierheilig H (2007) Mycorrhizal characterization of the boojum tree, Fouquieria columnaris, an endemic ancient tree from the Baja California Peninsula, Mexico. Trees Struct Funct 21:329–335
Bendavid-Val R, Rabinowitch HD, Katan J, Kapulnik Y (1997) Viability of VA-mycorrhizal fungi following soil solarization and fumigation. Plant Soil 195:185–193
Bever JD, Schultz PA, Pringle A, Morton JB (2001) Arbuscular mycorrhizal fungi: more diverse than meets the eye, and the ecological tale of why. Bioscience 51:923–931
Boddington CL, Dodd JC (2000) The effect of agricultural practices on the development of indigenous arbuscular mycorrhizal fungi. I. Field studies in an Indonesian ultisol. Plant Soil 218:137–144
Bolan NS, Robson AD, Barrow NJ (1984) Increasing phosphorus supply can increase the infection of plant roots by vesicular-arbuscular mycorrhizal fungi. Soil Biol Biochem 16:419–420
Börstler B, Renker C, Kahmen A, Buscot F (2006) Species composition of arbuscular mycorrhizal fungi in two mountain meadows with differing management types and levels of plant biodiversity. Biol Fertil Soils 42:286–298
Bouamri R, Dalpé Y, Serrhini MN, Bennani A (2006) Arbuscular mycorrhizal fungi species associated with rhizosphere of Phoenix dactylifera L. in Morocco. Afr J Biotechnol 5:510–516
Brundrett M, Melville L, Peterson L (1994) Practical methods in mycorrhiza research. Mycologue Publications, Sidney
Chapman HD, Pratt PF (1961) Methods of analysis for soils, plants and water. University of California, Berkeley
Cookson P (1996) Soil research in Oman: opportunities and priorities. SQU J Sci Res Agric Sci 1:25–32
Corradi N, Croll D, Colard A, Kuhn G, Ehinger M, Sanders IR (2007) Gene copy number polymorphisms in an arbuscular mycorrhizal fungal population. Appl Environ Microbiol 73:366–369
Cui M, Nobel PS (1992) Nutrient status, water uptake and gas exchange for three desert succulents infected with mycorrhizal fungi. New Phytol 122:643–649
Daniels BA, Skipper HD (1982) Methods for the recovery and quantitative estimation of propagules from soil. In: Schenck NC (ed) Methods and principles of mycorrhizal research. The American Phytopathological Society, St. Paul, pp 29–35
Dreyer B, Morte A, Pérez-Gilabert M, Honrubia M (2006) Autofluorescence detection of arbuscular mycorrhizal fungal structures in palm roots: an underestimated experimental method. Mycol Res 110:887–897
Dutton RW (1988) The scientific results of The Royal Geographical Society’s Oman Wahiba Sands Project 1985–1987. Journal of Oman Studies: Special Report No. 3. Diwan of Royal Court, Muscat, Sultanate of Oman
FAO (2004) Global date palm production at risk due to pests and diseases. In: FAO Newsroom. Food and Agriculture Organization of the United Nations, Rome. http://www.fao.org/newsroom/en/news/2004/48147/index.html. Accessed 12 Nov 2009
FAO (2009) FAOSTAT data. Food and Agriculture Organization of the United Nations, Rome. http://faostat.fao.org/. Accessed 12 Nov 2009
Fisher M, Membery DA (1998) Climate. In: Fisher M, Membery DA (eds) Geobotany: vegetation of the Arabian Peninsula. Kluwer Academic, Dordrecht, pp 5–38
Fitter AH (2005) Darkness visible: reflections on underground ecology. J Ecol 93:231–243
Ghazanfar SA, Fisher M (1998) Geobotany: vegetation of the Arabian Peninsula. Kluwer Academic, Dordrecht
Glennie KW, Singhvi AK (2002) Event stratigraphy, paleoenvironment and chronology of SE Arabian deserts. Quat Sci Rev 21:853–869
Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW (1998) Ploughing up the wood-wide web? Nature 394:431
Hijri I, Sýkorová Z, Oehl F, Ineichen K, Mäder P, Wiemken A, Redecker D (2006) Communities of arbuscular mycorrhizal fungi in arable soils are not necessarily low in diversity. Mol Ecol 15:2277–2289
Jaiti F, Meddich A, El Hadrami I (2007) Effectiveness of arbuscular mycorrhizal fungi in the protection of date palm (Phoenix dactylifera L.) against bayoud disease. Physiol Mol Plant Pathol 71:166–173
Jaiti F, Kassami M, Meddich A, El Hadrami I (2008) Effect of arbuscular mycorrhization on the accumulation of hydroxycinnamic acid derivatives in date palm seedlings challenged with Fusarium oxysporum f. sp. albedinis. J Phytopathol 156:641–646
Johnson NC (1993) Can fertilization of soil select less mutualistic mycorrhizae. Ecol Appl 3:749–757
Khaliel AS (1989) Mycorrhizal status of some desert plants and correlation with edaphic factors. Nippon Kingakukai Kaiho 30:231–238
Koide RT, Li MG (1990) On host regulation of the vesicular-arbuscular mycorrhizal symbiosis. New Phytol 114:59–64
Koske RE (1987) Distribution of VA mycorrhizal fungi along a latitudinal temperature gradient. Mycologia 79:55–68
Koske RE, Tessier B (1983) A convenient permanent slide mounting medium. Mycol Soc Am Newsl 34:59
Kovács GM, Balázs T, Pénzes Z (2007) Molecular study of arbuscular mycorrhizal fungi colonizing the sporophyte of the eusporangiate rattlesnake fern (Botrychium virginianum, Ophioglossaceae). Mycorrhiza 17:597–605
Li LF, Li T, Zhao ZW (2007) Differences of arbuscular mycorrhizal fungal diversity and community between a cultivated land, an old field, and a never-cultivated field in a hot and arid ecosystem of southwest China. Mycorrhiza 17:655–665
Ministry of Agriculture and Fisheries (1989) Effect of nitrogen fertilization and variety on bread wheat production under sprinkler irrigation. Agricultural Research Annual Report, Ministry of Agriculture and Fisheries, Sultanate of Oman, pp 86–98
Ministry of Agriculture and Fisheries (1991) The effect of different levels of nitrogen, phosphorus and potasch fertilizer on the growth and productivity of three perennial grass species. Agricultural Research Annual Report, Ministry of Agriculture and Fisheries, Sultanate of Oman, pp 43–54
Muthukumar T, Udaiyan K (2002) Arbuscular mycorrhizal fungal composition in semi-arid soils of Western Ghats, southern India. Curr Sci 82:624–628
Neumann E, George E (2004) Colonisation with the arbuscular mycorrhizal fungus Glomus mosseae (Nicol. & Gerd.) enhanced phosphorus uptake from dry soil in Sorghum bicolor (L.). Plant Soil 261:245–255
Newsham KK, Fitter AH, Watkinson AR (1995) Multi-functionality and biodiversity in arbuscular mycorrhizas. Trends Ecol Evol 10:407–411
Oehl F, Sieverding E, Ineichen K, Mäder P, Boller T, Wiemken A (2003) Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of Central Europe. Appl Environ Microbiol 69:2816–2824
Oehl F, de Souza FA, Sieverding E (2008) Revision of Scutellospora and description of five new genera and three new families in the arbuscular mycorrhiza-forming Glomeromycetes. Mycotaxon 106:311–360
Öpik M, Moora M, Liira J, Zobel M (2006) Composition of root-colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J Ecol 94:778–790
Preusser F, Radies D, Matter A (2002) A 160,000-year record of dune development and atmospheric circulation in Southern Arabia. Science 296:2018–2020
Radies D, Preusser F, Matter A, Mange M (2004) Eustatic and climatic controls on the development of the Wahiba Sand Sea, Sultanate of Oman. Sedimentology 51:1359–1385
Radies D, Hasiotis ST, Preusser F, Neubert E, Matter A (2005) Paleoclimatic significance of Early Holocene faunal assemblages in wet interdune deposits of the Wahiba Sand Sea, Sultanate of Oman. J Arid Environ 62:109–125
Redecker D (2000) Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza 10:73–80
Redecker D, Hijri I, Wiemken A (2003) Molecular identification of arbuscular mycorrhizal fungi in roots: perspectives and problems. Folia Geobot 38:113–124
Rillig MC, Mummey DL (2006) Mycorrhizas and soil structure. New Phytol 171:41–53
Rosendahl S, Stukenbrock EH (2004) Community structure of arbuscular mycorrhizal fungi in undisturbed vegetation revealed by analyses of LSU rDNA sequences. Mol Ecol 13:3179–3186
Sabet Y (1940) Mycorrhizal habit in the date palm (Phoenix dactylifera linn.). Nature 145:782–783
Sanders I, Alt M, Groppe K, Boller T, Wiemken A (1995) Identification of ribosomal DNA polymorphisms among and within spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol 130:419–427
Schenck NC, Perez Y (1990) Manual for the identification of VA mycorrhizal fungi, 3rd edn. Synergistic Publications, Gainesville
Schubert A, Hayman DS (1986) Plant growth responses to vesicular-arbuscular mycorrhiza.XVI. Effectiveness of different endophytes at different levels of soil phosphate. New Phytol 103:79–90
Schwarzott D, Walker C, Schüßler A (2001) Glomus, the largest genus of the arbuscular mycorrhizal fungi (Glomales), is nonmonophyletic. Mol Phylogenet Evol 21:190–197
Shi ZY, Chen ZC, Zhang LY, Feng G, Christie P, Tian CY, Li XL (2007) Diversity and zonal distribution of arbuscular mycorrhizal fungi on the northern slopes of the Tianshan Mountains. Sci China Ser D Earth Sci 50:135–141
Sieverding E, Oehl F (2006) Revision of Entrophospora and description of Kuklospora and Intraspora, two new genera in the arbuscular mycorrhizal Glomeromycetes. J Appl Bot Food Qual 80:69–81
Smith SE, Gianinazzi-Pearson V (1988) Physiological interactions between symbionts in vesicular-arbuscular mycorrhizal plants. Annu Rev Plant Physiol Plant Mol Biol 39:221–244
Spain JL, Sieverding E, Oehl F (2006) Appendicispora: a new genus in the arbuscular mycorrhiza-forming Glomeromycetes, with a discussion of the genus Archaeospora. Mycotaxon 97:163–182
Stahl PD, Christensen M (1982) Mycorrhizal fungi associated with Bouteloua and Agropyron in wyoming sagebrush-grasslands. Mycologia 74:877–885
Stutz JC, Morton JB (1996) Successive pot cultures reveal high species richness of arbuscular endomycorrhizal fungi in arid ecosystems. Can J Bot 74:1883–1889
Swofford DL (2001) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland
Sýkorová Z, Wiemken A, Redecker D (2007) Cooccurring Gentiana verna and Gentiana acaulis and their neighboring plants in two Swiss upper montane meadows harbor distinct arbuscular mycorrhizal fungal communities. Appl Environ Microbiol 73:5426–5434
Tchabi A, Coyne D, Hountondji F, Lawouin L, Wiemken A, Oehl F (2008) Arbuscular mycorrhizal fungal communities in sub-Saharan Savannas of Benin, West Africa, as affected by agricultural land use intensity and ecological zone. Mycorrhiza 18:181–195
Tchabi A, Burger S, Coyne D, Hountondji F, Lawouin L, Wiemken A, Oehl F (2009) Promiscuous arbuscular mycorrhizal symbiosis of yam (Dioscorea spp.), a key staple crop in West Africa. Mycorrhiza 19:375–392
Toljander JF, Santos-Gonzalez JC, Tehler A, Finlay RD (2008) Community analysis of arbuscular mycorrhizal fungi and bacteria in the maize mycorrhizosphere in a long-term fertilization trial. FEMS Microbiol Ecol 65:323–338
Trouvelot A, Kough J, Gianinazzi-Pearson V (1986) Evaluation of VA infection levels in root systems. Research for estimation methods having a functional significance. In: Gianinazzi-Pearson V, Gianinazzi S (eds) Physiological and genetical aspects of mycorrhizae. INRA Press, France, pp 217–221
Uhlmann E, Görke C, Petersen A, Oberwinkler F (2004) Arbuscular mycorrhizae from semiarid regions of Namibia. Can J Bot 82:645–653
UNESCO (2006) World Heritage, Aflaj: irrigation systems of Oman. United Nations Educational, Scientific and Cultural Organization. http://whc.unesco.org/en/list/1207. Accessed 2 May 2010
Walker C (2008) Ambispora and Ambisporaceae resurrected. Mycol Res 112:297–298
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–322
Wubet T, Weiß M, Kottke I, Oberwinkler F (2006) Two threatened coexisting indigenous conifer species in the dry Afromontane forests of Ethiopia are associated with distinct arbuscular mycorrhizal fungal communities. Can J Bot 84:1617–1627
Xavier LJC, Germida JJ (1997) Growth response of lentil and wheat to Glomus clarum NT4 over a range of P levels in a Saskatchewan soil containing indigenous AM fungi. Mycorrhiza 7:3–8
Zaid A, de Wet PF (1999) Origin, geographical distribution and nutritional values of date palm. In: Zaid A, Arias-Jimenez EJ (eds) Date palm cultivation. Food and Agriculture Organization (FAO) (FAO Plant Production and Protection Paper No. 156). FAO, Rome, pp 29–44
Zaid A, de Wet PF, Djerbi M, Oihabi A (1999) Diseases and pests of date palm. In: Zaid A, Arias-Jimenez EJ (eds) Date palm cultivation. Food and Agriculture Organization (FAO) (FAO Plant Production and Protection Paper No. 156). FAO, Rome, pp 223–278
Acknowledgments
The authors would like to thank Kurt Ineichen and Maria Teresa Della Beffa for their advices in setting up and maintaining the trap culture systems; Sarah Symanczik and Sally Koegel for their assistance in the isolation of the AMF spores; Valeria Bianciotto and Massimiliano Del Pero for the general useful advices; in Oman, Ahmad Al-Bakri for the formal and logistical support; Hamood Al-Hashmi for his support during the sampling trips; and Annette Patzelt (Oman Botanic Garden) for the plant identification. The work was supported by complementary grants from the Universities of Torino and of Basel, the Ministry of Agriculture of Oman and the Swiss National Science Foundation, which are gratefully acknowledged.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Al-Yahya’ei, M.N., Oehl, F., Vallino, M. et al. Unique arbuscular mycorrhizal fungal communities uncovered in date palm plantations and surrounding desert habitats of Southern Arabia. Mycorrhiza 21, 195–209 (2011). https://doi.org/10.1007/s00572-010-0323-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-010-0323-5