Abstract
Correct anther development is essential for male fertility and subsequently agricultural yield. Defects in anther development range from the early stage of stamen formation until the late stage of tapetum degeneration. In particular, the specification of the four distinct somatic layers and the inner sporogenous cells need perfect orchestration relying on precise cell–cell communication. Up to now, several signals, which coordinate the anther´s developmental program, have been identified. Among the known signals are phytohormones, environmental conditions sensed via glutaredoxins, several receptor-like kinases triggered by ligands like MAC1, and small RNAs such as miRNAs and the monocot-prevalent reproductive phasiRNAs. Rather than giving a full review on anther development, here we discuss anther development with an emphasis on mobile elements like ROS/oxygen, secreted proteins and small RNAs (only briefly touching on phytohormones), how they might act and interact, and what the future of this research area might reveal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthers are intricately formed reproductive structures which enable the creation and dispersal of male gametophytes. Anthers are thus an important factor for fertility and agronomic yield. Since anthers and anther defects are easily visible particularly in maize, early observations identified multiple classical genes required for anther development and male meiosis (partly re-analyzed and summarized by Timofejeva et al. 2013).
In general, studies provided descriptive insights about the signals orchestrating timely formation of certain anther layers, their redifferentiation, and about synchronized development of the different layers and the archesporial cells which develop into meiocytes and finally pollen in the center of the anthers. Key players and developmental programs for early anther development have been extensively studied and described (reviewed in: Timofejeva et al. 2013; Kelliher et al. 2014; Walbot and Egger 2016; van der Linde and Walbot 2019) (Table 1). Other studies gave insight into late anther and pollen development (for example, see: Dickinson and Bell 1976; Schreiber et al. 2004), and by now also provide a sound understanding about involved transcription factors and components for the tapetal cell death and pollen wall formation (for example, Huo et al. 2020; Xiong et al. 2020; Uzair et al. 2020; Lu et al. 2020). A recurring observation points to the high importance of the tapetum layer for successful pollen maturation and release (Cigan et al. 2001; Ito and Shinozaki 2002; Albrecht et al. 2005; Colcombet et al. 2005).
For very detailed insight into especially early anther development, we refer to previous reviews (Walbot and Egger 2016; van der Linde and Walbot 2019). What is currently poorly addressed is how the already known and partly characterized signals, namely transcription factors, ROS, peptides, and small RNAs, interact with and depend on each other. Here, we shortly lay out steps in formation of anther morphology (Fig. 1a), recapitulate knowledge about mobile elements (Table 1), and then offer some conclusions. The main focus of this article is to provide testable hypotheses (Figs. 2, 3) plus further questions with emphasis on ROS, secreted peptides, and small RNAs which could help to direct future research in the field.
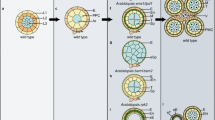
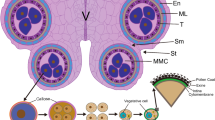
Schematic developmental progression of maize anthers and key mobile elements involved in the process. a Schematic cross sections or half-cross section of different anther developmental stages, ranging from before lobe specification, through mitotic proliferation to microgametogenesis. All anther cells date back to L1-d, L2-d cells and in dicots L3-d cells. L1-d become epidermis, while L2-d/L3-d cells form the vasculature, connective tissue and all lobe tissues. b Schematic timeline of anther lobe development. c Mobile elements known to occur during anther development. The approximate timing and levels of occurrence are depicted schematically by filled areas. Circles represent anther lobes, with gradients or layer-specific location shown by corresponding colors. Colors used are selected due to the colors in panel b, with mobile elements arising or occurring in the respective color-coded layers. AR: archesporial, EN: endothecium, EPI: epidermis, TP: tapetum
Schematic overview on proposed signal mobility and specificity in cell–cell communication. Some signals can freely diffuse (O2), while others migrate through plasmodesmata, e.g., transcription factors and small RNAs. Also, small RNAs might be transported in vesicles between cells. Proteins and ligands are secreted by unconventional and conventional secretion into the plant apoplast. There they can interact, for example, with receptor-like kinases or might be taken up by other cells. All these modes of mobility can result in signal gradient formation along tissues. Mobility can be limited by size exclusion of plasmodesmata or capture in certain tissues. Specificity of signals can be achieved by expression of response molecules in certain cell layers
Speculative interconnection of mobile signals during anther development. a, b 21nt phasiRNAs are generated in the epidermis early in lobe differentiation. Consecutively, they will migrate toward the lobe center, load onto AGO18, and presumably degrade mRNAs for, e.g., a RLKs or b transcription factors in the emerging cell layers. c, d 24nt phasiRNAs emerge later, when all cell layers are formed, and derive from the tapetum. Both 21nt and 24nt phasiRNAs accumulate in PMCs and the surrounding tapetum. In the tapetum, they might be involved in timely initiation of PCD via gene silencing, by, e.g., DNA methylation of the HXK promoter, and via degradation of RNA for, e.g., RLKs involved in PCD initiation. Once phasiRNAs decrease, PCD could then occur. In PMCs, phasiRNAs seem to mediate DNA methylation, and thus/or independently promote early meiosis initiation with DSBs. DSB: double strand break, AR: archesporial, EN: endothecium, SP: secondary parietal, ML: middle layer, TP: tapetum, PMCs: pollen mother cells; ROS: reactive oxygen species, PCD: programmed cell death, HXK: HEXOKINASE
Morphology of anther development
In most angiosperm anthers, four lobes surround the connective tissue and the central vasculature. Within each lobe, layers of sub-epidermal somatic cell types (endothecium, middle layer, and tapetum) encircle the centrally located archesporial cells (AR cells), which later become pollen mother cells (PMCs; “meiocytes” during meiosis) and then pollen (Fig. 1a, b). The somatic cell layers are often only one cell thick, and all four layers are needed for successful male germline development, pollen maturation, and dispersal in most angiosperms.
Anther development can be separated into two distinct stages: (1) establishment of anther layers, followed by meiosis, and (2) pollen grain differentiation with accompanying re-differentiation and programmed cell death of certain anther layers (Goldberg et al. 1993). All anther cells trace back to the floral meristem and are L1-derived (L1-d) for the epidermis and plant-type-dependent L-2d and/or L-3d for all sub-epidermal cells (Goldberg et al. 1993; Kelliher and Walbot 2011). Based on stamen primordium location, initial adaxial-abaxial stamen patterning is established. This pattern is then rearranged in the anther resulting in a cross pattern of domains, where outgrowths between domains differentiate into lobes. At lobe inception, L2-d cells are pluripotent (Fig. 1a, b). A subset of L2-d cells differentiates into archesporial (AR) cells first. AR cells stimulate their neighboring L2-d cells to adopt a somatic fate through periclinal cell division. This generates two layers, the endothecium (EN) and secondary parietal cells (SPCs), establishing the somatic niche. The SPCs start to divide periclinally, forming two additional layers, the middle layer (ML) and tapetum (TP). This step is followed by cell proliferation, and differentiation of AR cells into pollen mother cells (PMCs) (for detailed review on premeiotic anther development see Walbot and Egger 2016). Meiosis (microsporogenesis) begins with the expansion of the individual PMCs which then undergo two rounds of cell division producing tetrads of haploid microspores (for review see Mercier et al. 2015). Callose deposition starts to isolate PMCs from each other later in meiosis, ending with microspores enclosed by a unique callose cell wall. Also, at the end of meiosis, the ML is mechanically crushed or degraded, and then, there is a short relapse phase when the callose wall is degraded by enzymes secreted from the tapetum to release the individual microspores. At this stage, the developing pollen is coated by exine components secreted by the tapetum, namely lipase and glycine-rich proteins (Lu et al. 2020) Afterward, the tapetum undergoes programmed cell death (PCD), with its cell remnants forming the pollen kitt on the maturing pollen. A first mitotic division of the microspore without cytokinesis results in a vegetative and a generative nucleus. A second mitotic division of the generative cell gives rise to two sperm cells. Dependent on the plant species, pollen mitosis II occurs within the anther or later during pollen tube growth, resulting in tricellular or bicellular pollen, respectively. For pollen release, first the septum between the two lobes within a theca and then the epidermis are degraded (for review see Hafidh et al. 2016). Obviously, all these developmental steps need a high degree of spatiotemporal coordination to allow proper development of the anther and the pollen within it.
Challenges of anther development
Stamens lack a general central organization unit, i.e., meristem, and cells must self-organize. Curiously, stamens are considered modified leaves, with the same need to generate adaxial-abaxial patterns though in a more complicated fashion (reviewed in Walbot and Egger 2016). The lack of a meristem is reminiscent of the situation in both plant and animal embryos where internal polarization and positional cues serve to create basal/apical or anterior/posterior ends, respectively, with support by asymmetric cell divisions in self-organization (reviewed in Peris et al. 2010; White et al. 2018). In contrast, plant meristems contain stem cells that initiate and organize their surrounding cell populations as in animal stem cells (reviewed in Heidstra and Sabatini 2014). However, similar to meristem-free organ development, key molecules include secreted peptides, their receptors, and transcription factors.
Since anther development requires proper timing of events, in correct order and often synchronized between layers, signals need to coordinate anther development in a spatiotemporal fashion: (1) giving each step in development sufficient time while moving on to the next step as soon as possible, (2) ensuring the formation and development of separate layers, and (3) coordinating simultaneous progress in, e.g., tapetal development and PMCs (pollen mother cells).
Mobile elements in anther development
Multiple mobile elements or their gradients along the anther radius shape anther development: Here, we focus on oxygen, secreted peptides and small RNAs (Fig. 1c).
Plant hormones are suitable candidates for shaping anther development as well, and seem to act especially very early and later than the core morphology-forming stages. A recent review covers the roles of gibberellin and jasmonate in stamen development (Marciniak and Przedniczek 2019). For example, gibberellic acid and jasmonic acid play roles in pollen formation and release via anther dehiscence (Zhao and Ma 2000; Cheng et al. 2004). Similarly, ethylene production peaks at degeneration of tapetum and middle layer, and at pollen maturation and dispersal (Kovaleva et al. 2011). Auxin participates in anther development, dehiscence, pollen maturation and filament elongation via ARFs (AUXIN RESPONSE FACTORS) (Nagpal et al. 2005; Wu et al. 2006; reviewed in Cardarelli and Cecchetti 2014) which will be further mentioned as small RNA targets below. Furthermore, Brassinosteroids have also been described in connection with ROS regulation in anther development as detailed below (Yan et al. 2020). In contrast, during early anther development, signaling is not mainly based on phytohormones (Walbot and Skibbe 2010; Zhang et al. 2014; van der Linde and Walbot 2019) but rather on environmental clues and other small mobile signals which orchestrate cell–cell communication (Kelliher and Walbot 2012; Zhang and Yang 2014; van der Linde et al. 2018b).
Oxygen
In developing monocot anthers, hypoxic conditions are a natural result of rapid cell proliferation in flower organs and of the flowers being tightly encased by a whorl of not-yet-photosynthetic leaves. Consequently, an oxygen gradient is formed within the anther (Fig. 1c). In maize, this oxygen gradient can be manipulated by flushing the tassel buried in the leave whorl with O2 or N2 (Kelliher and Walbot 2012). Treatment with nitrogen shifted the hypoxia gradient more toward the outer cells of the lobes, where then ectopic AR cells specified. AR formation also dislocated by manipulation of redox status within the anther through treatment with reducing or oxidizing agents. Thus, oxidizing agents bias AR formation to the connective and vascular tissues, whereas reducing agent treatment results in subepidermal or epidermal ectopic AR cells (Kelliher and Walbot 2012).
The maize mutant with the earliest developmental defect after anther primordial formation is Zmmsca1 (male sterile converted anther1) (Chaubal et al. 2003; Albertsen et al. 2011; Kelliher and Walbot 2012). In this mutant, L2-d cells fail to progress to archesporial fate and instead undergo longitudinal divisions and differentiate as vasculature but can be rescued by treatment with reductive agents (Kelliher and Walbot 2012). ZmMSCA1 encodes a CC-type glutaredoxin, and homologs of ZmMSCA1 have been identified in rice (MICROSPORELESS1, OsMIL1) and the dicot plant Arabidopsis thaliana (AtROXY1 and AtROXY2) (Xing and Zachgo 2008; Hong et al. 2012a, b). Osmil1 mutant development is disrupted after archesporial, endothecium, and secondary parietal layer formation (Hong et al. 2012b). Deletion of Atroxy1 and Atroxy2 prevents formation of normal parietal cells as well as pollen release, while meiosis is still induced (Xing and Zachgo 2008). It thus seems that the mutant defects occur at different developmental times and differentiation steps, maybe due to differences in the spatiotemporal redox gradients among species. Several studies provide evidence that CC-type glutaredoxins modify bZIP transcription factors of the TGACG (TGA) motif-binding family, thereby regulating their activity and downstream processes (Xing et al. 2005; Ndamukong et al. 2007; Li et al. 2009, 2011; Murmu et al. 2010; Hong et al. 2012b).
While oxygen and subsequently ROS levels are extremely low during early anther development, and by that trigger AR formation, they increase strongly when the middle layer and tapetum are formed (Yang et al. 2018), and are elevated when tapetum cell death occurs (Fig. 1c). Even though ROS have functions in signaling, at this stage their destructive potential toward proteins, DNA, and lipids seems to ultimately cause tapetal cell death (for detailed review about ROS in rice anther development see Yu and Zhang 2019). Tapetal cell death and thus ROS production need to be precisely timed to allow proper pollen formation. This seems to be achieved by at least two kinds of mobile elements, small RNAs and a phytohormone: ROS-mediated tapetal PCD is brassinosteroids (BR) and most likely small RNA dependent by modulation of ROS producing enzyme expression. Tomato BRASSINAZOLE RESISTANT 1 (SlBZR1), a BR signaling regulator, directly binds to the promoter of SlRBOH1 (RESPIRATORY BURST OXIDASE HOMOLOG) and by that increases SlRBOH1-dependent ROS production (Yan et al. 2020). Besides SlBZR1, the argonaute OsAGO2 was shown to modulate expression of an enzyme involved in tapetal ROS generation. OsAGO2 facilitates DNA methylation in the promoter region of OsHXK (hexokinase) and by that subsequent ROS homeostasis is controlled. The Osago2 mutant is defective in anther development, and downregulation of OsAGO2 causes ROS overaccumulation and premature PCD of the TP (Zheng et al. 2019b).
Taken together, anther development seems to make good use of the naturally occurring spatiotemporal oxygen/ redox gradient across anther lobes. In the beginning, when the anther primordia and surrounding leaf whorl are very dense, only low oxygen levels exist, which enable signaling. Later, oxygen and ROS levels increase in the anther, and ROS (likely specifically produced at certain layers like the tapetum) can then trigger PCD to release pollen from the anther.
Small, secreted proteins
Besides the above-mentioned signals that induce ROS accumulation, tapetal degeneration seems to be also regulated via signaling by ligand–receptor interactions, as there are two rice RLKs (receptor-like kinases), OsTMS10 and OsTMS10L (THERMO-SENSITIVE GENIC MALE STERILE 10 and ~ -LIKE) that are needed for tapetal degeneration and male fertility, especially at high temperature (Fig. 1c) (Yu et al. 2017). Several more RLK mutants have defects in anther development, resulting in male sterility (for reviews see Zhao 2009; Kelliher et al. 2014; Cai and Zhang 2018). These are Ater/erl1/erl2 (erecta/ ~ like1/2) triple mutants with absent or defective anthers (Hord et al. 2008), Atserk1/serk2 (somatic embryogenesis receptor-like kinase1/2) double mutants with excess AR cells and absent tapetum (Colcombet et al. 2005), Atrpk2 (receptor-like kinase2) which lacks the ML (Mizuno et al. 2007), and Atbam1/bam2 (barely any meristem 1/2) double mutants where mutant anthers have larger but fewer L2-d cells, only form AR-like cells and no internal somatic cells, and have shriveled appearance (Hord et al. 2006). AtSERK1 and 2 were proposed to form heterodimeric receptors with AtEMS1/EXS since their mutant phenotypes are the same (Colcombet et al. 2005). Proteomic data revealed more RLKs in developing anther cells (Ye et al. 2015, 2016), and their ligands are assumed to be small secreted proteins, but not yet identified. Potential ligand candidates can be found in the CLE (CLAVATA3/ESR-RELATED) group, for example. This idea is based on GUS-expression reporter lines in A. thaliana, which found six AtCLEs to be expressed in anthers: AtCLE1 in the tapetum and in pollen grains, AtCLE7 in anther lobes, AtCLE11 in mature pollen, AtCLE12 in all stages of pollen development, AtCLE13 in young anthers, and AtCLE25 throughout anther development in lobes (Jun et al. 2010).
The only ligand–receptor pair identified in anther development so far has been reported in both monocots and dicots: AtTPD1 (TAPETUM DETERMINANT (1): AtEMS1/EXS (EXCESS MALE SPOROCYTES 1/ EXTRA SPOROGENOUS CELLS) in A. thaliana, OsTDL1A (TPD1-LIKE 1A): OsMSP1 (MULTIPLE SPOROCYTE1) in rice, ZmMAC1 (MULTIPLE ARCHESPORIAL CELLS 1): ZmMSP1 in maize (Fig. 1c). Although cytological phenotypes differ among plant species, all receptor knockout mutants are male sterile (Sheridan et al. 1999; Nonomura et al. 2003; Yang et al. 2003, 2016; Wang et al. 2012a; Huang et al. 2016; van der Linde et al. 2018a). In maize, newly specified AR cells secret ZmMAC1 in its 218 aa form, which is recognized via the RLK ZmMSP1 of the surrounding L2-d cells (Wang et al. 2012a; van der Linde et al. 2018a). L2-d cells undergo a periclinal division and acquire the cell fate to form EN and SPCs when receiving the ZmMAC1 signal (van der Linde et al. 2018a). Notably, each individual L2-d cell has to receive the ZmMAC1 signal to facilitate this step as it was shown by microinjecting ZmMAC1 into the anther cell apoplast using the Trojan horse method (van der Linde et al. 2018a,b; Fiedler et al. 2019). This might mean that target cells are either not connected with each other, or that downstream signals and components are not transmitted via plasmodesmata (Fig. 2).
Small RNAs
Peptides are well known as morphogens, acting by forming a gradient. Small RNAs (sRNAs) might play similar roles, as suggested by Benkovics and Timmermans (2014). Not many specific sRNAs regulating anther development are known but is clear from the characterized miRNAs (see below) that they play important roles. Similar to miRNAs, tasiRNAs (trans-acting small interfering RNAs) have specific target genes which they regulate by mRNA degradation. tasiRNAs are a subclass of phasiRNAs, which are named according to their biogenesis since they are processed from longer dsRNA precursors into consecutive fragments of 21 or 24nt length. Another phasiRNA subclass, i.e., highly abundant reproductive phasiRNAs in mainly monocots, has the potential to include further yet undescribed tasiRNAs supporting anther development (Zhai et al. 2014).
miRNAs
A few key miRNAs for anther development have been studied, but for most of them it is unknown whether they are mobile between cells. The general importance of miRNAs for anther development is evident through mutants of the miRNA biogenesis factor DCL1 (DICER-like1). The maize Zmdcl1 mutant “fuzzy tassel” is defective in miRNA biogenesis and is male sterile due to defects in late anther maturation and dehiscence (Field and Thompson 2016). The Arabidopsis Atdcl1 mutant has still viable pollen but partly absent or abnormal anthers with less than four lobes (Jacobsen et al. 1999). Specific miRNAs generated by AtDCL1 might thus play roles in early anther formation and later anther maturation and dehiscence, for example, miR165/6, miR167 and miR156 as described below.
miR165/6 is a specific miRNA with roles in anther development, and its expression patterns suggest that it is mobile. Before lobe initiation, miR165/6 accumulates in lateral-adaxial regions of stamen and then in the four newly formed corners and afterward—but before TP differentiation—in the four lobe centers (Li et al. 2019a). MiR165/6 overexpression in Arabidopsis highlights the early role of this miRNA in shaping the anther structure through the formation of internal boundaries since only two instead of four lobes emerge (Li et al. 2019a). Additionally, studies in Arabidopsis showed that MiR165/6 targets AtPHB (PHA-BU-LO-SA) which in turn binds to the promoter of AtSPL/NZZ (SPOROCYTELESS/NOZZLE) to activate it for its role in microsporogenesis (Li et al. 2019a). In Atspl/nzz mutants, EN and TP cannot form, and AR cells do not progress to meiosis (Yang et al. 1999; Liu et al. 2009b).
Mobility between cells has been suggested for AtmiR167, which seems to be expressed in vascular cells, but retained in connective cells (Wu et al. 2006). If its targets, AtARF6 and 8 (AUXIN RESPONSIVE FACTOR) are immune to downregulation by miR167, connective cells become very large, locules do not break, and no dehiscence occurs (Wu et al. 2006). The same is the case for AtmiR167 null mutants, and together with other data, this led to the model where miR167 arrests growth to permit for more time for anther dehiscence (Zheng et al. 2019a).
Another miRNA in early anther development, AtmiR156, targets transcription factors of the SPL (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE) family which in turn regulate genes for cell division, differentiation and specification (Xing et al. 2010). Expression of AtmiR156 arises dynamically from multiple loci, but mobility and source and sink cells have not yet been addressed.
tasiRNAs
Anthers need to initially establish adaxial-abaxial patterns, which are known to be shaped by gradients of specific tasiARFs during leaf development (Chitwood et al. 2009). Similarly, a rice mutant of the tasiRNA production enzyme OsSHL2 (SHOOTLESS 2, an RNA-dependent RNA polymerase) has adaxial-abaxial anther patterning defects (Toriba et al. 2010), pointing to the use of tasiRNA gradients as a general developmental strategy in different organs, including anthers.
Until very recently, no specific tasiRNAs have been implied in reproductive processes, but vegetative phasi/tasiRNA targets indicated that reproductive phasi/tasiRNAs might be good candidates to regulate processes in anther development as well: (1) TasiRNAs from disease-related NB-LRR loci regulate NB-LRRs and thus subsequent PCD (Zhai et al. 2011; reviewed in Cui et al. 2015), (2) tasiRNAs target transcription factors of the NAC and MYB classes (reviewed in Liu et al. 2020), and, most importantly, (3) a recent study in tomato revealed tasiRNAs/non-reproductive phasiRNAs to target multiple RLKs (Luan et al. 2020). In reproductive tissues, most studies could either not predict or prove that phasiRNAs target and degrade other mRNAs (Song et al. 2012; Zhai et al. 2015; Patel et al. 2018). One study in pummelo predicted and verified targets in trans with gene ontologies of broad terms but also including PCD and pollen development (Fang et al. 2020). By now, two pioneering study in rice meiocytes finally found indeed RNA degradation of target genes of reproductive 21nt-phasiRNAs (Jiang et al. 2020; Zhang et al. 2020c). In rice meiocytes, target genes were enriched for carbohydrate biosynthesis and metabolism, and also included OsDMC1B, a meiotic gene (Jiang et al. 2020). Another study gained insight on genes targeted for degradation in rice anthers through degradome data from whole rice spikelets of WT and of mutants in MEL1, the rice AGO5 which binds phasiRNA. Intriguingly, many RLKs were identified as phasiRNA target genes, among them MSP1 (Zhang et al. 2020c). This re-defines part of the reproductive phasiRNAs as tasiRNAs, with specific target genes with roles and/or high abundance in meiosis and anther development (Dukowic-Schulze et al. 2014; Wang et al. 2016).
Reproductive phasiRNAs
Reproductive phasiRNAs are expressed at very high abundance in mainly monocot anthers, and arise from several hundred genomic locations (PHAS loci). Their biogenesis and occurrence throughout anther development and phyla have been addressed in several studies (Johnson et al. 2009; Zhai et al. 2015), and the current knowledge on phasiRNA has been comprehensively reviewed in Liu et al. (2020). In short, there are two waves of phasiRNA expression, distinguishable by their size (21 vs. 24nt), peak in abundance (premeiotic vs meiotic), and biogenesis factors (miR2118 vs. miR2275; DCL4 vs DCL5) as seen in rice and maize (Johnson et al. 2009; Song et al. 2012; Zhai et al. 2015). Interestingly, phasiRNAs from an individual PHAS locus do not have the same abundance, which is likely due to stabilization of only functionally relevant phasiRNAs by, e.g., AGO proteins. Indeed, OsMEL1 (MEIOSIS ARRESTED AT LEPTOTENE 1; rice AGO5) binds 21nt phasiRNAs with mainly 5′C in PMCs (Komiya et al. 2014), and ZmAGO18b binds 21nt phasiRNAs with mainly 5′U (Sun et al. 2019) which occur preferentially in anther wall cells as shown in rice (Araki et al. 2020). Whether AGO proteins bind to these phasiRNAs at the place of action or whether they migrate as bound complexes is unresolved. The fact that 21nt phasiRNAs seem to bind to different AGOs implies that different molecular mechanisms such as PTGS (posttranscriptional gene silencing) or RdDM (RNA-dependent DNA methylation) could be mediated by phasiRNAs.
Arikit et al. (2013) suggested that 21nt phasiRNAs might be involved in PTGS (see “tasiRNA” section above) and 24nt phasiRNAs in silencing by RdDM. DNA methylation has indeed been shown in cis at maize PHAS loci, albeit at both 21 and 24nt PHAS loci, and recently been confirmed with extensive analysis of maize phasiRNA mutants (Zhang et al. 2020b). Since increased DNA methylation was detected in meiocytes, a role in meiotic chromosome dynamics has been suggested (Dukowic-Schulze et al. 2016), similar to fission yeast lncRNAs that are involved in recognition and pairing of homologous chromosomes (recently reviewed by Hiraoka 2020). In line with a function of phasiRNAs in chromosome dynamics, a role for the phasiRNA-binding rice ARGONAUTE OsMEL1 in meiotic chromosome structure has been suggested, in addition to a role in male sporophyte development (Komiya et al. 2014). Rice Osmel1 mutants have defects in development of tapetum and PMCs (pollen mother cells), arresting early due to problems during premeiotic mitosis or male meiosis where chromosome condensation and synapsis is defective (Komiya et al. 2014). Furthermore, OsMEL1 localization was detected in the nucleus in meiosis and in the cytoplasm otherwise (Komiya et al. 2014).
Means to achieve mobility and specificity
As apparent from Fig. 2, three major ways of signal mobility have been reported in anther development: (1) diffusion, (2) migration through plasmodesmata and cytomictic channels, and (3) via the apoplast. PMCs during microsporogenesis are connected by cytomictic channels through which even nuclear material, i.e., chromosomes, can migrate (reviewed in Mursalimov et al. 2013). These special cellular connections have been observed for over a century, and also have been reported between TP cells during pollen maturation; the smaller and more widespread plasmodesmata, on the other hand, connect multiple somatic anther cells with each other. Plasmodesmata between PMCs and TP cells exist and become closed when pollen mature, so that TP cells are isolated when they undergo PCD (reviewed in Sager and Lee 2014). Plasmodesmata also connect EN cells with each other, and with EPI and ML cells (Mamun et al. 2005).
A beneficial property for elements to be mobile is their relatively small size. Small molecules like O2 move across cell membranes via diffusion resulting in gradient formation, which is dependent on factors like temperature, and consumption in certain tissues as it was proposed in anther development (Kelliher and Walbot 2012). In contrast to this, ROS have a strongly limited mobility because of their short lifespan and by that can function mostly at their origin as signals.
A limiting factor for symplastic migration is plasmodesmata size which is, for example, modulated via redox regulation (Benitez-Alfonso et al. 2009; reviewed in Maule et al. 2011), and can change depending on the developmental state. Immature cells seem to have increased transport rates, for example, in Arabidopsis embryos up to the torpedo stage when 0.5 kDa tracers can still pass but 10 kDa tracers are excluded (Kim et al. 2002; reviewed in Brunkard et al. 2013). Intriguingly, the above-mentioned RLKs AtBAM1/BAM2 have been implied in increasing plasmodesmata size in the context of RNAi spread as part of antiviral defense (Rosas-Diaz et al. 2018).
Proteins and peptides can be secreted into the plant apoplast via conventional or unconventional secretion. So far, only one apoplastic ligand was identified in anther development, namely AtTPD/OsTDL1/ZmMAC1, which contains a signal peptide sequence for conventional secretion (Wang et al. 2012a). ZmMAC1 was shown to be secreted by onion cells, and later experiments confirmed that ZmMAC1 acts in the maize anther apoplast (Wang et al. 2012a; van der Linde et al. 2018a).
The ability of tasiRNA/phasiRNAs to migrate across cell layers seems to be a core property for reproductive phasiRNAs. 21nt phasiRNAs are generated in outer layers like the epidermis and end up later at highest concentrations in tapetal cells and PMCs (Fig. 3a, b, d), similar to 24nt phasiRNAs that are generated later in the TP (Fig. 3c, d) (Huang et al. 2020). How phasiRNAs migrate from cell to cell within the anther remains elusive. In Arabidopsis leaves, phasi and tasiRNAs have been found in extracellular vesicles (Baldrich et al. 2019), while in the plant developmental context, sRNAs were described to travel through plasmodesmata (Dunoyer et al. 2013).
The spatiotemporal specificity of mobile signals is an essential factor in cell–cell communication and dependent on multiple aspects, for example, on pace: tasiRNAs seem to be faster in their cell-to-cell-movement than miRNAs (Felippes et al. 2011), maybe because tasiRNAs are generated in the cytoplasm but miRNAs mainly in the nucleus, with the nuclear export rate as a limiting factor (Jouannet et al. 2012; reviewed in Rogers and Chen 2013). Developmental sRNAs seem to migrate less far than somatic sRNAs (3–6 vs 10–15 cells) (reviewed in Benkovics and Timmermans 2014). While sRNA gradient formation might just be an unavoidable result of their mode of migration within an organ, their tissue-specific signaling function could dependent on either the “Threshold Model” or the “Gradual Model” (reviewed in Benkovics and Timmermans 2014). On the other hand, specificity at that point might be achieved by binding to a tissue-specific AGO protein with preference for 5′ uridine 21nt phasiRNAs in anther cell wall (Araki et al. 2020) or 5′ cytosine 21nt phasiRNAs in PMCs (Komiya et al. 2014). Binding to a partner such as an AGO protein is a prominent way to prevent further spreading of sRNAs, as reported for AtAGO10 binding to miR166 stopping it from moving to the embryonic meristem (Liu et al. 2009a; Zhu et al. 2011). These partners could also be lncRNAs, so-called ceRNA (competitive endogenous RNAs) whose purpose is to capture sRNA at certain times and position before or after their actual action (Li et al. 2019b). In turn, sRNAs could act in supporting mRNA decay of genes before or after their time of need, presumably key factors such as transcription factors, and RLKs and by this support spatiotemporal specificity of other mobile elements (Fig. 3c).
Thus, mobile elements should always be considered together with the presence of their potential partners, and this applies to tasi/phasiRNA-target pairs as well as peptide ligand–receptor pairs, and redox-sensitive transcription factors.
Open questions (Q) and hypotheses (H)
Proper anther and pollen formation are essential for fertilization and by that a key factor in agriculture. Several mobile signals that govern anther development have been identified (e.g., see Table 1), but many questions about their functions remain open.
Why are anther developmental modules conserved but not consistently so?
-
Q1: Why do conserved signaling modules (such as OsMIL1/ZmMSCA1/AtROXY1/AtROXY2 and AtTPD/OsTDL1/ZmMAC1, together with their interactors) have different effects in anther development according to their mutant phenotypes? Specifically, the mutant phenotypes of Zmmsca1, Osmil1 and Atroxy1/2 occur early in AR specification, later after EN/SP differentiation, at TP/SPC differentiation and pollen release, respectively.
-
H1: Answers to differences due to OsMIL1/ZmMSCA1/AtROXY1/AtROXY2 might lie in small changes in the respective oxygen gradients and migration properties. For example, Arabidopsis, rice and maize anthers are of different size and in vastly different surroundings during the formation. Besides differences in environmental conditions, expression patterns of the glutaredoxins or their targets might differ or their mobility between tissues. For the secreted peptide homologs AtTPD/OsTDL1A/ZmMAC1, their spatiotemporal presence influenced by mobility and turnover could differ.
-
-
Q2: Why do reproductive phasiRNAs seem to be highly conserved in monocots but less so in angiosperms (Xia et al. 2019; Feng et al. 2019)?
-
H2: At the moment, it can only be speculated that phasiRNAs are either not essential, or that their functions are taken over by other small RNAs that follow a different biosynthesis path but have similar functions.
-
How do reproductive phasiRNAs contribute to anther development?
-
Q3: How can the class of phasiRNAs mediate different outcomes? Is DNA methylation in cis a side effect or does it have a real biological function (Fig. 3d)?
-
H3: The first part is already partly resolved, i.e., by binding to different AGO proteins at certain layers and time. The second part needs further examination.
-
-
Q4: What are the functions of genes targeted by phasiRNAs for degradation, as identified now in rice (Jiang et al. 2020; Zhang et al. 2020c)? Are there further targets in other cells, at other times? Are phasiRNAs maybe involved in the processes mediated by AGO2 in tapetal PCD, or by DCL1 in early anther formation or late anther dehiscence? Why might the regulation of targets be needed, and most so when the developmental timing is altered?
-
H4: Undoubtedly, monocot-prevalent reproductive phasiRNAs do play a supportive, but maybe non-essential role in anther development and/or meiosis, since mutants lacking the otherwise vast abundance of 21 or 24 nt phasiRNAs have phenotypes including defects in meiosis and anther morphology, as well as environment-sensitive male sterility (Liu and Nonomura 2016; Sun et al. 2018; Ono et al. 2018; Teng et al. 2020). This can occur through temperature or photoperiod, depending on the kind and amount of phasiRNAs missing or misregulated, and species and genetic background (Ding et al. 2012a, b; Zhu and Deng 2012; Fan et al. 2016; Teng et al. 2020). Common to both photoperiod and temperature changes is the modification of developmental timing—higher temperature as well as long-day conditions lead to faster anther development, meiosis and microsporogenesis (Zhang et al. 2020a; Zhu et al. 2020; Teng et al. 2020). One hypothesis regarding the biological function of phasiRNAs is a role in destabilization of mRNA that are not needed anymore, as by animal piRNAs (Gou et al. 2014). Given enough time, this RNA decrease will occur naturally, but in conditions that accelerate development, phasiRNAs might be needed.
-
-
Q5: Could phasiRNAs coordinate anther development and meiosis simultaneously, enabling proper and coordinated timing of, e.g., tapetum redifferentiation and PMC stage?
-
H5: Reproductive tasi/phasiRNAs are indeed good candidates for supporting anther development by any of the following possibilities which remain to be verified: (1) supporting decay of mRNAs not needed anymore, by (2) building transcription factor gradients, by (3) removing other anther layer specifying factors like receptors or oxygen sensors at a certain time and/or from certain layers, and/or by (4) changing the chromatin state of meiotic chromosomes to support pairing or other processes.
-
-
Q6: Are phasiRNAs influencing DSBs (double strand breaks) which are initiated in meiosis on purpose? This question stems from the observations that the usual consequences of DSBs, γH2AX marks on chromosomes, and high expression of the DSB repair mediator ATM (ATAXIA TELANGIECTASIA MUTATED) are decreased if 21nt phasiRNA pathways are defective (Liu and Nonomura 2016; Teng et al. 2020).
How do known mobile elements in AR cell formation act exactly?
-
Q7: Why (or how) do L1-d cells have such a high degree of plasticity that they can become AR cells under certain redox-altering conditions?
-
H7: The spatiotemporal location of ZmMSCA1 might be an aspect and needs further detailed investigation.
-
-
Q8: Why do pluripotent L2-d cells surrounding the AR cell column acquire a somatic fate, in spite of a mild hypoxia and ZmMSCA1 expression (both of which promote AR cell formation)?
-
H8: Here, two possible scenarios come to mind: (1) The required hypoxia threshold is reached first in future AR cells, rapid differentiation and subsequent secretion of ZmMAC1 hinders surrounding cells. (2) Even though L2-d cells are morphologically identical (Kelliher et al. 2014), they might differ in expression and by that in responsiveness toward hypoxia. Initial patterning of stamen, anthers and anther lobes is based on expression domains within subsets of L2-d cells (Toriba et al. 2010) and might continue during AR and somatic cell differentiation.
-
-
Q9: How does ZmMAC1 limit the number of AR cells?
-
H9: There might be yet undiscovered, subsequent signaling from the newly specified EN/SP cells involved. On the other hand, ZmMAC1 might interact with a receptor on the AR surface, maybe even ZmMSP1, and by that control AR cell number.
-
-
Q10: Why is there ZmMAC1 expression detectable before and after (even during meiosis) AR cell initiation?
-
H10: While a function of ZmMAC1 before AR differentiation is not promoted by mutant analysis, one might speculate that ZmMAC1 functions as a reoccurring signal later in anther development. The strong, initial AR and somatic layer phenotype observed in mutants may cover-up later phenotypes. Inducible mutants or further studies using the Trojan horse approach on different stages of anther development might help to answer this question.
-
-
Q11: How are mobile signals transmitted toward transcription factors and vice versa?
-
H11: One way might be by direct interaction of signal receptor with transcription factors. It has been suggested that the naturally occurring oxygen gradient is sensed by ZmMSCA1/OsMIL1 within the anther, accompanied by reduction of its disulfide bridge (see Fig. 2). In turn, ZmMSCA1/OsMIL1 could reduce and by that activate TGA-transcription factors (Fig. 2), resulting in lobe cell specification.
-
Which yet unidentified factors and functions are we still missing?
-
Q12: How many apoplastic peptide/protein signaling pathways are yet to be discovered in anther development?
-
o
H12: A variety of proteomic analysis identified RLKs and potential ligands in developing anther cells (Wang et al. 2012b; Zhang et al. 2014; Ye et al. 2015, 2016)) but, only a few have been studied yet, and ligands for most known receptors remain unknown. Another indication for the impact of apoplastic proteins/signals in anther development is the importance of a signal peptide peptidase (SPP) during pollen development in Arabidopsis. SPPs are endoplasmatic reticulum localized proteases and cleave signal peptides from proteins entering the secretory pathway. The Atssp mutant shows poor pollen germination, and nuclear morphology is abnormal at the tricellular pollen stage. AtSSP expression is highest in root, shoot apex, carpel, and stamen/pollen as well as in stage 6 and 7 seeds (Han et al. 2009).
-
o
-
Q13: How many other cell–cell communications pathways are we still missing in anther development?
-
H13: Apoplastic vesicles containing RNA (Baldrich et al. 2019; for review see Cai et al. 2019), direct uptake of proteins from the apoplast (for review see Petre and Kamoun 2014) and transport of bigger proteins through plasmodesmata (reviewed in Stahl and Simon 2013) have been discovered and shown to play pivotal roles in other cell–cell communication scenarios and could possible play roles in anther development as well.
-
How are the mobile elements interconnected?
Figure 3 sums up suggested interactions between transcription factors and mobile signals (phasiRNAs, ROS, RLKs and secreted peptides), which, however, remain to be demonstrated. Two specific thoughts are the following ones:
-
Q14: Are mobile signals connected with each other?
-
H14: A first indication for the connection of different mobile signals might come from mutant analysis. The maize Zmocl4 (outer cell layers 4) mutant lacks 21nt phasiRNAs and has abnormal and partly duplicated endothecium layers, attributed to defective epidermal signaling (Wang et al. 2012a). If the 21nt phasiRNAs include tasiRNAs that target ZmMSP1 or ZmMAC1 for RNA degradation, Zmocl4 mutants might have prolonged presence of ZmMSP1 or ZmMAC1, resulting in more cells to become EN (Fig. 3a).
-
-
Q15: Which anther cell types produce the mobile signals (brassinosteroids and smallRNAs) that influence transcription of ROS production enzymes and when, and by that ultimately regulate ROS levels in the TP?
Summary and outlook
Taken together, the triad of oxygen, secreted proteins and sRNAs are key players beside phytohormones governing the spatiotemporal program of anther development (Fig. 4). They arise naturally, or emerge from newly formed cell layers and can migrate from their location of origin along the lobe radius. While O2 naturally forms a gradient across the anther, the allocation of other mobile signals remains more elusive. Specific sRNA might form gradients, while other sRNAs might be retained in certain anther tissues. Mobile sRNAs could support timely and position-specific presence of components like transcription factors and RLKs and thereby interconnect with other mobile element pathways. Whether this turns out to be the case will need to be experimentally validated by using, e.g., sequencing approaches including AGO-immunoprecipitations to identify target genes, and phasiRNA-defective lines for cytological approaches such as (F) ISH to study regulation of these target genes. Proteomic data of anthers, among other published data, indicate that there are more protein signals to be found in the future. Spatiotemporal, functional and conservational characterization of those will help to understand their mobility, how they gain spatiotemporal specificity, and how mobile element pathways mingle to orchestrate anther development together. We look forward to future discoveries in the field of mobile elements whether they support our partly speculative hypotheses here or dispute them to move on to novel ideas.
Authors contribution statement
SDS and KL equally contribute to this manuscript.
Abbreviations
- AR:
-
Archesporial
- PPCs/SPCs:
-
Primary/secondary parietal cells
- EPI:
-
Epidermis
- EN:
-
Endothecium
- ML:
-
Middle layer
- TP:
-
Tapetum
- PMC:
-
Pollen mother cell
- RLK:
-
Receptor like kinase
- ROS:
-
Reactive oxygen species
References
Albertsen MC, Fox T, Trimnell M, Wu Y (2011) MSCA1 nucleotide sequences impacting plant male fertility and method of using same. Patent EP2631243A2
Albrecht C, Russinova E, Hecht V et al (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17:3337–3349. https://doi.org/10.1105/tpc.105.036814
Araki S, Le NT, Koizumi K et al (2020) miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat Commun 11:3115. https://doi.org/10.1038/s41467-020-16637-3
Arikit S, Zhai J, Meyers BC (2013) Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr Opin Plant Biol 16:170–179. https://doi.org/10.1016/j.pbi.2013.01.006
Baldrich P, Rutter BD, Karimi HZ et al (2019) Plant extracellular vesicles contain diverse small RNA species and are enriched in 10–17-nucleotide “tiny” RNAs. Plant Cell 31:315–324. https://doi.org/10.1105/tpc.18.00872
Benitez-Alfonso Y, Cilia M, Roman AS et al (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. PNAS 106:3615–3620. https://doi.org/10.1073/pnas.0808717106
Benkovics AH, Timmermans MC (2014) Developmental patterning by gradients of mobile small RNAs. Curr Opin Genet Dev 27:83–91. https://doi.org/10.1016/j.gde.2014.04.004
Brunkard JO, Runkel AM, Zambryski PC (2013) Plasmodesmata dynamics are coordinated by intracellular signaling pathways. Curr Opin Plant Biol 16:614–620. https://doi.org/10.1016/j.pbi.2013.07.007
Cai W, Zhang D (2018) The role of receptor-like kinases in regulating plant male reproduction. Plant Reprod 31:77–87. https://doi.org/10.1007/s00497-018-0332-7
Cai Q, He B, Weiberg A et al (2019) Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog 15:e1008090. https://doi.org/10.1371/journal.ppat.1008090
Cardarelli M, Cecchetti V (2014) Auxin polar transport in stamen formation and development: how many actors? Front Plant Sci. https://doi.org/10.3389/fpls.2014.00333
Chaubal R, Anderson JR, Trimnell MR et al (2003) The transformation of anthers in the msca1 mutant of maize. Planta 216:778–788. https://doi.org/10.1007/s00425-002-0929-8
Cheng H, Qin L, Lee S et al (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131:1055–1064. https://doi.org/10.1242/dev.00992
Chitwood DH, Nogueira FTS, Howell MD et al (2009) Pattern formation via small RNA mobility. Genes Dev 23:549–554. https://doi.org/10.1101/gad.1770009
Cigan AM, Unger E, Xu R et al (2001) Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex Plant Reprod 14:135–142. https://doi.org/10.1007/s004970100099
Colcombet J, Boisson-Dernier A, Ros-Palau R et al (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17:3350–3361. https://doi.org/10.1105/tpc.105.036731
Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66:487–511. https://doi.org/10.1146/annurev-arplant-050213-040012
de Felippes FF, Ott F, Weigel D (2011) Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucl Acids Res 39:2880–2889. https://doi.org/10.1093/nar/gkq1240
Dickinson HG, Bell PR (1976) The changes in the tapetum of Pinus banksiana accompanying formation and maturation of the pollen. Ann bot 40:1101–1109. https://doi.org/10.1093/oxfordjournals.aob.a085219
Ding J, Lu Q, Ouyang Y et al (2012a) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. PNAS 109:2654–2659. https://doi.org/10.1073/pnas.1121374109
Ding J, Shen J, Mao H et al (2012b) RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol Plant 5:1210–1216. https://doi.org/10.1093/mp/sss095
Dukowic-Schulze S, Sundararajan A, Mudge J et al (2014) The transcriptome landscape of early maize meiosis. BMC Plant Biol 14:118. https://doi.org/10.1186/1471-2229-14-118
Dukowic-Schulze S, Sundararajan A, Ramaraj T et al (2016) Novel meiotic miRNAs and indications for a role of phasiRNAs in meiosis. Front Plant Sci 7:762. https://doi.org/10.3389/fpls.2016.00762
Dunoyer P, Melnyk C, Molnar A, Slotkin RK (2013) Plant mobile small RNAs. Cold Spring Harb Perspect Biol 5:a017897. https://doi.org/10.1101/cshperspect.a017897
Fan Y, Yang J, Mathioni SM et al (2016) PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. PNAS 113:15144–15149. https://doi.org/10.1073/pnas.1619159114
Fang Y-N, Yang X-M, Jiang N et al (2020) Genome-wide identification and expression profiles of phased siRNAs in a male-sterile somatic cybrid of pummelo (Citrus grandis). Tree Genet Genomes 16:46. https://doi.org/10.1007/s11295-020-01437-z
Feng L, Xia R, Liu Y (2019) Comprehensive characterization of miRNA and PHAS loci in the diploid strawberry (Fragaria vesca) genome. Hortic Plant J 5:255–267. https://doi.org/10.1016/j.hpj.2019.11.004
Fiedler I-C, Weiberg A, van der Linde K (2019) Guidelines for using Ustilago maydis as a Trojan horse for in situ delivery of maize proteins. J Vis Exp 144:e58746. https://doi.org/10.3791/58746
Field S, Thompson B (2016) Analysis of the maize dicer-like1 mutant, fuzzy tassel, implicates microRNAs in anther maturation and dehiscence. PLoS ONE 11:e0146534. https://doi.org/10.1371/journal.pone.0146534
Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5:1217–1229. https://doi.org/10.1105/tpc.5.10.1217
Gou L-T, Dai P, Yang J-H et al (2014) Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 24:680–700. https://doi.org/10.1038/cr.2014.41
Hafidh S, Fíla J, Honys D (2016) Male gametophyte development and function in angiosperms: a general concept. Plant Reprod 29:31–51. https://doi.org/10.1007/s00497-015-0272-4
Han S, Green L, Schnell DJ (2009) The signal peptide peptidase is required for pollen function in Arabidopsis. Plant Physiol 149:1289–1301. https://doi.org/10.1104/pp.108.130252
Heidstra R, Sabatini S (2014) Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol 15:301–312. https://doi.org/10.1038/nrm3790
Hiraoka Y (2020) Phase separation drives pairing of homologous chromosomes. Curr Genet. https://doi.org/10.1007/s00294-020-01077-9
Hong L, Tang D, Shen Y et al (2012a) MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol 196:402–413. https://doi.org/10.1111/j.1469-8137.2012.04270.x
Hong L, Tang D, Zhu K et al (2012b) Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 24:577–588. https://doi.org/10.1105/tpc.111.093740
Hord CLH, Chen C, Deyoung BJ et al (2006) The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18:1667–1680. https://doi.org/10.1105/tpc.105.036871
Hord CLH, Sun Y-J, Pillitteri LJ et al (2008) Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol Plant 1:645–658. https://doi.org/10.1093/mp/ssn029
Huang J, Zhang T, Linstroth L et al (2016) Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet 12:e1006147. https://doi.org/10.1371/journal.pgen.1006147
Huang K, Demirci F, Batish M et al (2020) Quantitative, super-resolution localization of small RNAs with sRNA-PAINT. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa623
Huo Y, Pei Y, Tian Y et al (2020) IRREGULAR POLLENEXINE2 encodes a GDSL lipase essential for male fertility in maize. Plant Physiol 184:1438–1454. https://doi.org/10.1104/pp.20.00105
Ito T, Shinozaki K (2002) The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol 43:1285–1292. https://doi.org/10.1093/pcp/pcf154
Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126:5231–5243. https://dev.biologists.org/content/126/23/5231.long
Jiang P, Lian B, Liu C et al (2020) 21-nt phasiRNAs direct target mRNA cleavage in rice male germ cells. Nat Commun 11:5191. https://doi.org/10.1038/s41467-020-19034-y
Johnson C, Kasprzewska A, Tennessen K et al (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19:1429–1440. https://doi.org/10.1101/gr.089854.108
Jouannet V, Moreno AB, Elmayan T et al (2012) Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. The EMBO Journal 31:1704–1713. https://doi.org/10.1038/emboj.2012.20
Jun J, Fiume E, Roeder AHK et al (2010) Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol 154:1721–1736. https://doi.org/10.1104/pp.110.163683
Kelliher T, Walbot V (2011) Emergence and patterning of the five cell types of the Zea mays anther locule. Dev Biol 350:32–49. https://doi.org/10.1016/j.ydbio.2010.11.005
Kelliher T, Walbot V (2012) Hypoxia triggers meiotic fate acquisition in maize. Science 337:345–348. https://doi.org/10.1126/science.1220080
Kelliher T, Egger RL, Zhang H, Walbot V (2014) Unresolved issues in pre-meiotic anther development. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00347
Kim I, Hempel FD, Sha K et al (2002) Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development 129:1261–1272. https://dev.biologists.org/content/129/5/1261
Komiya R, Ohyanagi H, Niihama M et al (2014) Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J 78:385–397. https://doi.org/10.1111/tpj.12483
Kovaleva LV, Dobrovolskaya A, Voronkov A, Rakitin V (2011) Ethylene is involved in the control of male gametophyte development and germination in Petunia. J Plant Growth Regul 30:64–73. https://doi.org/10.1007/s00344-010-9168-6
Li S, Lauri A, Ziemann M et al (2009) Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21:429–441. https://doi.org/10.1105/tpc.108.064477
Li S, Gutsche N, Zachgo S (2011) The ROXY1 C-terminal L**LL motif is essential for the interaction with TGA transcription factors. Plant Physiol 157:2056–2068. https://doi.org/10.1104/pp.111.185199
Li X, Lian H, Zhao Q, He Y (2019a) MicroRNA166 monitors SPOROCYTELESS/NOZZLE for building of the anther internal boundary. Plant Physiol 181:208–220. https://doi.org/10.1104/pp.19.00336
Li Z, An X, Zhu T et al (2019b) Discovering and constructing ceRNA-miRNA-target gene regulatory networks during anther development in maize. Int J Mol Sci 20:3480. https://doi.org/10.3390/ijms20143480
Liu H, Nonomura K-I (2016) A wide reprogramming of histone H3 modifications during male meiosis I in rice is dependent on the Argonaute protein MEL1. J Cell Sci 129:3553–3561. https://doi.org/10.1242/jcs.184937
Liu Q, Yao X, Pi L et al (2009a) The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J 58:27–40. https://doi.org/10.1111/j.1365-313X.2008.03757.x
Liu X, Huang J, Parameswaran S et al (2009b) The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 151:1401–1411. https://doi.org/10.1104/pp.109.145896
Liu Y, Teng C, Xia R, Meyers BC (2020) Phased secondary small interfering RNAs (phasiRNAs) in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell. https://doi.org/10.1105/tpc.20.00335
Lu J-Y, Xiong S-X, Yin W et al (2020) MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis. J Exp Bot 71:4877–4889. https://doi.org/10.1093/jxb/eraa219
Luan W, Dai Y, Li X-Y et al (2020) Identification of tRFs and phasiRNAs in tomato (Solanum lycopersicum) and their responses to exogenous abscisic acid. BMC Plant Biol 20:320. https://doi.org/10.1186/s12870-020-02528-8
Mamun EA, Cantrill LC, Overall RL, Sutton BG (2005) Cellular organisation in meiotic and early post-meiotic rice anthers. Cell Biol Int 29:903–913. https://doi.org/10.1016/j.cellbi.2005.08.001
Marciniak K, Przedniczek K (2019) Comprehensive insight into gibberellin- and jasmonate-mediated stamen development. Genes 10:811. https://doi.org/10.3390/genes10100811
Maule AJ, Benitez-Alfonso Y, Faulkner C (2011) Plasmodesmata – membrane tunnels with attitude. Curr Opin Plant Biol 14:683–690. https://doi.org/10.1016/j.pbi.2011.07.007
Mercier R, Mézard C, Jenczewski E et al (2015) The molecular biology of meiosis in plants. Annu Rev Plant Biol 66:297–327. https://doi.org/10.1146/annurev-arplant-050213-035923
Mizuno S, Osakabe Y, Maruyama K et al (2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50:751–766. https://doi.org/10.1111/j.1365-313X.2007.03083.x
Murmu J, Bush MJ, DeLong C et al (2010) Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol 154:1492–1504. https://doi.org/10.1104/pp.110.159111
Mursalimov SR, Sidorchuk YV, Deineko EV (2013) New insights into cytomixis: specific cellular features and prevalence in higher plants. Planta 238:415–423. https://doi.org/10.1007/s00425-013-1914-0
Nagpal P, Ellis CM, Weber H et al (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132:4107–4118. https://doi.org/10.1242/dev.01955
Ndamukong I, Abdallat AA, Thurow C et al (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J 50:128–39. https://doi.org/10.1111/j.1365-313X.2007.03039.x
Nonomura K-I, Miyoshi K, Eiguchi M et al (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15:1728–1739. https://doi.org/10.1105/tpc.012401
Ono S, Liu H, Tsuda K et al (2018) EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet 14:e1007238. https://doi.org/10.1371/journal.pgen.1007238
Patel P, Mathioni S, Kakrana A et al (2018) Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytol 220:851–864. https://doi.org/10.1111/nph.15349
Peris CIL, Rademacher EH, Weijers D (2010) Green beginnings: pattern formation in the early plant embryo. In: Timmermans MCP (ed) Current topics in developmental biology. Academic Press, Cambridge, pp 1–27. https://doi.org/10.1016/s0070-2153(10)91001-6
Petre B, Kamoun S (2014) How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol 12:e1001801. https://doi.org/10.1371/journal.pbio.1001801
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399. https://doi.org/10.1105/tpc.113.113159
Rosas-Diaz T, Zhang D, Fan P et al (2018) A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. PNAS 115:1388–1393. https://doi.org/10.1073/pnas.1715556115
Sager R, Lee J-Y (2014) Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. J Exp Bot 65:6337–6358. https://doi.org/10.1093/jxb/eru365
Schreiber DN, Bantin J, Dresselhaus T (2004) The MADS box transcription factor ZmMADS2 is required for anther and pollen maturation in maize and accumulates in apoptotic bodies during anther dehiscence. Plant Physiol 134:1069–1079. https://doi.org/10.1104/pp.103.030577
Sheridan WF, Golubeva EA, Abrhamova LI, Golubovskaya IN (1999) The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics 153:933–941. https://www.genetics.org/content/153/2/933.long
Skopelitis DS, Husbands AY, Timmermans MC (2012) Plant small RNAs as morphogens. Curr Opin Cell Biol 24:217–224. https://doi.org/10.1016/j.ceb.2011.12.006
Song X, Li P, Zhai J et al (2012) Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J 69:462–474. https://doi.org/10.1111/j.1365-313X.2011.04805.x
Song S, Qi T, Huang H, Xie D (2013) Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant 6:1065–1073. https://doi.org/10.1093/mp/sst054
Stahl Y, Simon R (2013) Gated communities: apoplastic and symplastic signals converge at plasmodesmata to control cell fates. J Exp Bot 64:5237–5241. https://doi.org/10.1093/jxb/ert245
Sun W, Xiang X, Zhai L et al (2018) AGO18b negatively regulates determinacy of spikelet meristems on the tassel central spike in maize. J Integr Plant Biol 60:65–78. https://doi.org/10.1111/jipb.12596
Sun W, Chen D, Xue Y et al (2019) Genome-wide identification of AGO18b-bound miRNAs and phasiRNAs in maize by cRIP-seq. BMC Genom 20:656. https://doi.org/10.1186/s12864-019-6028-z
Teng C, Zhang H, Hammond R et al (2020) Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat Commun 11:2912. https://doi.org/10.1038/s41467-020-16634-6
Timofejeva L, Skibbe DS, Lee S et al (2013) Cytological characterization and allelism testing of anther developmental mutants identified in a screen of maize male sterile lines. G3 (Bethesda) 3:231–49. https://doi.org/10.1534/g3.112.004465
Toriba T, Suzaki T, Yamaguchi T et al (2010) Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22:1452–1462. https://doi.org/10.1105/tpc.110.075291
Uzair M, Xu D, Schreiber L et al (2020) PERSISTENT TAPETAL CELL2 is required for normal tapetal programmed cell death and pollen wall patterning. Plant Physiol 182:962–976. https://doi.org/10.1104/pp.19.00688
van der Linde K, Walbot V (2019) Pre-meiotic anther development. In: Grossniklaus U (ed) Current topics in developmental biology. Academic Press, San Diego, pp 239–256. https://doi.org/10.1016/bs.ctdb.2018.11.001
van der Linde K, Timofejeva L, Egger RL et al (2018a) Pathogen Trojan horse delivers bioactive host protein to alter maize (Zea mays) anther cell behavior in situ. Plant Cell 30:528–542. https://doi.org/10.1105/tpc.17.00238
van der Linde K, Timofejeva L, Egger RL, Walbot V (2018b) Application of the pathogen Trojan horse approach in premeiotic maize (Zea mays) anther development. Plant Signal Behav 13:e1547575. https://doi.org/10.1080/15592324.2018.1547575
Walbot V, Egger RL (2016) Pre-meiotic anther development: Cell fate specification and differentiation. Annu Rev Plant Biol 67:365–395. https://doi.org/10.1146/annurev-arplant-043015-111804
Walbot V, Skibbe DS (2010) Maize host requirements for Ustilago maydis tumor induction. Sex Plant Reprod 23:1–13. https://doi.org/10.1007/s00497-009-0109-0
Wang C-JR, Nan G-L, Kelliher T et al (2012a) Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139:2594–2603. https://doi.org/10.1242/dev.077891
Wang D, Adams CM, Fernandes JF et al (2012b) A low molecular weight proteome comparison of fertile and male sterile 8 anthers of Zea mays. Plant Biotechnol J 10:925–935. https://doi.org/10.1111/j.1467-7652.2012.00721.x
Wang H, Hu Q, Tang D et al (2016) OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol 171:230–241. https://doi.org/10.1104/pp.16.00167
White MD, Zenker J, Bissiere S, Plachta N (2018) Instructions for assembling the early mammalian embryo. Dev Cell 45:667–679. https://doi.org/10.1016/j.devcel.2018.05.013
Wu M-F, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133:4211–4218. https://doi.org/10.1242/dev.02602
Xia R, Chen C, Pokhrel S et al (2019) 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat Commun 10:627. https://doi.org/10.1038/s41467-019-08543-0
Xing S, Zachgo S (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53:790–801. https://doi.org/10.1111/j.1365-313X.2007.03375.x
Xing S, Rosso MG, Zachgo S (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132:1555–1565. https://doi.org/10.1242/dev.01725
Xing S, Salinas M, Höhmann S et al (2010) miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22:3935–3950. https://doi.org/10.1105/tpc.110.079343
Xiong S-X, Zeng Q-Y, Hou J-Q et al (2020) The temporal regulation of TEK contributes to pollen wall exine patterning. PLoS Genet 16:e1008807. https://doi.org/10.1371/journal.pgen.1008807
Yan M-Y, Xie D-L, Cao J-J et al (2020) Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J 102:931–947. https://doi.org/10.1111/tpj.14672
Yang W-C, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13:2108–2117. https://doi.org/10.1101/gad.13.16.2108
Yang S-L, Xie L-F, Mao H-Z et al (2003) Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15:2792–2804. https://doi.org/10.1105/tpc.016618
Yang L, Qian X, Chen M et al (2016) Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiol 171:2085–2100. https://doi.org/10.1104/pp.16.00016
Yang X, Li G, Tian Y et al (2018) A rice glutamyl-tRNA synthetase modulates early anther cell division and patterning. Plant Physiol 177:728–744. https://doi.org/10.1104/pp.18.00110
Ye J, Zhang Z, Long H et al (2015) Proteomic and phosphoproteomic analyses reveal extensive phosphorylation of regulatory proteins in developing rice anthers. Plant J 84:527–544. https://doi.org/10.1111/tpj.13019
Ye J, Zhang Z, You C et al (2016) Abundant protein phosphorylation potentially regulates Arabidopsis anther development. J Exp Bot 67:4993–5008. https://doi.org/10.1093/jxb/erw293
Yu J, Zhang D (2019) Molecular control of redox homoeostasis in specifying the cell identity of tapetal and microsporocyte cells in rice. Rice 12:42. https://doi.org/10.1186/s12284-019-0300-3
Yu J, Han J, Kim Y-J et al (2017) Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc Natl Acad Sci USA 114:12327–12332. https://doi.org/10.1073/pnas.1705189114
Zhai J, Jeong D-H, De Paoli E et al (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25:2540–2553. https://doi.org/10.1101/gad.177527.111
Zhai L, Sun W, Zhang K et al (2014) Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. J Integr Plant Biol 56:1042–1052. https://doi.org/10.1111/jipb.12205
Zhai J, Zhang H, Arikit S et al (2015) Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA 112:3146–3151. https://doi.org/10.1073/pnas.1418918112
Zhang D, Yang L (2014) Specification of tapetum and microsporocyte cells within the anther. Curr Opin Plant Biol 17:49–55. https://doi.org/10.1016/j.pbi.2013.11.001
Zhang H, Egger RL, Kelliher T et al (2014) Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3 (Bethesda, Md) 4:993–1010. https://doi.org/10.1534/g3.113.009738
Zhang C, Xu T, Ren M-Y et al (2020a) Slow development restores the fertility of photoperiod-sensitive male-sterile plant lines. Plant Physiol. https://doi.org/10.1104/pp.20.00951
Zhang M, Ma X, Wang C et al (2020b) CHH DNA methylation increases at 24-PHAS loci depend on 24-nt phasiRNAs in maize meiotic anthers. New Phytol. https://doi.org/10.1111/nph.17060
Zhang Y-C, Lei M-Q, Zhou Y-F, et al (2020c) Reproductive phasiRNAs regulate reprogramming of gene expression and meiotic progression in rice. Nat Commun 11:6031. https://doi.org/10.1038/s41467-020-19922-3
Zhao D (2009) Control of anther cell differentiation: a teamwork of receptor-like kinases. Sex Plant Reprod 22:221–228. https://doi.org/10.1007/s00497-009-0106-3
Zhao D, Ma H (2000) Male fertility: a case of enzyme identity. Curr Biol 10:R904–R907
Zheng L, Nagpal P, Villarino G et al (2019a) miR167 limits anther growth to potentiate anther dehiscence. Development. https://doi.org/10.1242/dev.174375
Zheng S, Li J, Ma L et al (2019b) OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. PNAS 116:7549–7558. https://doi.org/10.1073/pnas.1817675116
Zhu D, Deng XW (2012) A non-coding RNA locus mediates environment-conditioned male sterility in rice. Cell Res 22:791–792. https://doi.org/10.1038/cr.2012.43
Zhu H, Hu F, Wang R et al (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145:242–256. https://doi.org/10.1016/j.cell.2011.03.024
Zhu J, Lou Y, Shi Q-S et al (2020) Slowing development restores the fertility of thermo-sensitive male-sterile plant lines. Nat Plants. https://doi.org/10.1038/s41477-020-0622-6
Acknowledgements
The authors would like to thank Thomas Dresselhaus for comments on a draft of the manuscript. This work was supported by SFB924 project A14 of the DFG.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Hugh G. Dickinson.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dukowic-Schulze, S., van der Linde, K. Oxygen, secreted proteins and small RNAs: mobile elements that govern anther development. Plant Reprod 34, 1–19 (2021). https://doi.org/10.1007/s00497-020-00401-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-020-00401-0