Abstract
Key message
New gametic homozygous mutants.
In angiosperms, a haploid male gamete (sperm cell) fuses with a haploid female gamete (egg cell) during fertilization to form a zygote carrying paternally and maternally derived chromosomes. Several fertilization-defective mutants in Arabidopsis thaliana, including a generative cell-specific 1 (gcs1)/hapless 2 mutant, the sperm cells of which are unable to fuse with female gametes, can only be maintained as heterozygous lines due to the infertile male or female gametes. Here, we report successful generation of a gcs1 homozygous mutant by heat-inducible removal of the GCS1 transgene. Using the gcs1 homozygous mutant as male, the defect in gamete fusion was observed with great frequency; in our direct observation by semi-in vivo fertilization assay using ovules, 100 % of discharged sperm cells in culture failed to show gamete fusion. More than 70 % of ovules in the pistil received a second pollen tube as attempted fertilization recovery. Moreover, gcs1 mutant sperm cells could fertilize female gametes at a low frequency in the pistil. This strategy to generate homozygous fertilization-defective mutants will facilitate novel approaches in plant reproduction research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In angiosperms, male and female gametes are generated by meiosis from pollen and embryo sac mother cells, respectively. The number of sets of chromosomes is reduced to half the original number during meiotic division, resulting in haploid reproductive cells. The male haploid cells, the microspores, divide asymmetrically to form vegetative cells containing the generative cell and develop into male gametophytes, pollen. In angiosperms with tricellular pollen, including Arabidopsis thaliana, the generative cell undergoes mitosis to generate two male gametes, i.e., sperm cells, within the pollen. On the other hand, the female haploid cell, the megaspore, undergoes three rounds of mitosis and cellularization to form the embryo sac containing two female gametes, i.e., an egg cell and a central cell. The two sperm cells are delivered by a pollen tube elongated from the pollen by tip growth and released into a female gametophyte. One sperm cell fertilizes the egg cell and the other fertilizes the central cell to form a zygote and a nutritious endosperm, respectively (for a review, see Hamamura et al. 2012). The number of sets of chromosomes is doubled in the zygote by fusion of the paternal and maternal chromosomes of the sperm and egg cells.
Successful double fertilization requires various male–female interactions. Precise pollen tube guidance and reception are critical for sperm cell delivery. These steps are controlled by accessory cells of the female gametes, i.e., two synergid cells, which attract the pollen tube to the embryo sac by diffusible attractant peptides (for a review, see Takeuchi and Higashiyama 2011), recognize pollen tube arrival, and induce pollen tube rupture (for a review, see Kessler and Grossniklaus 2011). After pollen tube discharge, two sperm cells remain in place between the egg and central cells, and these are activated in response to egg-secreted EC1 peptides (Sprunck et al. 2012). Sperm cells possess the plasma membrane proteins GAMETE EXPRESSED 2 (GEX2) for gamete adhesion (Mori et al. 2014) and GENERATIVE CELL-SPECIFIC 1 (GCS1)/HAPLESS 2 (HAP2) (Mori et al. 2006; von Besser et al. 2006) for gamete fusion. Successful molecular recognition between male and female gametes results in double fertilization, and two male nuclei begin to move precisely toward the nuclei of their respective female target cells (Hamamura et al. 2011). Calcium spikes (Hamamura et al. 2014; Denninger et al. 2014) and actin-based male nuclear migration (Ohnishi et al. 2014; Kawashima et al. 2014) in the female gametes are also involved in double fertilization. Once double fertilization is completed in both female gametes of an ovule, pollen tube guidance to the ovule stops in order to block polytubey (Kasahara et al. 2012; Beale et al. 2012; Maruyama et al. 2013; Völz et al. 2013). When gcs1/hap2 sperm cells are delivered but fail in gamete fusion, a second pollen tube is additionally attracted (Kasahara et al. 2012; Beale et al. 2012). Fertilization recovery by the second pollen tube was shown to increase the fertilization success rate (Kasahara et al. 2012).
To determine the mechanisms of plant fertilization, including the fertilization recovery system, physiological analyses using fertilization-defective mutants have been performed. However, several gametophytic mutants never generate homozygous offspring because half of the haploid male or female gametophytes carrying a mutant allele are unable to complete the reproductive process. These mutants must therefore be maintained as heterozygous lines. For example, no homozygous mutants of gcs1/hap2 have been reported due to the lack of paternal transmission (Mori et al. 2006; von Besser et al. 2006). The male germ cell-specific transcription factor DUO POLLEN 1 (DUO1) is crucial for regulating sperm cell differentiation and function (Durbarry et al. 2005; Rotman et al. 2005) by activating the expression of many sperm-expressed genes, including GCS1 (Brownfield et al. 2009; Borg et al. 2011). Therefore, homozygous duo1 mutants have not been obtained by self-crossing. The lack of the availability of homozygous mutants for essential genes in fertilization interferes with analyses of double-fertilization mechanisms and the fertilization recovery process, as the mutant phenotype in heterozygous mutants is always mixed with the wild-type phenotype. Moreover, large-scale gene expression analyses, such as transcriptomics or proteomics, which are powerful means of identifying novel fertilization-related factors (Qin et al. 2009; Grobei et al. 2009; Okuda et al. 2013), are hampered due to the presence of the wild-type gametophytes. Generation of homozygous mutants would significantly contribute to plant reproduction research.
In mammals, conditional knockout, such as Cre-loxP-mediated site-specific DNA recombination, which has also been shown to function in Arabidopsis (Russell et al. 1992), is applicable to bypass embryonic lethality by removable gene rescue (for a review, see Kos 2004). Therefore, we examined the application of this system to generate homozygous mutants with defects in fertilization by bypassing the process of fertilization with the expression of a heat-induced removable transgene. Here, we report the generation of gcs1 homozygous (gcs1/−) plants by the heat-inducible Cre-loxP recombination system in A. thaliana. The phenotypes of gcs1 homozygous plants were examined to determine the potential utility of this mutant and the associated technology for plant reproduction research.
Materials and methods
Plant materials and growth conditions
The A. thaliana Columbia (Col-0) accession was used as a wild-type control. Transgenic plants possessing HTR10p::HTR10:mRFP (Ingouff et al. 2007) and RPS5Ap::H2B:GFP gene, which express histone H2B (At1g07790) fused with GFP by RIBOSOMAL PROTEIN SUBUNIT 5A (RPS5A; At3g11940; Adachi et al. 2011) promoter, were used to visualize sperm cell nuclei and female gametophytic cell nuclei, respectively. Sperm cell nuclei of gcs1 heterozygous mutants (gcs1/+; SALK_135496; Mori et al. 2006) were labeled with the HTR10p::HTR10:mRFP transgene.
A. thaliana seeds were sterilized with a solution containing 2 % Plant Preservative Mixture™ (Cosmo Bio, Japan), 50 µg/mL magnesium sulfide, and 0.1 % Tween overnight. The seeds were sown on MS medium consisting of 1 × Murashige and Skoog salt (Wako, Osaka, Japan), 2 % sucrose, 1 × Gamborg’s vitamin solution (Sigma, St. Louis, MO), and 0.3 % Gelrite (Wako) and adjusted to pH 5.7 with KOH. For the selection of gcs1/+ seedlings, MS medium containing 50 mg/L kanamycin sulfate and 0.8 % Bacto™ Agar (Becton, Dickinson and Company, Le Pont de Claix Cedex, France) instead of Gelrite was used. Plants were germinated and grown in a growth chamber at 22 °C with continuous lighting after cold treatment at 4 °C for 2–3 days. Two-week-old seedlings were transferred to soil and grown at 22 °C with continuous lighting. Hyponex 6-10-5 (Hyponex Japan, Osaka, Japan) was used as fertilizer (1:1,000 dilution).
Generation of constructs and plant transformation
To rescue the infertility of sperm cells in gcs1/+ mutants, a construct was designed as shown in Fig. 1 (see Table S1 for primers used for construction). Three fragments for Hsp::Cre, loxP-H2B:tdTomato, and RPS5Ap::loxP were amplified from a vector (DKv137; pMDC100/Hsp::Cre-RPS5Ap::loxP-GUS-loxP-H2B-tdTomato-NosT), which was a kind gift from Dr. Daisuke Kurihara (Nagoya University), and subcloned into pT7Blue vector (Novagen, Madison, WI) or pCR-Blunt II-TOPO vector (Invitrogen, Carlsbad, CA). The Hsp::Cre fragment consisted of the promoter region of the gene encoding soybean heat-shock protein [Gmhsp 17.3-B (Schöffl et al. 1984; Kurup et al. 2005); hereinafter called Hsp], an intron-containing sequence of the Cre-int gene (Zuo et al. 2001), and a Nos-terminator sequence. The loxP-H2B:tdTomato fragment consisted of a loxP sequence followed by the histone H2B coding sequence of A. thaliana, the tandem dimeric Tomato (tdTomato) gene sequence, and the Nos-terminator sequence. The RPS5Ap::loxP fragment consisted of the RPS5A promoter and a loxP sequence. The fragments RPS5Ap::loxP, Hsp::Cre, and loxP-H2B:tdTomato were cloned into the pMDC99 cloning vector (Curtis and Grossniklaus 2003) in this order using SpeI/PacI sites, AscI/KpnI sites, and SbfI/PmeI sites, respectively, resulting in pMDC99/rescue vector. A genomic sequence containing 1,002 bp of upstream and coding regions of GCS1 gene (At4g11720) was amplified using wild-type genomic DNA as a template. The sequence was cloned into the pENTR™/D-TOPO vector (Invitrogen), and the Nos-terminator sequence was added downstream of the GCS1 gene using NotI and NcoI sites. The GCS1 fragment was introduced between RPS5Ap::loxP and Hsp::Cre sequences in the pMDC99/rescue vector by LR reaction using Gateway LR Clonase II Enzyme mix (Invitrogen), resulting in pMDC99/rescue-GCS1 vector.
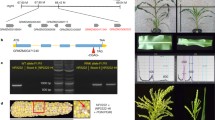
Generation of gcs1 homozygous mutant rescued by heat-inducible removal of the GCS1 transgene. a Flow chart of pMDC99/rescue-GCS1 vector construction. The GCS1p::GCS1 genomic fragment was introduced into the pMDC99/rescue vector by LR reaction. LB, left border region of T-DNA; RB, right border region of T-DNA; Hyg R, hygromycin resistance gene; ccdB, ccdB resistance gene; Cm R, chloramphenicol resistance gene. Purple arrows with numbers indicate different primer sets used for genomic PCR of the transgenic plants. b Genomic PCR of T1 plants. The upper and lower images show the gel images of the PCR analysis with non-heat-treated and heat-shocked leaves, respectively. The numbers under each lane indicate each T1 line number from which genomic DNA was extracted. The numbers 1 and 2 on the right side of the gel images indicate the fragments RPS5Ap-GCS1p and RPS5Ap-H2B, respectively, amplified with primer sets 1 and 2, described in Fig. 1a. c Sequence analysis of the recombined RPS5Ap-H2B gene. The fragment amplified with primer set 2 in Fig. 1b was confirmed to have the expected RPS5Ap-H2B sequence with one loxP sequence remaining. A part of the analyzed sequence is shown. The letters under each wave indicate DNA base sequences. d Genomic PCR of T2 plants to check whether gcs1 homozygous plants are produced by the introduced pMDC99/rescue-GCS1 vector. In the left image, the sites of primers are shown by red arrows with numbers. Primer set 3 amplifying the wild-type GCS1 gene was designed in intron 16 and the 3′ UTR region to amplify only the endogenous GCS1 gene. Primer set 4 was designed in the LB (left border) region of T-DNA to detect T-DNA insertion disrupting the GCS1 gene (gcs1 allele). The right image shows a part of the gel image of genomic PCR of the T2 #19 siblings. The yellow arrow indicates the band pattern of the gcs1/− plant
The construct was introduced into Agrobacterium tumefaciens stain GV3101 (pMP90) by electroporation. The T-DNA construct was transformed into gcs1/+ plants by the floral dip method, and transformed seeds were selected on MS medium containing 25 mg/L hygromycin B and 50 mg/L kanamycin sulfate. To confirm the introduction of the T-DNA into the resistant T1 plants, genomic PCR was performed to amplify part of the Cre sequence using genomic DNA of each T1 plant as the template. T2 seeds were selected on MS medium containing 25 mg/L hygromycin B or 50 mg/L kanamycin sulfate. The ratio of kanamycin-resistant and kanamycin-sensitive seedlings was counted and subjected to Chi-squared analysis based on an expectation of 50 %. The genomic DNA from hygromycin-resistant T2 plants was extracted and checked by genomic PCR using the primer sets listed in Table S1 to determine whether gcs1 homozygous mutant plants were produced by the introduced T-DNA. The primers were designed to target the endogenous gcs1 mutant allele and wild-type GCS1 allele without amplifying the GCS1 gene on the introduced T-DNA (Fig. 1d).
Heat-shock treatment for induction of the Cre-loxP system
For T1 plants, heat treatment was performed by exposing leaves collected in 1 mL of sterilized water of a 48-well plate to a temperature of 37 °C for 3 h. The heat-shocked leaves were then incubated for 24 h in a growth chamber at 22 °C, and genomic DNA was extracted from the leaves. To determine whether the Cre-loxP system was induced by heat-shock treatment, DNA sequences specific to non-excision (RPS5Ap-GCS1p) or excision (RPS5Ap-H2B) of a region between two loxP sites were amplified from genomic DNA with the primer sets listed in Table S1.
For heat-shock treatment of T3 plants, which were confirmed to be homozygous for the gcs1 mutant allele, seeds in 1.5-mL tubes containing sterilization solution or 2- or 7-day-old seedlings sown on MS medium were placed in an incubator at 37 °C for 6 h at each stage. The plants were then returned to the growth chamber at 22 °C and transferred to soil 2 weeks after sowing.
Expression analysis of GCS1 by RT-PCR
Total RNA was isolated from the anthers of 30 flowers for both wild-type and T4 gcs1/− plants as described below (Fig. 4) using an RNAqueous-Micro Kit (Life Technologies, Gaithersburg, MD) according to the manufacturer’s protocol. Reverse transcription reaction was conducted with a High Capacity RNA-to-cDNA Kit (Life Technologies) according to the manufacturer’s protocol. The amount of cDNA was approximately equalized between the cDNA from wild-type and gcs1/− based on the expression level of TUBULIN BETA CHAIN 4 (TUB4; At5g44340). The primers were designed to amplify the region upstream of the T-DNA insert and the region sandwiching the T-DNA insert. The primers used are listed in Table S1.
Semi-in vivo fertilization assay
Semi-in vivo fertilization assay was performed as described previously (Palanivelu and Preuss 2006; Hamamura et al. 2014). Briefly, 150 µL of pollen growth medium was poured into a silicon rubber well on a coverslip. Flowers were emasculated 12–20 h before hand pollination. The pistils of the RPS5Ap::H2B:GFP transgenic plants were pollinated by transgenic plants with HTR10p::HTR10:mRFP. The hand-pollinated pistils were cut with a 27-gauge needle at the junction between the style and ovary, and the stigmas were then placed horizontally on the pollen growth medium under a stereomicroscope. Ovules from the remaining ovaries were picked out and placed on the medium. Five to six ovules were arranged around the cut end of the stigmas with an insect pin 0.5 mm in diameter. The growth medium in the well was covered with another coverslip and incubated at 22 °C in the dark for about 6 h.
Microscopy settings and image processing for live-cell imaging of semi-in vivo fertilization were performed as described previously (Maruyama et al. 2013). Briefly, about 6 h after pollination (HAP), time-lapse and z-plane images were acquired every 5 min and seven planes (3-μm intervals) using a confocal microscope system. Sequential images were acquired after every 100 ms of exposure time. Images were processed with Metamorph version 7.7.7.0 (Universal Imaging Corp., Downingtown, PA) to create maximum-intensity projection images and to add color. Adobe Photoshop CS6 (Adobe Systems, Inc., San Jose, CA) was used to adjust the images. The images and movies were edited using MacBiophotonics ImageJ software (http://www.macbiophotonics.ca/).
Physiological analyses of ovules and pollen tubes
To examine seed development in pistils pollinated with wild-type or gcs1/− pollen, the pistils were cleared after the removal of the ovary wall. The pistils with ovules were dipped into a drop of clearing solution (8:1:3 w/v mixture of chloral hydrate, glycerol, and water) on glass slides and covered with a coverslip. The glass slides with samples were placed into a dark chamber at 4 °C for 3 days and observed using an upright microscope (Axio Imager. A2; Zeiss, Oberkochen, Germany).
To visualize pollen tubes entering the micropyle of ovules in the pistil, aniline blue staining was performed 24 HAP by the method described previously (Hülskamp et al. 1995). Briefly, one side of the ovary wall was removed, and the pistil was fixed on the glass slide with grease. The bare ovules were opened and fixed to each side with grease. The growing pollen tubes on the septum surface and ovules were stained by drops of aniline blue solution (5:8:7 v/v mixture of 2 % aniline blue, 1 M glycerol, pH 9.5, and water). Then, the glass slide was covered with a coverslip, and the sample was immediately observed using a confocal microscope (LSM780-DUO-NLO; Zeiss) or an upright microscope (DP71; Olympus).
Results
Generation of gcs1 homozygous mutant rescued by heat-induced removable GCS1 transgene
To obtain homozygous loss-of-function mutants for genes essential for gametophytic functions, we generated a rescue construct, pMDC99/rescue, in which introduced sequences of interest can be removed by the heat-induced Cre-loxP recombination system (Fig. 1a). This construct was designed to be applicable for the introduction of variable sequences using different entry vectors and site-specific recombination using GATEWAY technology. The GCS1p::GCS1 genomic fragment was integrated into the pMDC99/rescue vector to complement the gcs1 defect, resulting in pMDC99/rescue-GCS1 vector containing GCS1p::GCS1 and Cre recombinase genes between the loxP sites (Fig. 1a). We introduced the T-DNA region of the pMDC99/rescue-GCS1 vector into gcs1/+; HTR10p::HTR10:mRFP plants and obtained 19 T1 plants. Next, genomic PCR for heat-treated leaves from the T1 plants was performed to investigate whether the Cre-loxP recombination system in this construct could work in a heat-inducible manner (Fig. 1b). PCR using primer pairs specific to non-excision (RPS5Ap-GCS1p; primer set 1 in Fig. 1a) or excision (RPS5Ap-H2B; primer set 2 in Fig. 1a) of a region between two loxP sites showed that all of the heat-shocked T1 leaves had the RPS5Ap-H2B sequence produced after loxP recombination (Fig. 1b). We also confirmed that the fragment amplified with primer set 2 corresponded to the RPS5Ap-H2B sequence with the loxP sequence between the RPS5Ap and H2B sequences as expected (Fig. 1c). In 14 of 19 T1 lines, amplification of the recombinant RPS5Ap-H2B sequence was detected in genomic DNA from non-heat-treated leaves, indicating that leaky expression of Cre recombinase by Hsp promoter led to spontaneous recombination. Amplification of the RPS5Ap-GCS1p sequence was decreased by heat treatment in almost all T1 plants, indicating that the Cre-loxP recombination system in the construct functioned properly under the control of the Hsp promoter in leaves.
To confirm whether the introduced construct rescued the infertile phenotype of gcs1 sperm cells, we performed segregation analysis of T2 seeds obtained from 12 T1 lines using the kanamycin resistance cassette linked to the gcs1 mutant allele (Mori et al. 2006). We found a significant increase in kanamycin-resistant plants compared to the expected value (50 %) from self-pollinated heterozygous gcs1/+ (Table 1), indicating that the gcs1 allele was paternally transmitted by the rescue of the infertile phenotype in T1 plants. T2 lines with single insertion of the introduced T-DNA were also chosen by segregation analysis of hygromycin resistance (Table 1). Finally, we selected plants homozygous for the gcs1 allele by genomic PCR in T2 hygromycin-resistant plants from T1 lines #3, 6, 12, 16, and 19 (Fig. 1d).
Infertile transgenic gcs1 mutant plants can be induced by heat-shock treatment
To generate plants lacking both endogenous and transferred GCS1 by the heat-inducible Cre-loxP recombination system, we attempted to define the optimal heat treatment conditions using T3 seeds that were confirmed homozygous for gcs1 in the T2 generation (Table 1). As the heat-inducible recombination system was not expected to function effectively within the shoot apical meristem forming the aerial part of the plant body, seeds and/or seedlings were heat-treated several times (see Methods). When T3 seedlings were treated at 0, 2, and 7 days old for 6 h at 42 °C in accordance with the method reported previously (Kurup et al. 2005), most treated seedlings whitened and no longer grew, although some of the heat-shocked seedlings survived and showed infertility (Table 1). Therefore, we modified the heat treatment conditions to 37 °C twice at 2 and 7 days old to reduce heat damage. Some plants from lines #3, 16, and 19 had markedly short siliques with no or few seeds (Fig. 2a; Table 1). However, most showed partial infertility with long fertile siliques in some branches. When exposed to 37 °C treatment at 0 days old (i.e., seed stage) in addition to 2 and 7 days old, the majority of the heat-shocked plants exhibited the fully infertile phenotype, in which all branches had sterile siliques, whereas wild-type plants did not show this infertile phenotype (Table 1). Based on these results, we concluded that three rounds of 37 °C heat treatment of transgenic plant seedlings at 0, 2, and 7 days old efficiently generated heat-shock-dependent fully infertile plants.
Genotype and phenotype analyses of heat-shocked infertile plants. a Comparison of the heat-shocked wild-type plant (left) with the heat-shocked T3 #3-1 plant (right). Insets show magnifications of the fertile (1, 3) and infertile (2) siliques in areas marked by squares with numbers. Scale bar, 10 cm. b Genomic PCR of T3 plants to confirm complete removal of GCS1. The endogenous GCS1 (upper) and exogenous GCS1 on introduced T-DNA (lower) were amplified with primer sets 3 and 4 shown in Fig. 1d and primer sets 1′ and 2 shown in Fig. 1a, respectively. Genomic DNA was extracted from wild-type (WT), gcs1/+, and #3-1 siblings. Genomic DNA of #3-1 siblings was extracted from leaves of the non-heat-treated plant (None), infertile (Heat_I), and fertile (Heat_F) branches of the heat-shocked plant. c RT-PCR analysis of the gcs1/− plants. Total RNA isolated from wild-type plant or T4 gcs1/− plant (Fig. 4) and genomic DNA were used as PCR templates. Target genes are indicated on the right side of each gel image. GCS1_up and GCS1_int indicate the region upstream of the T-DNA insert and the region interrupted by the T-DNA insert in the gcs1 allele, respectively. d Seven days after pollination (DAP) wild-type siliques crossed with untreated (1) or heat-shocked (2) wild-type plants and untreated (3) or heat-shocked (4) T3 #3-1 plants. Scale bar, 5 mm. e Seed development in each silique shown in Fig. 2d. Scale bar, 1 mm. f Quantitative analysis of the proportion of developed seeds in the seven DAP siliques pollinated by untreated and heat-shocked wild-type and independent T3 lines (#3-1, #16-10, #19-12). Error bars indicate standard deviation. n, Number of siliques examined. g Reciprocal crossing of heat-shocked #16-10 plant with wild-type plant. The proportion of developed seeds in the seven DAP siliques was counted. Error bars indicate standard deviation. n, Number of siliques examined
To confirm whether the infertility resulted from complete removal of the GCS1 transgene, we performed genomic PCR using cauline leaves from non-heat-shocked fertile and heat-shocked infertile #3-1 sibling plants (Fig. 2a, b). As we found a partially fertile branch in the heat-shocked #3-1 plant (Fig. 2a; insert 3), a cauline leaf of the fertile branch was also assessed. As confirmed in the T2 generation, #3-1 plant siblings were homozygous for gcs1 (Fig. 2b). Although a pre-recombined sequence containing the GCS1 gene on the introduced T-DNA was detected from the non-heat-shocked fertile plant (Fig. 1a, amplified with primer set 1′), no amplification was detected from the heat-shocked, fully infertile branch (Fig. 2b). Interestingly, a partially fertile branch in the heat-shocked plant had the pre-recombined sequence (Fig. 2b), suggesting that heat-inducible Cre-loxP recombination was partially non-functional in the stem cell forming this branch. Such partially fertile branches were frequently observed in transgenic plants with heat-inducible fully infertile branches (Table 1). These results showed that the heat-shock-induced fully infertile phenotype was associated with complete loss of the GCS1 gene in the transgenic plants. We also performed RT-PCR analysis of the infertile gcs1/− plant and confirmed that transcripts of GCS1 were barely expressed in the gcs1/− plant, although slight expression of the region upstream of the T-DNA insert was detected (Fig. 2c).
We then checked the infertile phenotype by hand pollination experiments. The proportion of developed seeds was nearly 100 % when pollen from non-heat-treated #3-1 plants as well as heat-shocked or untreated wild-type plants was crossed to wild-type pistils (Fig. 2d, e, f). The proportion of developed seeds decreased significantly, to nearly 0 %, when crossed with pollen from the heat-shocked #3-1 plant (Fig. 2d, e, f). The same results were obtained using the untreated or heat-shocked plants of additional lines #16-10 and #19-12 (Fig. 2f). Normal female fertility of the heat-shocked #16-10 plant was confirmed by reciprocal crossing (Fig. 2g). Consequently, we achieved generation of male sterile plants without the GCS1 gene.
All pollen tubes from infertile plants contained gcs1 sperm cells and showed the lowest fertilization rate as male mutant
We examined whether the heat-shocked infertile plants showed the defective gcs1 sperm cell phenotype. The gcs1 sperm cells were reported to have a defect in membrane fusion with the egg cell and central cell during double fertilization, resulting in sperm cells remaining within the ovule receiving a pollen tube (Mori et al. 2006). We observed signals of discharged sperm cells and a vegetative nucleus in ovules crossed with the heat-shocked infertile plants (Fig. 3a). Most of the ovules contained at least one pair of unfertilized sperm cells (Fig. 3a). We then counted the number of red fluorescent signals remaining in the ovules using non-heat-treated or heat-shocked wild-type and #16-10 plants as pollen donors. In the case of the heat-shocked #16-10 plant, two to three signals and four to six signals were considered to be one and two pairs of sperm cells, respectively. Heat-induced H2B-tdTomato expression was sometimes too weak to detect the vegetative nucleus. The proportion of ovules with red fluorescent signals was significantly increased in crosses using pollen from the heat-shocked #16-10, with one pair of sperm cells in 50.3 % and two pairs in 25.6 % of ovules, whereas the signals of unfertilized sperm cells were rarely observed in the ovules crossed with the wild-type and untreated #16-10 plants (Fig. 3b). Although about half of the remaining 24 % of ovules also had one signal, we were unable to conclude whether it was from unfertilized sperm cells or a vegetative nucleus due to the weakness of the signal. For the same reason, the estimated proportion of ovules with two sperm cell pairs may have been lower than the actual level.
Observation of fertilization-defective sperm cells of the heat-shocked infertile plants. a Observation of the phenotype of gcs1 sperm cells in vivo. The wild-type ovules crossed with the heat-shocked #16-10 plant were observed at 12 HAP by confocal laser microscopy. The maximum projections of optical sections are stacked. The arrowheads and arrows indicate the signals of the unfertilized sperm cell nuclei (HTR10-mRFP) and vegetative nuclei (H2B-tdTomato), respectively. Insets show magnifications of the ovules with one pair (1) or two pairs (2) of unfertilized sperm cells in areas marked by squares with numbers. Scale bar, 100 µm. b Quantitative analysis of the proportion of ovules with unfertilized sperm cell pairs in wild-type pistils at 24 HAP. The number of red fluorescent signals as shown in Fig. 3a was counted. For ovules crossed with wild-type pollen possessing HTR10p::HTR10:mRFP gene, zero, one to two, and three or more signals were considered as zero, one, and two pairs of unfertilized sperm cells, respectively. For ovules crossed with #16-10 pollen possessing both HTR10p::HTR10:mRFP gene and RPS5Ap::H2B:tdTomato gene with leaky expression, zero to one, two to three, and four to six signals were considered as zero, one, and two pairs of unfertilized sperm cells, respectively. The white and black bars indicate the proportion of one pair and two pairs of sperm cells, respectively. Error bars indicate standard deviation. n, Number of siliques examined. c Time series images (maximum projections of optical sections) of semi-in vivo fertilization. Sperm nuclei and nuclei of female gametophytic cells were labeled by mRFP and GFP, respectively. The upper image is a montage of double fertilization process of wild-type sperm cells observed with gcs1/+ plants. The yellow and white arrowheads indicate the sperm cell nuclei moving toward the central cell nucleus and the egg cell nucleus, respectively. The asterisk indicates sperm cells in the other pollen tube. The lower image is a montage of images in which released sperm cell nuclei (arrowheads) of a heat-shocked #3-1 plant did not move toward the egg cell and central cell nuclei. The arrow indicates the vegetative nucleus labeled by H2B-tdTomato, which was expressed after recombination induced by heat treatment. Time indicates elapsed time; 0 min indicates the last frame just before pollen tube discharge. CCN central cell nucleus, ECN egg cell nucleus, SYN synergid cell nuclei. Scale bars, 20 µm. d Histogram of the number of cases observed by semi-in vivo assay with heat-shocked gcs1/+ and T3 #16-10 plants. The black and white bars indicate the number of ovules in which sperm cells showed wild-type and gcs1-like phenotypes, respectively. The numbers over each bar indicate the number of ovules observed
To examine the phenotype of the gcs1 sperm cells directly from the start of sperm cell release, we performed live-cell imaging of double fertilization by semi-in vivo fertilization assay (Hamamura et al. 2011). When crosses were performed with the pollen from a gcs1/+ plant, the behavior of wild-type sperm cells was observed as reported previously (Hamamura et al. 2011). The sperm cells labeled with HTR10-mRFP moved rapidly to the region between the egg cell and the central cell. Then, sperm nuclei moved toward the nuclei of their own fertilization targets (Fig. 3c, upper; Movie S1). In contrast, sperm cells of the heat-shocked #3-1 plant were discharged normally, but always remained motionless between the egg cell and the central cell (Fig. 3c, lower; Movie S2). In the case of gcs1/+, successful double fertilization occurred in about half of the ovules (5/11; Fig. 3d), and the gcs1 sperm cell phenotype was observed in the other ovules (6/11; Fig. 3d). On the other hand, when the heat-shocked #16-10 plant was used as a pollen donor, the gcs1 sperm cell phenotype was observed in all ovules (12/12; Fig. 3d). Finally, we concluded that the infertile plants induced by heat treatment were gcs1/− plants.
Homozygous gcs1 mutants can be obtained by infrequent fertilization of gcs1 sperm cells
About 80 % (59/73, n = 73 for T3_hs1; 48/61, n = 61 for T3_hs2) of the short siliques of auto-pollinated infertile plants contained a few aborted and/or developed seeds (Fig. 4a, b). As other mutants defective in sperm cell function such as gamete expressed 2 (gex2) and kokopelli were shown to produce aborted seeds resulting from single fertilization (Mori et al. 2014; Ron et al. 2010), we hypothesized that single fertilization of the egg cell or the central cell may be responsible for the aborted seeds in heat-inducible gcs1 homozygous plants. By clearing ovules accepting the gcs1 pollen tube, we observed ovules with only embryo or endosperm development, indicating single fertilization events (Fig. 4c), as observed in ovules fertilized by sperm cells complemented with partially functional GCS1 variants (Wong et al. 2010). Additionally, we found a few ovules with both embryo and endosperm in a silique, consistent with the presence of developed seeds (Fig. 4a). To determine whether the developed seeds were produced by successful double fertilization, we sowed the seeds on MS medium and analyzed the phenotype and genotype. The seeds germinated normally and grew into mature plants (Fig. 4d). Genomic PCR of the endogenous GCS1 allele and the transgene indicated that most of the growing plants had no functional GCS1 sequence similar to the heat-shocked T3 infertile parent (Fig. 4e), although some plants had either of the functional GCS1 genes, probably due to unintended cross-pollination of fertile pollen grains (data not shown). In agreement with these results, the plants without the GCS1 gene exhibited an infertile phenotype when crossed to wild-type pistils (Fig. 4f, g, h). The aborted and developed seeds were also observed in this T4 plant derived from the developed seeds in the heat-shocked T3 plant (Fig. 4b), suggesting that seed development in the gcs1/− plants was not due to the effect of heat treatment. Briefly, the gcs1/− plants could be maintained without heat treatment. Based on these results, we expected that gcs1 sperm cells may occasionally fertilize at a low frequency.
Analyses of seed development in heat-shocked infertile siliques and gcs1 homozygous mutant plants obtained from the developed seeds. a Aborted (arrowhead) and developed (arrow) seeds generated in the heat-shocked infertile silique. Scale bar, 1 mm. b Average numbers of aborted and developed seeds per silique of heat-shocked infertile plants (T3_hs1, 2) and T4 plants. T4 siblings were obtained from the T3_hs2 line. n, Number of siliques examined. c Cleared wild-type ovules at 3 DAP crossed with wild-type (1) or gcs1/− (2–4) pollen. The arrow and yellow dashed line indicate the developed embryo and the outline of the developed endosperm, respectively. The black and white arrowheads indicate the unfertilized central cell nucleus and the egg cell, respectively. In the ovule accepting wild-type pollen shown in panel 1, normal embryo and endosperm development resulting from successful double fertilization were observed. In the ovules accepting gcs1 pollen shown in panels 2, 3, and 4, unfertilized egg and central cells, the developed endosperm without embryo, and the developed embryo without endosperm were observed, respectively. Scale bar, 50 µm. d Comparison of the wild-type plant (left) with the infertile T4 plants grown from the developed seed in the heat-shocked infertile T3 gcs1/− plant (right). e Genomic PCR of the T4 siblings obtained from the heat-shocked T3 infertile plant. The upper and lower images represent the results of PCR using primer sets 3 and 4 (Fig. 1d) for endogenous GCS1 and primer sets 1′ and 2 (Fig. 1a) for exogenous GCS1, respectively. The left images are positive controls of PCR using the genomic DNA used in Fig. 2b (for Heat, Heat_I was used). f Seven DAP wild-type siliques crossed with wild-type (1), gcs1/+ (2), or gcs1/− (3) plants. Scale bar, 5 mm. g Seed development in each silique shown in f. The asterisks indicate the undeveloped seeds in the silique pollinated by the gcs1/+ plant. The arrowheads indicate the developed seeds in the silique pollinated by the gcs1/− plant. Scale bar, 1 mm. h Quantitative analysis of the proportion of developed seeds in the seven DAP siliques crossed with the wild-type, gcs1/+, and gcs1/− plants. Error bars indicate standard deviation. n, Number of siliques examined. i Observation of pollen tube attraction of the wild-type ovules crossed with gcs1/− pollen by aniline blue staining. The pistil at 24 HAP was examined by confocal laser microscopy. The maximum projections of optical sections are stacked. The arrowheads indicate pollen tubes entering into the ovules. Scale bar, 100 µm. j Quantitative analysis of the proportion of ovules attracting zero, one, and two or more pollen tubes at 24 HAP. The numbers of pollen tubes entering into the wild-type ovules crossed with wild-type, gcs1/+, or gcs1/− pollen were counted by aniline blue staining. The white, gray, and black bars indicate the proportions of ovules with zero, one, and two or more pollen tubes, respectively. Note that while a few ovules attracting more than two pollen tubes were observed, it was difficult to provide an accurate count. ND not determined (blue bar). Error bars indicate standard deviation. n, Number of siliques examined
It was reported that failure of fertilization by gcs1 sperm cells in a gcs1/+ mutant caused attraction of a second pollen tube to an ovule accepting a gcs1 pollen tube (Kasahara et al. 2012; Beale et al. 2012). In the case of gcs1/+ plants, 50 % of the ovules first accept a pollen tube containing gcs1 sperm cells, and then ~80 % attract a second pollen tube for fertilization recovery (Kasahara et al. 2012; Beale et al. 2012). We postulated that the rate of ovules attracting a second pollen tube would increase by the use of gcs1/− plants because all of the ovules first receive a gcs1-pollen tube and most fail in double fertilization (Fig. 3). We counted the number of pollen tubes reaching the micropyle of wild-type ovules at 24 HAP by aniline blue staining using wild-type, gcs1/+, or gcs1/− plants as a pollen donor (Fig. 4i). The results indicated that only 17.9 % of the ovules attracted two or more pollen tubes when wild-type was used in the cross (Fig. 4j). In the case of gcs1/+, the proportion of ovules with two or more pollen tubes increased to 41.9 %, whereas 51.0 % of the ovules attracted one pollen tube. As expected, when gcs1/− was used as the pollen donor, 70.9 % of the ovules attracted two or more pollen tubes (Fig. 4j). These observations indicated that gcs1/− plants could facilitate the analysis of pollen tube attraction during fertilization recovery as a male mutant maximizing the number of ovules attracting a second pollen tube.
Discussion
Studying reproductive process is of fundamental importance for understanding the underlying molecular mechanisms and increasing crop production by manipulation of reproductive genes in angiosperms. One of the major problems limiting the functional analysis of reproductive genes is the inaccessibility of fully defective homozygous mutants of genes involved in the reproductive process, which cannot be obtained naturally due to gametophytic dysfunction. In this study, we succeeded in generating gcs1 homozygous plants in which all male gametophytes contained fertilization-defective sperm cells, which cannot be obtained by self-crossing of gcs1/+ mutants (Mori et al. 2006). The homozygous mutant-inducible system developed here will be useful for investigating the functions of well-known and unknown factors in addition to GCS1 during the reproductive process. A sequence of interest can be introduced easily into the pMDC99/rescue vector by LR reaction, which is unrestricted by restriction enzyme sites for cloning into the vector, to rescue mutants (Fig. 1a). Our method is applicable to essential genes that function specifically in male and/or female gametophytes. As all gametophytes of the homozygous mutant plants carry mutant gametes, analyses of fertilization-defective mutants will become more efficient, as shown in this study (Figs. 3, 4). For example, the efficiency of observation of the gcs1 phenotype was doubled using homozygous gcs1 mutant for semi-in vivo fertilization assay (Fig. 3d).
Identification of novel fertilization-related factors and analyses of known factors are necessary to understand the molecular mechanisms underlying double fertilization in Arabidopsis. To explore novel genes involved in double fertilization, comparative transcriptomics or proteomics analyses using wild-type and purely male/female mutant gametes will be informative. Although mutants showing a strong phenotype may be required for the detection of important factors in such differential analyses, it is difficult to obtain homozygous mutants (Mori et al. 2006; von Besser et al. 2006; Durbarry et al. 2005). Therefore, an efficient method to artificially produce homozygous mutants would be important for such studies. Our heat-inducible system will be a useful means of addressing the difficulty of generating homozygous mutants for various genes. For example, application of the system to generate the duo1 mutant, which shows defects in sperm cell specification and fertilization (Durbarry et al. 2005; Rotman et al. 2005), may accelerate identification of factors essential for sperm cell function, including cell–cell communication among male and female gametes during double fertilization. DUO1 is a transcription factor that regulates expression of many genes, including GCS1 and downstream transcription factors specifically expressed in sperm cells (Brownfield et al. 2009; Borg et al. 2011). Comparative transcriptomics/proteomics analyses using isolated duo1 sperm-like cells from homozygous mutants obtained after application of our system would be a good strategy to identify the molecular mechanism of double fertilization, which still remains largely unknown. As several methods for mass isolation of male gametes of A. thaliana (Borges et al. 2008, 2012) and transcriptomics/proteomics analyses using sperm cells in various plants have been reported (Borges et al. 2008; Abiko et al. 2013; Zhao et al. 2013), it would be feasible to conduct large-scale analyses in the future.
We observed a very few developed seeds that grew into gcs1/− plants (Fig. 4d), which might result from the fertilization of the gcs1 sperm cells and the egg and central cell (Fig. 4c). This occasional fertilization was not detected previously using heterozygous mutants of gcs1 (Mori et al. 2006) or hap2 (von Besser et al. 2006) alleles, although the male transmission of hap2 allele was suggested at a very low frequency (0.7 %; Johnson et al. 2004). GCS1/HAP2 is also an indispensable factor for fertilization in Chlamydomonas and Plasmodium (Liu et al. 2008; Hirai et al. 2008). It is possible that the truncated GCS1 protein with extracellular and transmembrane domains is responsible for this occasional fertilization, which is suggested by the expression of gcs1 transcript upstream of the T-DNA (Fig. 2c). This is supported by the observation that modified GCS1 variants rescued the gcs1/hap2 phenotype at different levels (Mori et al. 2010; Wong et al. 2010). To examine the possibility that the partial function of the truncated GCS1 protein is responsible for the low-frequency fertilization, analysis using other alleles, such as hap2 allele (von Besser et al. 2006), would provide insights into this possibility. The T-DNA insertion site of the hap2 allele is earlier in the GCS1 gene than gcs1 allele we used in this study. Generation of homozygous mutants by heat-inducible recombination is useful for examining whether a mutant allele is fully infertile. In this study, using this strategy, rate of successful fertilization and seed development by the gcs1 sperm cells was estimated to be 0.16–0.46 % (0.08–0.23 seeds per silique with 50 ovules; Fig. 4b). As the gcs1 homozygous mutant could be maintained by self-pollination, seeds of this mutant will be useful for plant fertilization research.
We also showed that the fertilization recovery system worked strictly in over 70 % of the ovules first accepting gcs1 sperm cells (Fig. 4h). Remarkably, not all of the ovules accepting a gcs1 pollen tube attracted a second pollen tube, consistent with a previous report using gcs1/+ (Kasahara et al. 2012). These results suggest that polytubey block, including the inactivation of the remaining synergid cell mediated by ethylene signaling and unidentified dual control from the egg cell and central cell (Völz et al. 2013; Maruyama et al. 2013), may be induced partially just by the reception of a pollen tube. On the other hand, we observed ovules attracting more than two pollen tubes corresponding to a previous report (Beale et al. 2012). However, we could not show whether reception of more than two pollen tubes was completed and how attraction of multiple pollen tubes occurred by the ovule with two synergid cells. The gcs1/− plants generated here may facilitate detailed analyses of the regulation of pollen tube attraction and polytubey block during fertilization recovery, including the secretory pattern of the pollen tube attractant LURE from the remaining synergid cell (Takeuchi and Higashiyama 2012). The gcs1/− plants are also expected to be useful for determining the changes in the ovule occurring after sperm cell release preceding double fertilization, which may include the secretion of EC1 peptides from the egg cell to activate the sperm cells (Sprunck et al. 2012). Analyses of the egg cell and central cell that have been exposed to the environment just before double fertilization are expected to be applied, in combination with methods for the isolation of the female gametes (for a review, see Wuest et al. 2013) from the ovules accepting the gcs1/− pollen. Comparative transcriptomics/proteomics between ovules receiving gcs1 sperm cells and unfertilized ovules may uncover the alterations of gene expression and protein expression that could separate the reproductive process before from that after sperm cell release.
Author contribution statement
SN and HT designed the experiments. SN carried out most of the experiments. HT carried out in vivo phenotype analysis with SN. TH supervised the project. SN wrote the manuscript, and HT and TH edited it.
References
Abiko M, Furuta K, Yamauchi Y, Fujita C, Taoka M, Isobe T, Okamoto T (2013) Identification of proteins enriched in rice egg or sperm cells by single-cell proteomics. PLoS ONE 8:e69578
Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, Kurihara D, Matsunaga S, Umeda M (2011) Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA 108:10004–10009
Beale KM, Leydon AR, Johnson MA (2012) Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Curr Biol 22:1090–1094
Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D (2011) The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 23:534–549
Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148:1168–1181
Borges F, Gardner R, Lopes T, Calarco JP, Boavida LC, Slotkin RK, Martienssen RA, Becker JD (2012) FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods 8:44
Brownfield L, Hafidh S, Borg M, Sidorova A, Mori T, Twell D (2009) A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet 5:e1000430
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469
Denninger P, Bleckmann A, Lausser A, Vogler F, Ott T, Ehrhardt DW, Frommer WB, Sprunck S, Dresselhaus T, Grossmann G (2014) Male-female communication triggers calcium signatures during fertilization in Arabidopsis. Nat. Commun 5:4645
Durbarry A, Vizir I, Twell D (2005) Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol 137:297–307
Grobei MA, Qeli E, Brunner E, Rehrauer H, Zhang R, Roschitzki B, Basler K, Ahrens CH, Grossniklaus U (2009) Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res 19:1786–1800
Hamamura Y, Saito C, Awai C, Kurihara D, Miyawaki A, Nakagawa T, Kanaoka MM, Sasaki N, Nakano A, Berger F, Higashiyama T (2011) Live-cell imaging reveals the dynamics of two sperm cells during double fertilization in Arabidopsis thaliana. Curr Biol 21:497–502
Hamamura Y, Nagahara S, Higashiyama T (2012) Double fertilization on the move. Curr Opin Plant Biol 15:70–77
Hamamura Y, Nishimaki M, Takeuchi H, Geitmann A, Kurihara D, Higashiyama T (2014) Live imaging of calcium spikes during double fertilization in Arabidopsis. Nat Commun 5:4722
Hirai M, Arai M, Mori T, Miyagishima SY, Kawai S, Kita K, Kuroiwa T, Terenius O, Matsuoka H (2008) Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr Biol 18:607–613
Hülskamp M, Schneitz K, Pruitt RE (1995) Genetic evidence for a long-range activity that directs pollen tube guidance in arabidopsis. Plant Cell 7:57–64
Ingouff M, Hamamura Y, Gourgue SM, Higashiyama T, Berger F (2007) Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17:1032–1037
Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, Preuss D (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168:971–982
Kasahara RD, Maruyama D, Hamamura Y, Sakakibara T, Twell D, Higashiyama T (2012) Fertilization recovery after defective sperm cell release in Arabidopsis. Curr Biol 22:1084–1089
Kawashima T, Maruyama D, Shagirov M, Li J, Hamamura Y, Yelagandula R, Toyama Y, Berger F (2014) Dynamic F-actin movement is essential for fertilization in Arabidopsis thaliana. Elife 3:e04501
Kessler SA, Grossniklaus U (2011) She’s the boss: signaling in pollen tube reception. Curr Opin Plant Biol 14:622–627
Kos CH (2004) Cre/loxP system for generating tissue-specific knockout mouse models. Nutr Rev 62:243–246
Kurup S, Runions J, Köhler U, Laplaze L, Hodge S, Haseloff J (2005) Marking cell lineages in living tissues. Plant J 42:444–453
Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, Grishin NV, Steele RE, Sinden RE, Snell WJ, Billker O (2008) The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev 22:1051–1068
Maruyama D, Hamamura Y, Takeuchi H, Susaki D, Nishimaki M, Kurihara D, Kasahara RD, Higashiyama T (2013) Independent control by each female gamete prevents the attraction of multiple pollen tubes. Dev Cell 25:317–323
Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T (2006) GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol 8:64–71
Mori T, Hirai M, Kuroiwa T, Miyagishima SY (2010) The functional domain of GCS1-based gamete fusion resides in the amino terminus in plant and parasite species. PLoS One 5:e15957
Mori T, Igawa T, Tamiya G, Miyagishima SY, Berger F (2014) Gamete attachment requires GEX2 for successful fertilization in Arabidopsis. Curr Biol 24:170–175
Ohnishi Y, Hoshino R, Okamoto T (2014) Dynamics of male and female chromatin during karyogamy in rice zygotes. Plant Physiol 165:1533–1543
Okuda S, Suzuki T, Kanaoka MM, Mori H, Sasaki N, Higashiyama T (2013) Acquisition of LURE-binding activity at the pollen tube tip of Torenia fournieri. Mol Plant 6:1074–1090
Palanivelu R, Preuss D (2006) Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol 6:7
Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5:e1000621
Ron M, Saez MA, Williams LE, Fletcher JC, McCormick S (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24:1010–1021
Rotman N, Durbarry A, Wardle A, Yang WC, Chaboud A, Faure JE, Berger F, Twell D (2005) A novel class of MYB factors controls sperm-cell formation in plants. Curr Biol 15:244–248
Russell SH, Hoopes JL, Odell JT (1992) Directed excision of a transgene from the plant genome. Mol Gen Genet 234:49–59
Schöffl F, Raschke E, Nagao RT (1984) The DNA sequence analysis of soybean heat-shock genes and identification of possible regulatory promoter elements. EMBO J 3:2491–2497
Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338:1093–1097
Takeuchi H, Higashiyama T (2011) Attraction of tip-growing pollen tubes by the female gametophyte. Curr Opin Plant Biol 14:614–621
Takeuchi H, Higashiyama T (2012) A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol 10:e1001449
Völz R, Heydlauff J, Ripper D, von Lyncker L, Groß-Hardt R (2013) Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block. Dev Cell 25:310–316
von Besser K, Frank AC, Johnson MA, Preuss D (2006) Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133:4761–4769
Wong JL, Leydon AR, Johnson MA (2010) HAP2(GCS1)-dependent gamete fusion requires a positively charged carboxy-terminal domain. PLoS Genet 6:e1000882
Wuest SE, Schmid MW, Grossniklaus U (2013) Cell-specific expression profiling of rare cell types as exemplified by its impact on our understanding of female gametophyte development. Curr Opin Plant Biol 16:41–49
Zhao X, Yang N, Wang T (2013) Comparative proteomic analysis of generative and sperm cells reveals molecular characteristics associated with sperm development and function specialization. J Proteome Res 12:5058–5071
Zuo J, Niu QW, Møller SG, Chua NH (2001) Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol 19:157–161
Acknowledgments
We thank Daisuke Kurihara for providing the vector (DKv137) used for construction and for critical comments on this work. We also thank Michitaka Notaguchi, Daisuke Maruyama, Minako Ueda, and Tomokazu Kawashima for discussion and technical advice. SN was supported by Grant number 9285 from the Japan Society for the Promotion of Science Fellowship. A part of this work was conducted at the Institute of Transformative Bio-Molecules (WPI-ITbM) at Nagoya University, supported by the Japan Advanced Plant Science Network. This work was supported by the Japan Science and Technology Agency (ERATO project to TH).
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Silvia Coimbra and Lucia Colombo.
A contribution to the special issue ‘From Gametes to Seeds’.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Nagahara, S., Takeuchi, H. & Higashiyama, T. Generation of a homozygous fertilization-defective gcs1 mutant by heat-inducible removal of a rescue gene. Plant Reprod 28, 33–46 (2015). https://doi.org/10.1007/s00497-015-0256-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-015-0256-4