Abstract
Nitrilases capable of performing hydroxylation of 2-chloroisonicotinonitrile to 2-chloroisonicotinic acid were screened, and ES-NIT-102 was the best nitrilase for said biotransformation. Nitrilase was immobilized as cross linked enzyme aggregates (nitrilase–CLEAs) by fractional precipitation with iso-propanol, and cross linked with glutaraldehyde. The nitrilase–CLEAs prepared with optimized 35 mM glutaraldehyde for 120 min cross linking time had 82.36 ± 4.45% residual activity, and displayed type-II structural CLEAs formation as confirmed by particle size, SEM, FTIR, and SDS–PAGE analysis. Nitrilase–CLEAs had superior pH and temperature stability, showed a shift in optimal temperature by 5 °C, and retained nearly 1.5 to 1.7 folds activity over free nitrilase at 50 °C and 55 °C after more than 9 h incubation. Nitrilase–CLEAs showed reduced affinity and decreased conversion of substrate as indicated by slightly higher Km values by 5.19% and reduced Vmax by 17%. Furthermore, these nitrilase–CLEAs showed 98% conversion, 94.72 g/L product formation, and 83.30% recovery after 24 h when used for hydroxylation of 2-chloroisonicotinonitrile to 2-chloroisonicotinic acid. Nitrilase–CLEAs were catalytically active for 3 cycles showcasing 81% conversion, 75.53 g/L product formation and 66.42% yield. The recovered product was confirmed by HPLC, FTIR, LC–MS, and 1H NMR, and displayed > 99% purity.
Article highlights
-
ES-NIT-102 was best nitrilase amongst screened for hydroxylation reaction
-
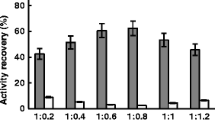
Nitrilase–CLEAs at 35 mM glutaraldehyde and 2 h crosslinking showed max activity recovery.
-
Nitrilase–CLEAs displayed good pH/thermal and kinetic stability and prominent reusability.
-
Nitrilase–CLEAs had good conversion, product formation and recovery for hydroxylation.
-
2-Chloroisonicotinonitrile and 2-chloroisonicotinic acid were confirmed by HPLC, FTIR, LCMS and NMR.










Similar content being viewed by others
References
Pavlova MV, Mikhalev AI, Kon’shin ME, Vasilyuk MV, Kotegov VP (2002) Synthesis and antiinflammatory activity of isonicotinic and cinchoninic acid derivatives. Pharm Chem J 36:425–427. https://doi.org/10.1023/A:1021258510259
Peters R, Althaus M, Diolez C, Rolland A, Manginot E, Veyrat M (2006) Practical formal total syntheses of the homocamptothecin derivative and anticancer agent diflomotecan via asymmetric acetate aldol additions to pyridine ketone substrates. J Org Chem 71(20):7583–7595. https://doi.org/10.1021/jo060928v
Fukunaga K, Uehara F, Aritomo K, Shoda A, Hiki S, Okuyama M, Usui Y, Watanabe K, Yamakoshi K, Kohara T, Hanano T, Tanaka H, Tsuchiya S, Sunada S, Saito KI, Eguchi JI, Yuki S, Asano S, Tanaka S, Mori A, Yamagami K, Baba H, Horikawa T, Fujimura M (2013) 2-(2-Phenylmorpholin-4-yl) pyrimidin-4 (3H)-ones; A new class of potent, selective and orally active glycogen synthase kinase-3β inhibitors. Bioorg Med Chem Lett 23(24):6933–6937. https://doi.org/10.1016/j.bmcl.2013.09.020
Haga Y, Sakamoto T, Shibata T, Nonoshita K, Ishikawa M, Suga T, Takahashi H, Takahashi T, Takahashi H, Ando M, Murai T, Gomori A, Oda Z, Kitazawa H, Mitobe Y, Kanesaka M, Ohe T, Iwaasa H, Ishii Y, Ishihara A, Kanatani A, Fukami T (2009) Discovery of trans-N-[1-(2-fluorophenyl)-3-pyrazolyl]-3-oxospiro[6-azaisobenzofuran-1(3H),1′-cyclohexane]-4′-carboxamide, a potent and orally active neuropeptide Y Y5 receptor antagonist. Bioorg Med Chem 17(19):6971–6982. https://doi.org/10.1016/j.bmc.2009.08.019
Schwender CF, Sunday BR, Herzig DJ (1979) 11-Oxo-11H-pyrido[2,1-b]quinazoline-8-carboxylic acid, an orally active antiallergy agent. J Med Chem 22(1):114–116. https://doi.org/10.1021/jm00187a026
Kost AN, Terent’ev PB, Golovleva LA, Stolyarchuk AA (1967) Synthesis of substituted pyridinecarboxylic acids and study of their properties. Pharm Chem J 1:235–240. https://doi.org/10.1007/BF00766422
Chapman J, Ismail AE, Dinu CZ (2018) Industrial applications of enzymes: recent advances, techniques, and outlooks. Catalysts 8(6):238. https://doi.org/10.3390/catal8060238
Shen JD, Cai X, Liu ZQ, Zheng YG (2021) Nitrilase: a promising biocatalyst in industrial applications for green chemistry. Crit Rev Biotechnol 41(1):72–93. https://doi.org/10.1080/07388551.2020.1827367
Asano Y, Kaul P (2012) Hydrolysis and reverse hydrolysis: selective nitrile hydrolysis using nitrilase and its related enzymes. Compr Chirality. https://doi.org/10.1016/b978-0-08-095167-6.00708-4
Cui J, Lin T, Feng Y, Tan Z, Jia S (2017) Preparation of spherical cross-linked lipase aggregates with improved activity, stability and reusability characteristic in water-in-ionic liquid microemulsion. J Chem Tech Biotechnol 92(7):1785–1793. https://doi.org/10.1002/jctb.5179
Teepakorn C, Zajkoska P, Cwicklinski G, De Berardinis V, Zaparucha A, Nonglaton G, Anxionnaz-Minvielle Z (2021) Nitrilase immobilization and transposition from a micro-scale batch to a continuous process increase the nicotinic acid productivity. Biotechnol J 16(10):2100010. https://doi.org/10.1002/biot.202100010
Federsel HJ, Moody TS, Taylor SJ (2021) Recent trends in enzyme immobilization—concepts for expanding the biocatalysis toolbox. Molecules 26(9):2822. https://doi.org/10.3390/molecules26092822
Peng H, Dong W, Chen Q, Song H, Sun H, Li R, Chang Y, Lu H (2022) Encapsulation of nitrilase in zeolitic imidazolate framework-90 to improve its stability and reusability. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-022-03890-z
Jin LQ, Guo DJ, Li ZT, Liu ZQ, Zheng YG (2016) Immobilization of nitrilase on bioinspired silica for efficient synthesis of 2-hydroxy-4-(methylthio) butanoic acid from 2-hydroxy-4-(methylthio) butanenitrile. J Ind Microbiol Biotechnol 43(5):585–593. https://doi.org/10.1007/s10295-016-1747-5
Qiu J, Su E, Wang W, Wei D (2014) Efficient asymmetric synthesis of D-N-formyl-phenylglycine via cross-linked nitrilase aggregates catalyzed dynamic kinetic resolution. Catal Commun 51:19–23. https://doi.org/10.1016/j.catcom.2014.03.019
Kaul P, Stolz A, Banerjee UC (2007) Cross-linked amorphous nitrilase aggregates for enantioselective nitrile hydrolysis. Adv Synth Catal 349(13):2167–2176. https://doi.org/10.1002/adsc.200700125
Cui J, Zhao Y, Feng Y, Lin T, Zhong C, Tan Z, Jia S (2017) Encapsulation of spherical cross-linked phenylalanine ammonia lyase aggregates in mesoporous biosilica. J Agric Food Chem 65(3):618–625. https://doi.org/10.1021/acs.jafc.6b05003
Bilal M, Cui J, Iqbal HMN (2019) Tailoring enzyme microenvironment: state-of-the-art strategy to fulfill the quest for efficient bio-catalysis. Int J Biol Macromol 130:186–196. https://doi.org/10.1016/j.ijbiomac.2019.02.141
Cui JD, Jia SR (2015) Optimization protocols and improved strategies of cross-linked enzyme aggregates technology: current development and future challenges. Crit Rev Biotechnol 35(1):15–28. https://doi.org/10.3109/07388551.2013.795516
Sheldon RA (2019) CLEAs, Combi-CLEAs and ‘Smart’ Magnetic CLEAs: biocatalysis in a bio-based economy. Catalysts 9(3):261. https://doi.org/10.3390/catal9030261
Behera S, Dev MJ, Singhal RS (2022) Cross-linked β-mannanase aggregates: preparation, characterization, and application for producing partially hydrolyzed guar gum. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-022-03807-w
Ren S, Li C, Jiao X, Jia S, Jiang Y, Bilal M, Cui J (2019) Recent progress in multienzymes co-immobilization and multienzyme system applications. Chem Eng J 373:1254–1278. https://doi.org/10.1016/j.cej.2019.05.141
Cui J, Zhao Y, Liu R, Zhong C, Jia S (2016) Surfactant-activated lipase hybrid nanoflowers with enhanced enzymatic performance. Sci Rep 6:27928. https://doi.org/10.1038/srep27928
Cui J, Lin T, Feng Y, Tan Z, Jia S (2017) Preparation of spherical cross-linked lipase aggregates with improved activity, stability and reusability characteristic in water-in-ionic liquid microemulsion. J Chem Technol Biotechnol 92:1785–1793. https://doi.org/10.1002/jctb.5179
Cui J, Tan Z, Han P, Zhong C, Jia S (2017) Enzyme shielding in a large mesoporous hollow silica shell for improved recycling and stability based on CaCO3 microtemplates and biomimetic silicification. J Agric Food Chem 65(19):3883–3890. https://doi.org/10.1021/acs.jafc.7b00672
Gupta K, Jana AK, Kumar S, Jana MM (2015) Solid state fermentation with recovery of amyloglucosidase from extract by direct immobilization in cross linked enzyme aggregate for starch hydrolysis. Biocatal Agricult Biotechnol 4:486–492. https://doi.org/10.1016/j.bcab.2015.07.007
Kulkarni NH, Muley AB, Bedade DK, Singhal RS (2020) Crosslinked enzyme aggregates of arylamidase from Cupriavidus oxalaticus ICTDB921: process optimization, characterization, and application for mitigation of acrylamide in industrial wastewater. Bioprocess Biosyst Eng 43(3):457–471. https://doi.org/10.1007/s00449-019-02240-4
Muley AB, Awasthi S, Bhalerao PP, Jadhav NL, Singhal RS (2021) Preparation of cross-linked enzyme aggregates of lipase from Aspergillus niger: process optimization, characterization, stability, and application for epoxidation of lemongrass oil. Bioprocess Biosyst Eng 44(7):1383–1404. https://doi.org/10.1007/s00449-021-02509-7
Chaudhari SA, Singhal RS (2017) A strategic approach for direct recovery and stabilization of Fusarium sp. ICT SAC1 cutinase from solid state fermented broth by carrier free cross-linked enzyme aggregates. Int J Biol Macromol 98:610–621. https://doi.org/10.1016/j.ijbiomac.2017.02.033
Kumar S, Mohan U, Kamble AL, Pawar S, Banerjee UC (2010) Cross-linked enzyme aggregates of recombinant Pseudomonas putida nitrilase for enantioselective nitrile hydrolysis. Bioresour Technol 101(17):6856–6858. https://doi.org/10.1016/j.biortech.2010.03.084
Kim MH, Park S, Kim YH, Won K, Lee SH (2013) Immobilization of formate dehydrogenase from Candida boidinii through cross-linked enzyme aggregates. J Mol Catal B Enzym 97:209–214. https://doi.org/10.1016/j.molcatb.2013.08.020
Miao C, Li H, Zhuang X, Wang Z, Yang L, Lv P, Luo W (2019) Synthesis and properties of porous CLEAs lipase by the calcium carbonate template method and its application in biodiesel production. RSC Adv 9(51):29665–29675. https://doi.org/10.1039/C9RA04365A
Shuddhodana GMN, Bisaria VS (2018) Effectiveness of crosslinked enzyme aggregates of cellulolytic enzymes in hydrolyzing wheat straw. J Biosci Bioeng 126(4):445–450. https://doi.org/10.1016/j.jbiosc.2018.04.007
Samoylova Y, Sorokina K, Piligaev A, Parmon V (2018) Preparation of stable cross-linked enzyme aggregates (CLEAs) of a Ureibacillus thermosphaericus esterase for application in malathion removal from wastewater. Catalysts 8(4):154. https://doi.org/10.3390/catal8040154
Yildirim D, Tükel SS, Alagoz D (2014) Crosslinked enzyme aggregates of hydroxynitrile lyase partially purified from Prunus dulcis seeds and its application for the synthesis of enantiopure cyanohydrins. Biotechnol Prog 30(4):818–827. https://doi.org/10.1002/btpr.1925
Zhang ZJ, Pan J, Li CX, Yu HL, Zheng GW, Ju X, Xu JH (2014) Efficient production of (R)-(−)-mandelic acid using glutaraldehyde cross-linked Escherichia coli cells expressing Alcaligenes sp. nitrilase. Bioprocess Biosyst Eng 37(7):1241–1248. https://doi.org/10.1007/s00449-013-1096-y
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt Biochem 72(1–2):248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Vergne-Vaxelaire C, Bordier F, Fossey A, Besnard-Gonnet M, Debard A, Mariage A, Pellouin V, Perret A, Petit JL, Stam M, Salanoubat M (2013) Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv Synth Catal 355(9):1763–1779. https://doi.org/10.1002/adsc.201201098
Thakur N, Sharma NK, Thakur S, Bhalla TC (2019) Bioprocess development for the synthesis of 4-aminophenylacetic acid using nitrilase activity of whole cells of Alcaligenes faecalis MTCC 12629. Catal Lett 149(10):2854–2863. https://doi.org/10.1007/s10562-019-02762-2
Cai W, Su E, Zhu S, Ren Y, Wei D (2014) Characterization of a nitrilase from Arthrobacter aurescens CYC705 for synthesis of iminodiacetic acid. J Gen Appl Microbiol 60(6):207–214. https://doi.org/10.2323/jgam.60.207
Zhang K, Pan T, Wang L, Wang H, Ren Y, Wei D (2022) Screening and characterization of a nitrilase with significant nitrile hydratase activity. Res Sq. https://doi.org/10.21203/rs.3.rs-1476795/v1
Chen Z, Chen H, Ni Z, Tian R, Zhang T, Jia J, Yang S (2015) Expression and characterization of a novel nitrilase from hyperthermophilic bacterium Thermotoga maritima MSB8. J Microbiol Biotechnol 25(10):1660–1669. https://doi.org/10.4014/jmb.1502.02032
Jiang W, Tao R, Yang Y, Xu Y, Kang H, Zhou X, Zhou Z (2017) Production of (R)-(-)-mandelic acid with nitrilase immobilized on D155 resin modified by l-lysine. Biochem Eng J 127:32–42. https://doi.org/10.1016/j.bej.2017.07.010
Xing X, Jia JQ, Zhang JF, Zhou ZW, Li J, Wang N, Yu XQ (2019) CALB immobilized onto magnetic nanoparticles for efficient kinetic resolution of racemic secondary alcohols: long-term stability and reusability. Molecules 24:490. https://doi.org/10.3390/molecules24030490
Muley AB, Mulchandani KH, Singhal RS (2020) Immobilization of enzymes on iron oxide magnetic nanoparticles: synthesis, characterization, kinetics and thermodynamics. Methods Enzymol 630:39–79. https://doi.org/10.1016/bs.mie.2019.10.016
Muley AB, Chaudhari SA, Bankar SB, Singhal RS (2019) Stabilization of cutinase by covalent attachment on magnetic nanoparticles and improvement of its catalytic activity by ultrasonication. Ultrason Sonochem 75:174–185. https://doi.org/10.1016/j.ultsonch.2019.02.019
Schoevaart R, Wolbers MW, Golubovic M, Ottens M, Kieboom APG, Van Rantwijk F, Sheldon RA (2004) Preparation, optimization, and structures, of cross-linked enzyme aggregates (CLEAs). Biotechnol Bioeng 87(6):754–762. https://doi.org/10.1002/bit.20184
Dennett GV, Blamey JM (2016) A new thermophilic nitrilase from an Antarctic hyperthermophilic microorganism. Front Bioeng Biotechnol 4(5):1–9. https://doi.org/10.3389/fbioe.2016.00005
De Martino MT, Tonin F, Yewdall NA, Abdelghani M, Williams DS, Hanefeld U, Rutjes FP, Abdelmohsen LK, Van Hest JC (2020) Compartmentalized cross-linked enzymatic nano-aggregates (c-CLEnA) for efficient in-flow biocatalysis. Chem Sci 11(10):2765–2769. https://doi.org/10.1039/C9SC05420K
Bedade DK, Muley AB, Singhal RS (2019) Magnetic crosslinked enzyme aggregates of acrylamidase from Cupriavidus oxalaticus ICTDB921 for biodegradation of acrylamide from industrial waste water. Bioresour Technol 272:137–145. https://doi.org/10.1016/j.biortech.2018.10.015
Malandra A, Cantarella M, Kaplan O, Vejvoda V, Uhnakova B, Stepankova B, Kubac D, Martinkova L (2009) Continuous hydrolysis of 4-cyanopyridine by nitrilases from Fusarium solani O1 and Aspergillus niger K10. Appl Microbiol Biotechnol 85(2):277–284. https://doi.org/10.1007/s00253-009-2073-x
Yang X, Huang A, Peng J, Wang J, Wang X, Lin Z, Li S (2014) Self-assembly amphipathic peptides induce active enzyme aggregation that dramatically increases the operational stability of nitrilase. RSC Adv 4(105):60675–60684. https://doi.org/10.1039/C4RA11236A
Vejvoda V, Kubac D, Davidova A, Kaplan O, Sulc M, Sveda O, Chaloupkova R, Martinkova L (2010) Purification and characterization of nitrilase from Fusarium solani IMI196840. Proc Biochem 45(7):1115–1120. https://doi.org/10.1016/j.procbio.2010.03.033
Martinkova L, Kren V (2010) Biotransformations with nitrilases. Curr Opin Chem Biol 14(2):130–137. https://doi.org/10.1016/j.cbpa.2009.11.018
Zou SP, Huang JW, Xue YP, Zheng YG (2018) Highly efficient production of 1-cyanocyclohexaneacetic acid by cross-linked cell aggregates (CLCAs) of recombinant E. coli harboring nitrilase gene. Proc Biochem 65:93–99. https://doi.org/10.1016/j.procbio.2017.10.014
Zhang XH, Liu ZQ, Xue YP, Wang YS, Yang B, Zheng YG (2018) Production of R-mandelic acid using nitrilase from recombinant E. coli cells immobilized with tris (hydroxymethyl) phosphine. Appl Biochem Biotechnol 184(3):1024–1035. https://doi.org/10.1007/s12010-017-2604-3
Toprak A, Tukel SS, Yildirim D (2021) Stabilization of multimeric nitrilase via different immobilization techniques for hydrolysis of acrylonitrile to acrylic acid. Biocatal Biotransform 39(3):221–231. https://doi.org/10.1080/10242422.2020.1869217
Sheldon RA, Basso A, Brady D (2021) New frontiers in enzyme immobilisation: robust biocatalysts for a circular bio-based economy. Chem Soc Rev 50(10):5850–5862. https://doi.org/10.1039/D1CS00015B
Chauhan V, Kaushal D, Dhiman VK, Kanwar SS, Singh D, Dhiman VK, Pandey H (2022) An insight in developing carrier-free immobilized enzymes. Front Bioeng Biotechnol 10:794411. https://doi.org/10.3389/fbioe.2022.794411
Talekar S, Nadar S, Joshi A, Joshi G (2014) Pectin cross-linked enzyme aggregates (pectin-CLEAs) of glucoamylase. RSC Adv 4(103):59444–59453. https://doi.org/10.1039/C4RA09552A
Nadar SS, Patil PD, Rohra NM (2020) Magnetic nanobiocatalyst for extraction of bioactive ingredients: a novel approach. Trends Food Sci Technol 103:225–238. https://doi.org/10.1016/j.tifs.2020.07.007
Acknowledgements
The authors are grateful to Dean, School of Basic & Applied Sciences, and Vice-Chancellor, Galgotias University, Greater Noida for providing required facilities to carry out this research work.
Author information
Authors and Affiliations
Contributions
Design of experiments: AGK and ABM; Performance of experiment: AGK; Analysis and interpretation of data: AGK, AKJ, and ABM; Manuscript draft and compilation: AGK, and ABM; Manuscript correction and revision: AGK, AKJ, and ABM.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not include any studies with human participants or animals performed by any authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khatik, A.G., Jain, A.K. & Muley, A.B. Preparation, characterization and stability of cross linked nitrilase aggregates (nitrilase–CLEAs) for hydroxylation of 2-chloroisonicotinonitrile to 2-chloroisonicotinic acid. Bioprocess Biosyst Eng 45, 1559–1579 (2022). https://doi.org/10.1007/s00449-022-02766-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-022-02766-0