Abstract
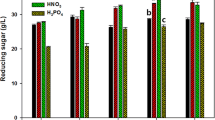
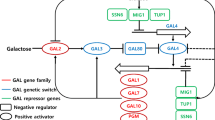
A total monosaccharide concentration of 47.0 g/L from 12% (w/v) Gracilaria verrucosa was obtained by hyper thermal acid hydrolysis with 0.2 M HCl at 140°C for 15 min and enzymatic saccharification with CTec2. To improve galactose utilization, we overexpressed two genes, SNR84 and PGM2, in a Saccharomyces cerevisiae CEN-PK2 using CRISPR/Cas-9. The overexpression of both SNR84 and PGM2 improved galactose utilization and ethanol production compared to the overexpression of each gene alone. The overexpression of both SNR84 and PGM2 and of PGM2 and SNR84 singly in S. cerevisiae CEN-PK2 Cas9 produced 20.0, 18.5, and 16.5 g/L ethanol with ethanol yield (YEtOH) values of 0.43, 0.39, and 0.35, respectively. However, S. cerevisiae CEN-PK2 adapted to high concentration of galactose consumed galactose completely and produced 22.0 g/L ethanol at a YEtOH value of 0.47. The overexpression of both SNR84 and PGM2 increased the transcriptional levels of GAL and regulatory genes; however, the transcriptional levels of these genes were lower than those in S. cerevisiae adapted to high galactose concentrations.







Similar content being viewed by others
References
Harun R, Yip JWS, Thiruvenkadam S, Ghani WA, Cherrington T, Danquah MK (2014) Algal biomass conversion to bioethanol—a step-by-step assessment. Biotechnol J 9:73–86
Meinita MDN, Marhaeni B, Winanto T, Setyaningsih D, Hong Y-K (2015) Catalytic efficiency of sulfuric and hydrochloric acids for the hydrolysis of Gelidium latifolium (Gelidiales, Rhodophyta) in bioethanol production. J Ind Eng Chem 27:108–114
Bellanger F, Verdus MC, Henocq V, Christiaen D (1990) Determination of the composition of the fibrillar part of Gracilaria verrucosa (Gracilariales, Rhodophyta) cell wall in order to prepare protoplasts. Hydrobiologia 204–205:527–531
Yanagisawa M, Kawai S, Murata K (2013) Strategies for the production of high concentrations of bioethanol from seaweeds: production of high concentrations of bioethanol from seaweeds. Bioengineered 4:224–235
Bensah EC, Mensah M (2013) Chemical pretreatment methods for the production of cellulosic ethanol: technologies and innovations. Int J Chem Eng 2013:1–21
Jönsson LJ, Alriksson B, Nilvebrant N-O (2013) Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6:1–16
Meinita MDN, Hong Y-K, Jeong G-T (2012) Detoxification of acidic catalyzed hydrolysate of Kappaphycus alvarezii (cottonii). Bioprocess Biosyst Eng 35:93–98
Ra CH, Nguyen TH, Jeong G-T, Kim S-K (2016) Evaluation of hyper thermal acid hydrolysis of Kappaphycus alvarezii for enhanced bioethanol production. Bioreour Technol 209:66–72
Ostergaard S, Olsson L, Johnston M, Nielsen J (2000) Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat Biotechnol 18:1283–1286
Hong K-K, Vongsangnak W, Vemuri GN, Nielsen J (2011) Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc Natl Acad Sci USA 108:12179–12184
Bro C, Knudsen S, Regenberg B, Olsson L, Nielsen J (2005) Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol 71:6465–6472
Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929–934
Ra CH, Kim YJ, Lee SY, Jeong G-T, Kim S-K (2015) Effects of galactose adaptation in yeast for ethanol fermentation from red seaweed, Gracilaria verrucosa. Bioprocess Biosyst Eng 38:1715–1722
Yu BJ, Park JC, Koo HM, Jin YS, Lee KS (2009) Vector and microorganism for increasing galactose catabolism and methods therefor. US patent 8,722,388 B2
Lee K-S, Hong M-E, Jung S-C, Ha S-J, Yu BJ, Koo HM, Park SM, Seo J-H, Kweon D-H, Park JC, Jin Y-S (2011) Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol Bioeng 108:621–631
Cunniff P (1995) AOAC (Association of Official Analytical Chemists), 16th edn. AOAC International, Washington DC
Cripwell R, Favaro L, Rose SH, Basaglia M, Cagnin L, Casella S, van Zyl W (2015) Utilisation of wheat bran as a substrate for bioethanol production using recombinant cellulases and amylolytic yeast. Appl Energy 160:610–617
Kubicek CP (1982) beta-Glucosidase excretion by Trichoderma pseudokoningii: correlation with cell wall bound beta-1.3-glucanase activities. Arch Microbiol 132:349–354
Mandels M, Andreotti R, Roche C (1976) Measurement of saccharifying cellulase. Biotechnol Bioeng Symp 6:21–33
Kim SR, Xu H, Lesmana A, Kuzmanovic U, Au M, Florencia C, Oh EJ, Zhang G, Kim KH, Jin Y-S (2015) Deletion of PHO13, encoding haloacid dehalogenase type IIA phosphatase, results in upregulation of the pentose phosphate pathway in Saccharomyces cerevisiae. Appl Environ Microbiol 81:1601–1609
Nguyen TH, Ra CH, Sunwoo IY, Jeong G-T, Kim S-K (2016) Evaluation of galactose adapted yeasts for bioethanol fermentation from Kappaphycus alvarezii hydrolyzates. J Microbiol Biotechnol 26:1259–1266
Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu J-H, Zhao H (2012) Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng 109:2082–2092
Xu H, Kim S, Sorek H, Lee Y, Jeong D, Kim J, Oh EJ, Yun EJ, Wemmer DE, Kim KH, Kim SR, Jin Y-S (2016) PHO13 deletion-induced transcriptional activation prevents sedoheptulose accumulation during xylose metabolism in engineered Saccharomyces cerevisiae. Metab Eng 34:88–96
EauClaire SF, Zhang J, Rivera CG, Huang LL (2016) Combinatorial metabolic pathway assembly in the yeast genome with RNA-guided Cas9. J Ind Microbiol Biotechnol 43:1001–1015
Ra CH, Choi JG, Kang C-H, Sunwoo IY, Jeong G-T, Kim S-K (2015) Thermal acid hydrolysis pretreatment, enzymatic saccharification and ethanol fermentation from red seaweed, Gracilaria verrucosa. Microbiol Biotechnol Lett 43:9–15
Jeong TS, Choi CH, Lee JY, Oh KK (2012) Behaviors of glucose decomposition during acid-catalyzed hydrothermal hydrolysis of pretreated Gelidium amansii. Bioresour Technol 116:435–440
Lee SY, Chang JH, Lee SB (2014) Chemical composition, saccharification yield, and the potential of the green seaweed Ulva pertusa. Biotechnol Bioprocess Eng 19:1022–1033
Avila-Gaxiola J, Velarde-Escobar OJ, Millan-Almaraz JR, Ramos-Brito F, Atondo-Rubio G, Yee-Rendon C, Avila-Gaxiola E (2018) Treatments to improve obtention of reducing sugars from agave leaves powder. Ind Crops Prod 112:577–583
Shi J, Wu D, Zhang L, Simmons BA, Singh S, Yang B, Wyman CE (2017) Dynamic changes of substrate reactivity and enzyme adsorption on partially hydrolyzed cellulose. Biotechnol Bioeng 114:503–515
Rodrigues AC, Haven MØ, Lindedam J, Felby C, Gama M (2015) Celluclast and Cellic® CTec2: Saccharification/fermentation of wheat straw, solid–liquid partition and potential of enzyme recycling by alkaline washing. Enzyme Microb Technol 79–80:70–77
Nguyen TH, Ra CH, Sunwoo IY, Sukwong P, Jeong G-T, Kim S-K (2018) Bioethanol production from soybean residue via separate hydrolysis and fermentation. Appl Biochem Biotechnol 184:513–523
Navarrete C, Nielsen J, Siewers V (2014) Enhanced ethanol production and reduced glycerol formation in fps1∆ mutants of Saccharomyces cerevisiae engineered for improved redox balancing. AMB Express 4:1–8
Mikkilineni V, Mitra RD, Merritt J, DiTonno JR, Church GM, Ogunnaike B, Edwards JS (2004) Digital quantitative measurements of gene expression. Biotechnol Bioeng 86:117–124
Oh D, Hopper JE (1990) Transcription of a yeast phosphoglucomutase isozyme gene is galactose inducible and glucose repressible. Mol Cell Biol 10:1415–1422
Hawkins KM, Smolke CD (2006) The regulatory roles of the galactose permease and kinase in the induction response of the GAL network in Saccharomyces cerevisiae. J Biol Chem 281:13485–13492
Bram RJ, Lue NF, Kornberg RD (1986) A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J 5:603–608
Lee H-J, Kim S-J, Yoon J-J, Kim KH, Seo J-H, Park Y-C (2015) Evolutionary engineering of Saccharomyces cerevisiae for efficient conversion of red algal biosugars to bioethanol. Bioresour Technol 191:445–451
Bae Y-H, Kweon D-H, Park Y-C, Seo J-H (2014) Deletion of the HXK2 gene in Saccharomyces cerevisiae enables mixed sugar fermentation of glucose and galactose in oxygen-limited conditions. Process Biochem 49:547–553
Rodríguez A, La Cera TD, Herrero P, Moreno F (2001) The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J 355:625–631
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2016R1D1A1A09918683), Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors indicate that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sukwong, P., Sunwoo, I.Y., Jeong, D.Y. et al. Enhancement of bioethanol production from Gracilaria verrucosa by Saccharomyces cerevisiae through the overexpression of SNR84 and PGM2. Bioprocess Biosyst Eng 42, 1421–1433 (2019). https://doi.org/10.1007/s00449-019-02139-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-019-02139-0