Abstract
We provide the first whole genome Copy Number Variant (CNV) study addressing Roma, along with reference populations from South Asia, the Middle East and Europe. Using CNV calling software for short-read sequence data, we identified 3171 deletions and 489 duplications. Taking into account the known population history of the Roma, as inferred from whole genome nucleotide variation, we could discern how this history has shaped CNV variation. As expected, patterns of deletion variation, but not duplication, in the Roma followed those obtained from single nucleotide polymorphisms (SNPs). Reduced effective population size resulting in slightly relaxed natural selection may explain our observation of an increase in intronic (but not exonic) deletions within Loss of Function (LoF)-intolerant genes. Over-representation analysis for LoF-intolerant gene sets hosting intronic deletions highlights a substantial accumulation of shared biological processes in Roma, intriguingly related to signaling, nervous system and development features, which may be related to the known profile of private disease in the population. Finally, we show the link between deletions and known trait-related SNPs reported in the genome-wide association study (GWAS) catalog, which exhibited even frequency distributions among the studied populations. This suggests that, in general human populations, the strong association between deletions and SNPs associated to biomedical conditions and traits could be widespread across continental populations, reflecting a common background of potentially disease/trait-related CNVs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Structural variants (SVs) are a class of genomic rearrangements, larger than 50 bp, comprising insertions, deletions, duplications, inversions and translocations, which are responsible for the largest fraction of base pair variation in the human genome (Weischenfeldt et al. 2013; Sudmant et al. 2015b). Within SVs, balanced mutations (inversions and translocations) do not alter the genomic dosage, while unbalanced rearrangements (insertions, duplications and deletions, the latter two also known collectively as Copy Number Variants, CNVs) involve losses or gains of genetic material. CNVs can exert their influence on gene expression, phenotypic traits, and diseases, and represent a main source of genetic variation on which natural selection can act upon (Stranger et al. 2007; Hurles et al. 2008; Perry et al. 2008; Handsaker et al. 2015; Audano et al. 2019; Collins et al. 2020; Hollox et al. 2022). Indeed, CNVs have been linked to a number of traits such as Crohn’s disease, osteoporosis, HIV susceptibility, body mass index, cancers and psoriasis (McCarroll et al. 2008; Yang et al. 2008; De Cid et al. 2009; Willer et al. 2009; Mohamad Isa et al. 2020; Dentro et al. 2021; Hamdan and Ewing 2022) and are intriguingly associated to neurodevelopmental disorders in humans (Sebat et al. 2007; Stefansson et al. 2008; Girirajan et al. 2013; Singh et al. 2017; Morris-Rosendahl and Crocq 2020; Sekiguchi et al. 2020; Kato et al. 2022).
Most of the studies addressing human population genetics have historically focused on SNPs to infer human population demography, such as changes in effective population size due to bottlenecks or founder events, or gene flow due to migration. This is also the case for the investigation of the mutation load, that is, the global contribution of deleterious mutations to disease. However, research using CNVs as markers in population genetics surveys, both in large worldwide comparisons and on finer scales, has been increasingly accumulating over the last two decades and confirmed their potential in this field, highlighting among/within group variability, the functional potential of the variants (including pathogenic effects) and their evolutionary relevance (Redon et al. 2006; Itsara et al. 2008; Gautam et al. 2012; Sudmant et al. 2015b; Hehir-Kwa et al. 2016; Urnikyte et al. 2016; Dennis et al. 2017; Almarri et al. 2020; Collins et al. 2020; Bergström et al. 2020). Attention has also been given to the study of CNVs in underrepresented or isolated populations, with a putative intriguing demographic history. Earlier reports mainly focused on isolates of European ancestry, such as Scottish islands of Orkney, the Italian South Tyrol region, the Croatian Vis island and the Finnish populations. These studies highlighted the general common sharing of CNVs among population as well as isolate-specific relatedness and the presence of novel variants, uncovering a previously hidden layer of CNV variation (Chen et al. 2011; Kanduri et al. 2013). Further analyses focusing on Asian and North African samples showed enrichment of structural variant events in biomedically relevant genes (e.g., drug and wound response) (Lou et al. 2015; Romdhane et al. 2021).
The Romani or Roma population (often referred to by the problematic misnomer Gypsies) nowadays forms the largest transnational minority ethnic group in Europe, numbering 10–15 million. their origin has been traced back to North-western India thanks to different sources of information. Linguistic studies and records from the populations that encountered the proto-Roma groups often suggest an Indian origin of this group, which left around 1000–1500 years ago and subsequently spread to Persia and Armenia (Boerger 1984; Fraser 1992; Liégeois 1994). Records from Greece, present-day Romania, and the Czech Republic account for putative Roma presence in these territories through the fourteenth century, and by the fifteenth–sixteenth centuries, additional historical evidence documents Roma movements in many West European countries (Fraser 1992; Liégeois 1994). The current distribution of Roma people throughout Europe can be attributed to such early fifteenth-century expansions from the Balkans and to later nineteenth-century dispersals (Fraser 1992; Liégeois 1994; Reyniers 1995; Gresham et al. 2001). In more recent historical times, the Roma population size and distribution in Europe is also the consequence of the genocide they suffered, carried out by the Nazi Germany regime (Milton 1991; Lutz 1995; Sridhar 2006). Finally, the fall of the communist regimes in Central and Eastern Europe facilitated westward economically driven migrations.
The European Roma groups, indeed, have had a complex history, both in terms of the movements and contacts with different populations. Population genetics studies traced back their South Asian-related ancestry, with subsequent European admixture, from autosomal and uniparental markers (Gresham et al. 2001; Moorjani et al. 2013; Font-Porterias et al. 2019; Ena et al. 2022). Their specific history also shaped the landscape of genetic diseases, as different deleterious mutations were detected at higher frequencies, while other mutations are absent or at lower frequencies compared to other non-Roma populations (Kalaydjieva et al. 2001; Morar et al. 2004; Mendizabal et al. 2013). Specifically, private disease-causing mutations, highlighting a scenario typically found in a founder population, have been identified also in the Roma. The traits associated to these mutations are, among others, polycystic kidney disease, congenital glaucoma, congenital myasthenia, galactokinase deficiency, different neuropathies and centronuclear myopathy (Kalaydjieva et al. 1996, 1999, 2001; Piccolo et al. 1996; Angelicheva et al. 1999; Morar et al. 2004; Cabrera-Serrano et al. 2018).
The whole genome sequence of 46 Roma individuals revealed a strong, early founder effect followed by a drastic reduction of ∼44% in effective population size (Ne) (Bianco et al. 2020). It is known that mutations reach fixation faster in small populations due to drift and, as a consequence, some deleterious mutations may rise in frequency and, under specific conditions (see Fig. 1 in Kimura et al. 1963), slightly deleterious variants can result in a larger load than more deleterious ones (Kimura et al. 1963; Kimura and Ohta 1969). In general, a rule of thumb is that drift will prevent the removal of deleterious mutations if Nes < 1, where s is the selection coefficient; still, this does not encompass the complexities of population growth and gene flow (Gazave et al. 2014; Lohmueller 2014). Different studies not only observed these phenomena in general populations as the Europeans, but also confirmed them in smaller and isolated groups which experienced more recent bottlenecks (i.e., Finnish, French-Canadians, Inuit and Ashkenazi Jewish) (Kaklamani et al. 2008; Lohmueller et al. 2008; Thaler et al. 2009; Casals et al. 2013; Lim et al. 2014; Pedersen et al. 2017). Moreover, disease-associated variants show specific haplotype ancestry backgrounds in Roma (European or South Asian), in line with the mutual contribution of these ancestries to Roma genetic makeup and, additionally, that the higher frequencies of SNPs mapping to drug-binding domains match the population higher proportion of diseases targeted by such drugs (Font-Porterias et al. 2021). This stresses how admixture dynamics, demographic history and the functional role of variants all contribute to the shaping of the extant diversity detectable nowadays in Roma.
In light of the information about Roma gathered so far, we hereby analyze for the first time CNVs in high-depth complete genomes from the underrepresented European Roma population to understand how their demographic history may have contributed (if at all) to their mutation spectrum and mutational load.
Materials and methods
Samples
Our study comprises 40 complete genomes of Roma people collected in five European countries (Spain, Lithuania, Hungary, Ukraine and Macedonia) and belonging to four major migrant groups: 15 North/Western, 5 Vlax, 10 Romungro and 10 Balkan as defined in a previous study (Bianco et al. 2020). Donors signed an informed consent and the project was approved by the Institutional Review Board of the Comitè Ètic d’Investigació Clínica-Institut Municipal d’Assistència Sanitària (CEIC-IMAS) in Barcelona, Spain, (2016/6,723/I). All participants self-identified as Roma and appropriate consent was obtained from all donors. The study was approved by our IRB (Comitè d’Ètica de la Investigació, Parc de Salut Mar, Barcelona) on June 7th 2016 (reference 2016/6723/I) and renewed on January 15th, 2020 (reference 2019/8900/I). Preliminary results were presented to the Roma community in a meeting on February 1st 2019 in Barcelona. All methods in this study were performed following the standard guidelines and regulations. Genome sequences were those analyzed in (Bianco et al. 2020), which fastq files had been deposited at the European Genome Archive with accession number EGAS00001004287. Reference samples with geographic origins matching the Roma diaspora comprised two main datasets: the Simons Genome Diversity Project (SGDP; samples from Europe, the Middle East and Pakistan) (Mallick et al. 2016) and Mondal et al. (Mondal et al. 2016) (samples from India) (see supplementary Table 5). Throughout this manuscript, when we use term European to refer to reference samples, we mean it as shorthand for non-Roma Europeans, and we do not imply that Roma should not be regarded as Europeans.
Structural variant calling
We selected a set of six different programs using algorithms based on different strategies to detect SVs from short read sequencing data, combining the strengths of each algorithm and integrating them. Our set is composed of CNVnator (version 0.4.1) (Abyzov et al. 2011), BreakDancer (version 1.4.5) (Chen et al. 2009), Pindel (version 0.2.5b8) (Ye et al. 2009), Tardis (version 1.0.4) (Soylev et al. 2017), Lumpy (version 0.2.13) (Layer et al. 2014), and GenomeSTRiP (version 2.00.1918) (Handsaker et al. 2011, 2015) callers, which implement read-depth, split-read and read-pair methods. See Supplementary Methods for the implementation of each method.
Data merging
We designed custom scripts to obtain the data both for the results for all callers for a single sample and among all samples. To do so, we first merged the output of the different software for each sample, specifically by merging those SVs residing on the same chromosome, deletions and duplications separately, with a reciprocal genomic coordinate overlap of at least 50% of their length. By doing so, we created clusters of overlapping pairs of calls and for each cluster (ranging from a pair of calls for two programs, up to the 15 possible pairs among six different callers) we selected the coordinates and the genotype of the most confident caller, based on the evaluation of caller performance in (Kosugi et al. 2019). Using this information for each cluster of calls mentioned above, we obtained a single call by retaining the best performing software for coordinates and genotype, respectively. To merge variants across samples, we proceeded in a similar manner as previously presented, where we joined all sample calls if variants of the same type resided on the same chromosome and reciprocally overlapped at least for 50% of their length. This allowed us to create a consensus set of calls listing the sharing of each variant among individuals.
Additional filters
We regenotyped the CNVs of each sample with a dedicated software, GraphTyper2 (version 2.5.1) (Eggertsson et al. 2019), to accurately recover more reliable genotypic information (see supplementary methods). We further filtered the results according to the best practices as described by the software authors, to retain only good quality genotypes. To additionally filter for false positives, we used the HardyWeinberg R package (version 1.7.2) (Graffelman 2015) to remove variants violating Hardy–Weinberg equilibrium. We computed the chi-squared test p-value for each CNV in each population and filtered out variants having a significant result after Bonferroni correction for multiple tests. Finally, we implemented an R package algorithm leveraging SNP data to infer reliable CNVs: CNVfilteR (version 1.8.0), which detects false positive heterozygous deletions and duplications by evaluating the frequencies of SNPs mapping to each variant (Moreno-Cabrera et al. 2021). We ran this software with default parameters and obtained a set of variants indicating false positive results that were subsequently filtered out from the dataset.
Statistical analysis
Principal component analysis was carried out using the smartpca algorithm within the Eigensoft package (version 6.0.1) (Patterson et al. 2006). Briefly, based on CNV genotypic calls, we coded biallelic deletions and duplications as zero, one, and two copy numbers and used those as input for the software to perform PCA on our samples. We additionally used another dimensionality reduction method, the uniform manifold approximation projection (UMAP) (McInnes et al. 2018) on copy number for deletions and duplications. Population structure was further assessed using ADMIXTURE (version 1.3) (Alexander et al. 2009), running 10 random seeds for each ancestral component (K: 2–10), to evaluate ancestry profiles among the studied samples. We filtered out variants with minor allele frequency < 0.01 and violating structure-aware Hardy–Weinberg equilibrium before running the analysis, as best practices described in previous studies (Narang et al. 2014; Hao and Storey 2019; Linck and Battey 2019). Pong (Behr et al. 2016) was used to visualize ADMIXTURE results by representing Q matrices for modes in each value of K. ANOVA test was performed with the R car package (version 3.0.10) (Fox and Weisberg 2011), while Kruskal–Wallis and Chi-squared tests were computed using the corresponding native R functions (R Core Team 2003). We estimated global differentiation values calculating FST statistics among pairwise populations using the StAMPP R package (Pembleton et al. 2013) and estimated p-values by performing 10,000 bootstraps. Taking advantage of the possibility to recapitulate population differentiation using CNVs data by means of the Vst statistic (Redon et al. 2006; Sudmant et al. 2015a), using a custom script, we implemented a variation of the formula described in a previous study (Serres-Armero et al. 2021), comparing directly copy number variance rather than log2 ratios from CGH array data. We applied the statistic in pairwise population comparisons computing the differentiation for each CNV individually.
Copy Number Variant annotation
We used the software AnnotSV (version 3.0.7) (Geoffroy et al. 2018, 2021) for multiple database annotation to retrieve the possible clinical or functional roles of the CNVs in our dataset. Since results from AnnotSV provided different information, we focused on: (1) the genes intersected by the CNV, (2) whether the intersection involved an intron, an exon, or both, (3) diseases associated to the intersected gene provided by OMIM catalog (Hamosh et al. 2005), (4) gene tolerance to loss of function. Specifically, the tolerance to loss of function for genes intersected by CNVs is ranked as Loss-of-function Observed/Expected Upper Fraction (LOEUF) bins (range 0–9) from genomAD database (Karczewski et al. 2020). The LOEUF metric refines over the widely used pLI (probability of Loss of function Intolerance), providing a continuous rather than a dichotomous scale (e.g., pLI < 0.9; pLI > 0.9). We carried out permutation tests to screen for possible intra-population higher/lower than expected abundance of deletions intersecting intronic portions of loss of function (LoF) intolerant genes. To do so, we downloaded the LOEUF information for each gene present in the gnomAD database and obtained those genes’ annotations via Ensembl database (version 86) (Cunningham et al. 2022) using the EnsDb.Hsapiens.v86 and ensembldb R packages (Rainer 2017; Rainer et al. 2019). For this list of genes we extracted the intronic coordinates using GenomicFeatures R package (Lawrence et al. 2013) of those genes with a LOEUF ≤ 4 (Lof intolerant) and LOEUF > 4 or not reported (LoF tolerant). Then, with our list of population-specific gene-intersecting deletions and introns coordinates of LoF tolerant/intolerant genes, we performed permutation tests separately in each population using the regioneR R package (Gel et al. 2016) performing 5000 permutations and estimating the numOverlaps and randomizeRegions as the evaluate and randomize functions.
Over-representation analysis
To assess putative significant enrichment in biological pathways for our gene-intersecting SVs, we interrogated the Gene Ontology Resource (Ashburner et al. 2000) using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) (Zhang et al. 2005; Liao et al. 2019), an online tool to interpret and analyze gene lists of specific interest. We tested whether the list of genes classified with a LOEUF score from 0 to 4 and hosting intronic variants was enriched in specific GO terms in each population. Accordingly, the inputs passed to the software were the above mentioned gene list as well as a reference set, namely all genes (regardless of their known intolerance level) having intronic deletions. We focused our analysis on biological and molecular function database categories, performing the analysis with default parameters and considering as significant the associations having an FDR < 0.05.
CNVs and GWAS catalog
We evaluated the level of association between our set of CNVs and diseases identified in the GWAS catalog (Buniello et al. 2019), using linkage disequilibrium (LD) with trait-associated SNPs as a proxy. The selected common variants underwent filtering using PLINK (version 1.9; www.cog-genomics.org/plink/1.9/) (Chang et al. 2015), removing individuals with a missing genotype rate > 0.1 and SNPs with missing call rate > 0.1, with minor allele frequency < 0.01 and those failing the Hardy–Weinberg equilibrium test. This set of filtered SNPs and our CNV set were merged together and phased using two programs, WhatsHap (version 1.1) (Patterson et al. 2015) and ShapeIt4 (version 4.1.3) (Delaneau et al. 2019), following procedures previously described (Valls-Margarit et al. 2022). The result provided the input for PLINK, where we computed LD between variants in our dataset (CNVs and SNPs) and those SNPs shared with the GWAS catalog, only including variants in high LD (r2 > 0.8) and mapping within 1 MB around the pathogenic SNP.
Results
Calling CNVs from whole genome sequences
We called CNVs in 40 genomes from already published Roma individuals (Bianco et al. 2020; García-Fernández et al. 2020) along with 98 samples from Europe, the Middle East and South Asia (Mallick et al. 2016; Mondal et al. 2016). Our calling pipeline comprised six programs (callers) for SV detection from WGS using GRCH38.p8 as reference genome (see “Methods” and supplementary text).
For our subsequent analyses, we included only deletions and duplications as some of the software used are unable to call insertions or inversions. We merged our data together by, first, creating a per-sample consensus among callers, finding 1484 ± 366 CNVs per sample on average (deletions: 1433 ± 352; duplications: 51 ± 23) and eventually by iteratively merging sample CNVs, obtaining calls for individuals sharing the same variant (see “Methods”). This step yielded a total number of 11,207 CNVs (9863 deletions and 1344 duplications) and an average of 1499 ± 352 CNVs per genome (deletions: 1449 ± 357; duplications: 50 ± 22).
Dataset characteristics and population structure
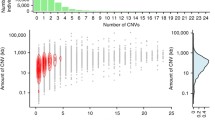
We grouped our 138 samples using a geographical rationale and divided the samples as follows: Roma (40 samples), Europe (22), Middle East (15), and South Asia (61). Initially, Principal Component Analysis (PCA) revealed that samples clustered by dataset of origin (Roma, Mondal et al. (2016) and SGDP) rather than by geographic affiliation (Supplementary Fig. 1). We addressed this batch effect by regenotyping each CNV and subsequently applying different filters based on quality (allele depth, read balance in heterozygotes), checking for consistency with SNP genotypes, and Hardy–Weinberg equilibrium (see Supplementary Methods). The final filtered dataset comprised 3660 CNVs (3171 deletions and 489 duplications). We controlled for possible structure within Roma using our set of CNVs and noticed no specific relationships within regional groups (Suppl. Fig. 2 shows deletion-based analysis). In an UMAP plot based on deletions (Fig. 1), Roma individuals cluster together and apart from the Europe–Middle East–South Asia continuum; see also similar patterns for PCA (Suppl. Figs. 3, 4) and ADMIXTURE (Suppl. Fig. 5). PCA and ADMIXTURE analysis on deletion genotypes showed similar patterns to those obtained with a random SNP sample of the same size (3171, Suppl. Figs. 6 and 7). On the contrary, when applying dimensionality reduction methods to duplications (Suppl. Figs. 8, 9), this pattern was fuzzier, probably because of the higher rate and bidirectionality on mutation in duplicated segments. Thus, population history has modeled deletion (and to a lesser extent duplication) genotype frequencies in the Roma.
Out of 3660 CNVs (supplementary Fig. 10), 1899 (52%), 329 (9%) and 459 (13%) are shared by four, three and two populations respectively. We additionally found 973 (27%) variants that were found in only one population (Roma: 257, Europe: 157, Middle East: 179 and South Asia: 380), most of which were singletons. Overall, our call set is composed of 2013 common (Allele Frequency, AF) > 0.05), 668 low frequency (0.01 ≤ AF ≤ 0.05) and 979 rare variants (AF < 0.01). Most common variants are shared preferentially by all four populations (four populations: 1792 (89%), three populations: 120 (6%), two populations 78 (4%), one population 23 (1%)) as expected in general populations. Low frequency variants are more evenly distributed (four populations: 107 (16%), three populations: 209 (31%), two populations 237 (36%), one population 115 (17%)) while rare variants, as expected, can be found only in one population or two at most (two populations: 144 (15%), one population: 835 (85%)) (supplementary Fig. 11). Note that these sharing proportions are underestimates, given the relatively low sample size, particularly in the Middle East. Within-population proportions of common, low frequency and rare variants change across populations, with South Asians having more variants across the frequency classes compared to the other populations and the Roma showing the same trend compared to Europe and Middle East (χ2 = 83.6, p-value = 6.25 × 10–16) (Table 1). Globally, South Asia and Roma retain a higher number of private CNVs and, evaluating the frequency profiles among populations, this pattern repeats within common, low-frequency and rare variant classes, demonstrating that the apportionment of private variants is not restricted to any specific frequency category.
Overall, the average FST (Fig. 2) among all pairs of populations was higher for deletions (0.0375) than for duplications (0.0272), which is consistent with repeat mutation at duplications counterbalancing population differentiation by drift. Thus, we will base our population inferences on deletions. The average FST between the Roma and each of the other populations was 0.0478, which is higher than for any other population. In particular, the Roma were slightly more distant from South Asia (0.0497) than from the Middle East (0.0473) or Europe (0.0465). South Asia is also equally distant from the Middle East (0.0363) and Europe (0.0383), while these two populations are close to each other (0.0067). This is the expected pattern as derived from nucleotide variation in arrays (Granot et al. 2016) or whole genomes (Mallick et al. 2016). Particularly for the Roma, these differentiation patterns are in line with previous studies based on genome-wide SNP data (Melegh et al. 2017) and could reflect the global landscape of CNVs in Roma, who had their own mutational history diverging from Northern India, ultimately admixing with Europeans and, in the process, accumulating genetic drift.
CNV annotation
Using the software AnnotSV (Geoffroy et al. 2018, 2021) we annotated variants leveraging different databases (Refseq, OMIM, ClinGen, gnomAD, among others) and gathered information about CNV localization within genes, their possible functional role and the pathogenic consequences of their presence in transcribed genome sequences. While more than half of the CNVs in our dataset, 2115 (58%), did not overlap any currently known gene, 1532 (42%) variants intersected transcribed sequences, of which 263 (7.2%) and 1268 (35%) resided within exons and introns respectively, in agreement with previous studies (Conrad et al. 2010; Mills et al. 2011; Valls-Margarit et al. 2022) (Supplementary Fig. 12). The remaining 13 CNVs intersected more than one gene, hitting multiple intronic and/or exonic locations. Overall, we found that genomic location and the type of CNV are dependent from each other (χ2 = 77.3, p-value < 2.2 × 10–16), with deletions representing the majority of variants within each genomic location (Table 2). It is interesting to notice that exons seem to tolerate duplications better than deletions: while 6.7% of deletions affect exons, this figure is 18.2% for duplications, likely due to the stronger selective constraints over deletions within genes (Sudmant et al. 2015a). Our dataset confirms what previous studies reported about the average frequencies apportionment of intergenic and genic variants and the easier-to-resolve deletion signal used by short reads structural variants software.
Geographic and genomic distribution of CNVs
We next tested for the number and length of CNVs carried by individuals. For duplications, we could not find any significant difference among the populations. As for deletions, Roma carry more events per individual (mean: 880 ± 24) with respect to all other populations, (Europe: 834 ± 16; Middle East: 828 ± 29; South Asia: 810 ± 26), (Anova p-value < 2.2 × 10–16). Testing for deletion location, we found out that the same pattern held true for intergenic (Kruskal–Wallis p-value < 2.2 × 10–16) and intronic (Kruskal–Wallis p-value = 10–14) events (Fig. 3). Regarding exonic deletions, Europeans carry significantly fewer variants compared to Roma, Middle East and South Asia populations (Anova, p-value = 0.007). In addition, variant length also differed among populations as, overall, deletions in the Roma are larger than those in Europeans, while deletions in South Asians are shorter compared to all other populations (Kruskal–Wallis p-value = 1.3 × 10–8) (Supplementary Fig. 13A) and, within South Asia, Indian group shows shorter variants than Pakistani (Kruskal–Wallis p-value = 0.023). In particular, Roma have larger variants only when considering intergenic deletions, while South Asian population shows shorter intergenic, intronic and exonic (Kruskal–Wallis p-values, intergenic = 8.2 × 10–10; exonic = 0.0016 and ANOVA p-value intronic = 0.0001), (Supplementary Fig. 13). Overall, the results of these first comparisons show that Roma carry more and longer intergenic/intronic deletions than other populations, but their intolerance to exonic deletions is similar.
Features of the highly differentiated CNVs
Next, we characterized the CNVs that were highly differentiated among populations by computing the VST statistic (Redon et al. 2006) for each CNV and pair of populations. For each variant, VST considers the variance of copy number in pairwise group comparisons; actually, for biallelic deletions (0 or 1 copies) or duplications (1 or 2 copies), VST is numerically identical to FST. The mean VST values are reported in Supplementary Table 1. We focused on highly differentiated CNVs by taking the top 20% VST values, for each pair of populations (Fig. 2D); the average VST values by genomic location in this highly differentiated set can be found in Supplementary Table 2. Intergenic deletions and duplications are at the top of the value distribution; indeed, as expected, these variants display fewer constraints in the mutation rates between populations and, thus, are freer to vary. Intronic and exonic variants follow in the distribution, showing lower values for the latter calls and pointing once again to a higher constraint on those deletions and duplications putatively having a higher disruptive power over genic sequences. Since pairs containing Roma exhibited higher values at the top of the distribution, we tested if any difference existed in VST values among pairs for variants intersecting genes. We found significant differences (Kruskal–Wallis, p-value < 2.2 × 10–16) for deletions in such pairs with respect to the others. In particular, pairs considering Roma had significantly higher values than pairs without and, dividing the analysis by variant location, we could find significant differences only for intronic events (Kruskal–Wallis, p-value < 2.2 × 10–16; mean values: Roma–Europe = 0.1316; Roma–Middle East = 0.1452; Roma–South Asia = 0.131; Europe–Middle East = 0.0952; Europe–South Asia = 0.0894; Middle East–South Asia = 0.0923). Estimating variant differentiation among pairs of populations highlighted how the major source of variability can be traced back to Roma individuals; nevertheless, when stratifying the analysis by genomic location of the variants, significant differences in differentiation scores can solely be found for intronic deletions.
Predicting the pathogenicity of CNVs
For each CNV we retrieved, whenever available, the OMIM (Online Mendelian Inheritance in Man) annotations (Hamosh et al. 2005) and the LOEUF (Loss-of-function Observed/Expected Upper Fraction) bin values (ranging in bins from 0 to 9) from gnomAD (Karczewski et al. 2020) when the variant overlapped a gene sequence. We compared the distribution of variants hitting genes having a linked OMIM entry among populations and Europeans showed a significantly lower number (Anova, p-value = 0.02) of deletions within OMIM genes compared to all other populations (mean number of deletions per genome: Roma = 87.8, Europe = 82.5, Middle East = 86.4, South Asia = 85.5; see supplementary Fig. 14). Duplications, instead, are significantly (Kruskal–Wallis, p-value = 0.007) more frequent in South Asian Indians than in South Asian Pakistani, Roma, Middle East and Europe populations (average of 7.5 for the former against 5.4, 5.7, 5.4, and 6.05 duplications per genome for the latter populations). These results could highlight a greater efficacy of natural selection removing deleterious mutations in Europeans compared to the other populations, probably due to their demographic history. Duplications within OMIM genes being more frequent in the South Asia compared to Roma and Middle East populations could reflect, to a certain degree, the increased recessive diseases specific to the group and the different selective pressures recorded for specific West Eurasian alleles, as highlighted in (Ayub and Tyler-Smith 2009; Nakatsuka et al. 2017). Roma individuals showed increased number of deletions in the 0, 1, 2 and 4 LOEUF bins and, upon stratification by location, only intronic events produced significant results for the same categories (bin 0: Anova, p-value = 2.8 × 10–7; bin 1: Kruskal–Wallis, p-value = 1.4 × 10–8; bin 2: Anova, p-value = 3.5 × 10–6; bin 4: Anova, p-value = 1.6 × 10–5). We assessed whether this higher number of deletions intersecting genes with low LOEUF values caused the overall increased number of intronic variants in Roma, as shown above. After removing these intolerant-gene deletions, Roma keep retaining a significantly higher number of intronic variants (Kruskal–Wallis, p-value = 5.2 × 10–10), demonstrating that the accumulation of these deletions at intolerant genes is an independent process that does not drive the general increase in intronic deletions.
Due to our findings of an increased number of deletions within introns of LoF-intolerant genes in Roma, we explored, separately for each population, the possibility that these mutations preferentially hit intronic coordinates while taking into account LoF tolerance. Permutation tests were performed using all genic deletions against intronic coordinates of genes either with a LOEUF ≤ 4 (intolerant) or LOEUF > 4—or for which the metrics was not available (tolerant). With these sets of regions we noticed that, while genic deletions intersect introns of tolerant genes more often than expected by chance (Permutation test, p-value = 0.0018–0.0004), the opposite is not true for the intersection with introns of intolerant genes (Permutation test, p-value > 0.05). This result points toward a general constraint for the accumulation of deletions, even at the intronic level, in intolerant genes within each population. In the context of the most differentiated variants described above, we looked at the distribution of frequencies and LOEUF values in pairwise populations containing Roma; we evaluated the frequencies in deletions showing larger differentiation, partitioning the variants across the most intolerant LOEUF classes (0–4). Despite the fact that the only significant result showed higher frequency in Roma compared to Middle Eastern population for deletions in the LOEUF 2 category (Kruskal–Wallis, p-value = 0.02), we noticed a general trend towards slightly higher frequencies in the Roma, across all LOEUF bins, compared to all other populations (Kruskal–Wallis, p-value = 0.0471). Nonetheless, pairwise group comparisons do not show significant results after multiple test correction. Following our previous results on the differentiation of intronic deletions in Roma, here we show an over-representation of such variants in this population that, together, highlight a pattern of recurring mutations occurring in untranslated genome portions. The differences in intolerant-gene deletions could highlight a lower constraint for Roma towards the accumulation of genic deletions residing outside the coding sequences but within genes whose function is more likely hampered by mutations.
CNVs and genetic associations
In Genome-Wide Association Studies (GWAS), genetic associations are established between specific diseases or traits, or sets of them, and genetic variants, usually SNPs. We wondered to which extent the CNVs we detected could be linked to pathogenic SNPs present in the GWAS catalog (Buniello et al. 2019). To do so, we downloaded the GWAS catalog dataset version 1.0.3 and identified common SNPs between this set and those previously found in our samples (Bianco et al. 2020); the intersection consisted of 74,009 variants. For these common SNPs, we estimated the associated CNVs by selecting, for each chromosome, only those CNVs in strong linkage disequilibrium (LD) (r2 > 0.8) and residing in a 1 MB window around the SNP. Following this procedure, we identified 78 unique deletions in LD (supplementary Table 4) with 125 disease-associated SNPs as reported in the GWAS catalog, while no duplication was in linkage disequilibrium with any SNP in the set. The identified deletions are in LD with one or more (up to eight) SNPs and, for each of them, we retrieved the information about deleteriousness using LOEUF scores. Among the traits in the GWAS catalog, we could identify different functional categories. The majority of the traits involves metabolic, neurodevelopmental/neurological, development and hematological–cardiovascular disorders. Looking at the genomic context of the linked deletions, 41 (53%) reside in intergenic loci, 32 (41%) intersect introns and only five (6%) within exons. While a direct role of intergenic variants upon the pathogenicity of linked SNPs is difficult to establish—but not a reason to exclude them a priori—intronic and exonic CNVs might act on the same genomic context of the SNP. Among the intronic variants, only eight deletions intersected genes having more tolerant LOEUF scores (> 5), six other gene-intersecting variants had no score information and the remaining 18 resided in genes with higher intolerance to LoF (scores 0–4). Among these latter deletions, four are in linkage with SNPs related to metabolic/inflammatory diseases (Type 2 diabetes, alanine transaminase levels, urate levels), four others link with GWAS traits related to heart, cardiovascular or hematological conditions (myocardial infarction, hemorrhoidal disease, red-cell width) and two variants link to colorectal cancer traits. For exonic variants, only one deletion intersects an intolerant gene (LOEUF bin 4) and is in LD with a SNP associated with metabolic disorders (total cholesterol/LDL levels); nevertheless, the deletion resides in a gene upstream the SNP and its involvement is unclear. The remaining four exonic deletions associate with inflammatory diseases, lung function, hematological and developmental features and all but one (lung function) affect the same gene of the linked SNP. Nonetheless, intolerance scores are either not available or point to a relaxation against LoF for exonic variants. Finally, when considering only the set of SNPs residing ~ 5000 bp around linked deletions, we noticed that intergenic events are the most frequent type of variants in the set (19 intergenic deletions, against nine intronic and one exonic deletions). This evidence, at least in part, might support the hypothesis of a possible influence, due to physical proximity (71 bp for the closest intergenic deletion), upon the genomic environment shared with the associated pathogenic SNP. In general, using data from the GWAS catalog, we were able to leverage SNPs information as a proxy for putative CNVs involvement in health-related traits, showing that either co-occurrence of a deletion and a SNP within the same gene or physical proximity may add novel information to both the traits and to the function of the structural variant under investigation.
Functions of the genes affected by deletions in the Roma
As previously shown, our analysis on deletion pathogenicity showed that the Roma retain a higher number of deletions intersecting LoF-intolerant genes, and specifically that intronic variants are responsible for this result. With this observation at hand, we wondered whether these more abundant intronic deletions in Roma had a specific influence on biological processes. We tested this hypothesis by performing an over-representation analysis separately in each population, using the online software GEne SeT AnaLysis Toolkit (WebGestalt) (Zhang et al. 2005; Liao et al. 2019), assessing whether LoF-intolerant genes (LOEUF bins: 0–4) intersected by intronic deletions were present more than expected in Gene Ontology (GO) terms (Ashburner et al. 2000). Results show significant enrichments in GO terms for the set of input genes in each population, with a marked prevalence of associations in Roma. Indeed, while Europe, Middle East and South Asian populations were significantly enriched for 24, 18 and 37 GO terms respectively, the Roma significant GO terms amounted to 187. For each term, using the available descriptions of related biological processes, we identified three recurrent functional categories, namely Nervous System, Signaling and Development, plus a catch-all Other category (Fig. 4). Overall, Roma showed higher number of GO terms among these classes compared to reference populations. The two most abundant categories in Roma were Signaling and Nervous System, which contained 61 and 55 GO terms, respectively. As a comparison, these two categories included 13/0, 0/2 and 6/11 terms in Europe, Middle East and South Asia, respectively. Furthermore, using a function within WebGestalt aiming at reducing possible redundancy for GO terms having similar gene sets, we obtained clusters of terms sharing related biological processes. Following this clusterisation, the Roma had 33 GO clusters, including 11 Signaling, 9 Nervous System, 5 Development and 8 comprising other processes such as chemotaxis, cell motility and cellular component organization. Europe, Middle East and South Asia had five, four and eight clusters with different proportions of the three major functional categories. We additionally checked for significant GO terms specifically found only in one population and noticed that Roma retain the highest number of private significant results, with 125 private terms against three, one and six found in Europeans, Middle Eastern and South Asian samples. Considering the deletions intersecting genes associated to the 125 private GO terms in Roma, we obtained 410 variants and retained only those overlapping a known pathogenic gene, either annotated in the OMIM or Deciphering Developmental Disorders (Firth and Wright 2011) (DDD) databases. The final filtered set included 168 deletions whose frequencies do not vary noticeably across populations; nevertheless, it is interesting to highlight that out of the 23 rare deletions, considering the global frequency in the whole dataset, 21 are indeed private to Roma. Within these Roma private deletions, more than half (15 variants) are singletons and reside in genes mainly associated with developmental/neurodevelopmental diseases and cancer. Of the remainder six deletions, four are doubletons and reside in genes associated to Cerebellofaciodental, Bardet-Biedl, Gillespie’s syndromes, spinocerebellar ataxia 15 and skeletal dysplasia with severe neurological disease, while the two more common variants intersect genes linked to Phelan-McDermid syndrome and -2-hydroxyglutaric aciduria. Overall, further investigation of intronic deletions in LoF-intolerant genes revealed significant enrichment in biological processes mainly related to signaling, nervous system and development, with a sharp accumulation of GO terms in Roma compared to the other populations. This supports our results of higher differentiation and abundance of intronic deletions within Roma, suggesting a possible relevance upon the functions of genes sets bearing such variants.
Discussion
In the current study, we analyzed CNVs in the Roma population using whole genome sequencing data with the dual purpose to provide the first published catalog of genome-wide unbalanced structural variants and, given previous knowledge of Roma demographic and genetic history, assess to which extent CNVs can inform us when used in a population genetics study of an underrepresented community. Comparing deletions and duplications from Roma and other reference populations (samples from Europe, Middle East and South Asia, covering the dispersal route Roma crossed in their diaspora) we estimated the main differences in the apportionment of events, the differentiation among populations and assessed the potential biomedical impact of the variants.
Deletions in Roma show a slight relaxation of natural selection
In our analysis, we have observed that the Roma carry more deletions than other European or Asian populations, that this additional load occurs in intergenic and intronic locations (but not in exons), that intergenic deletions in the Roma are longer, and that intronic deletions in the Roma are enriched for genes that are intolerant to loss of function (Lof) mutations. These results might be favored by population or sample-specific artifacts during deletions calling; however, variables that could affect the calling step, such as genome coverage, do not discriminate exclusively the Roma, as this latter population and the samples from SGDP share similar sequencing depth profiles. Differences in coverage among different batches, indeed, have been shown to affect CNVs calling in specific regions, but not overall (Khayat et al. 2021). Additionally, these spurious effects are unlikely to result in the apportionment observed in the Roma for deletions in introns and intergenic regions.
Although coding variation is the most obvious source of phenotypic differences, the evidence for introns and intergenic regions harboring functional variation has been accumulating (Vaz-Drago et al. 2017; Rigau et al. 2019; Telonis and Rigoutsos 2021; Keegan et al. 2022; Petersen et al. 2022). Thus, the additional intronic and intergenic deletions in the Roma point to a slight relaxation of natural selection; the effect of deletions in these regions is likely to be milder than in exons, which, in Roma, do not tolerate deletions at a higher rate than in other populations. The Roma present a unique combination of fragmentation, partial reproductive isolation, but also of admixture with their host populations. Founder events would have accumulated deletions at more tolerated locations with fewer constraints. Admixture, on the other hand, might have introduced new sources of variation in the population, while selection against deleterious mutations still acted to reduce the accumulation of harmful exonic deletions. As shown by reports on worldwide populations, usually selection acts against larger deletions in the genome (Sudmant et al. 2015a); however, in our case, this result could indicate that less efficient purifying forces may have taken place either because of the population history or because of the intergenic/intronic nature of the variants, bearing a presumably lower disrupting potential. In summary, the putative relaxed purifying selection in closed communities, which has been object of debate and addressed in finer detail by some reports (Fu et al. 2014; Balick et al. 2015; Gravel 2016; Henn et al. 2016), could be detectable only for low-impact mutations, such as intronic deletions in the Roma.
Highly differentiated CNVs in Roma intersect some genes of biomedical interest
Estimating the differentiation for shared CNVs in pairwise population comparisons by means of VST statistics, we found that intronic variants are significantly more differentiated in pairs with Roma, driving the overall trend. We could identify only one significant frequency difference, between the Roma and Middle East populations, for intronic variants when dividing for intolerant genes categories (LOEUF bin 2), showing Roma as the population with higher frequencies. Nonetheless, we also identified a significant difference in frequencies considering all intolerant categories together (LOEUF 0–4), with Roma exhibiting higher frequencies, even though pairwise populations comparisons did not pass multiple test correction.
Exploring further the possible deleterious nature of our variants, we assessed the levels of LD with known pathogenic SNPs from the GWAS catalog and identified 78 deletions in linkage with 125 trait-associated SNPs. Out of the whole set of these associated diseases, we could highlight four categories including most conditions: metabolic, neurodevelopmental/neurological, developmental and hematological–cardiovascular disorders. Although we acknowledge that only 33 tagged deletions reside on the same gene of the associated SNP (or SNPs), most deletions (41/45) with no common gene are intergenic variants which, among all linked deletions, are those residing in closer proximity to the linked SNP(s) and, thus, might exert a specific influence on the trait-related variant. As an example, the thirty closest deletions sharing no gene with the tagged SNP are all intergenic variants and range in distance from 71 bp to 18.8 kb. This evidence points at the importance of including intergenic variants in analyses assessing CNV function, as such mutations could be either actors or co-players, modifying their genomic neighborhood, participating to different scenarios, as already reported for specific diseases (Staehling-Hampton et al. 2002; Loots et al. 2005; Farrell et al. 2011; Uyan et al. 2013). Overall, SNPs in LD with intergenic deletions show associations with traits related to development, neurodevelopmental, metabolic and hematological conditions, as well as other traits such as height, smoking behavior and heart/cardiovascular ones. For genic deletions, it is expected, and probably more likely, that their influence over gene products or regulatory functions would be stronger than intergenic ones. Together, this set of deletions primarily associate to metabolic/inflammatory, cancer and neurodevelopmental/neurological traits. The collection of conditions related to metabolism mainly pertains to cholesterol levels, type 2 diabetes, alanine transaminase levels and obesity traits. Genes containing SNPs in LD with deletions had low reported LOEUF values, indicating their intolerance to loss of function (CCDC50, JAZF1, MYO9A, CNOT1 genes having, respectively, three, one, one and zero LOEUF bin scores). Intriguingly, a previous study showed how European Roma carried higher frequencies of SNPs involved in hyperlipidemia (Mendizabal et al. 2013); we found one deletion in RHCE gene in linkage with one cholesterol-associated SNP within the neighboring MACO1 gene (however, a direct functional effect upon the RHCE gene, which codes for a Rh-like red blood cell antigen, should not be dismissed), and indeed the deletion is higher in frequency within Roma.
Genes intersected by CNVs in Roma are enriched for central nervous system functions
We discovered that Roma carry a marked prevalence of GO terms associated to common functions subsets of inputted genes lists. Intriguingly, we could highlight marked differences only when using this type of gene sets, i.e., intolerant genes that contained intronic deletions, and not while using other sets, such as private deletions within populations or general classification based on genomic location. This is unlikely the result of a general higher number of deletions in Roma but rather the specific function of the affected genes. Roma show more biological process GO terms in each defined category (Nervous system, Signaling and Development categories plus “Other” containing general unrelated terms) compared to the other populations, and a strong difference can be noticed for the Nervous system and Signaling categories. We find these results of particular interest in light of the known private diseases specifically affecting Roma people in Europe. Indeed, among the different types of private disease-causing mutations described in the Roma, some involve neuropathies and neurological diseases such as hereditary motor and sensory neuropathy-Lom/Russe types, congenital cataracts facial dysmorphism neuropathy and limb-girdle muscular dystrophy type 2C (Kalaydjieva et al. 1996, 2001, 2005; Angelicheva et al. 1999; Morar et al. 2004). Nervous system-related GO terms often involved neurons connections organization, synaptic communication or brain development, highlighting the presence of putatively deleterious variants affecting physiological neuronal functions particularly in Roma, in line with previous reports of a higher rate of slightly deleterious variants, for other disorders, in Roma individuals (Mendizabal et al. 2013). Two examples of private intronic deletions in Roma (Supplementary Fig. 15), intersecting LoF genes implicated in central nervous system functions are in gene WDPCP (LOEUF score: 4), with GO terms related to CNS, such as “neuron differentiation”, “cell projection organization”, “cell morphogenesis involved in neuron differentiation”, “neuron development” and gene SHANK3 (LOEUF score: 0) with GO terms description include, among others, “regulation of nervous system development”, “telencephalon development”, “synaptic signaling”, “axongenesis”. Moreover, the disease-associated SNPs assessed in (Mendizabal et al. 2013) reside in genes belonging to biological processes associated to the significant GO terms we identified in our analysis, highlighting a possible action of different markers (deletions and SNPs) within same sets of genes, specifically affecting their functions. Lastly, as a general point, it is important to bear in mind that frequency spectra for clinically relevant variants differ among populations, with Roma showing their relatively low isolation by exhibiting highly represented deleterious alleles as well as near absence of others, whose ancestries are mainly related to South Asian and European haplotypes (Font-Porterias et al. 2021).
Isolated populations are an under-analyzed genomic resource, also for CNVs
Populations of non-European descent have traditionally been understudied in the context of genetic variation, particularly favoring GWAS research on more accessible cohorts of general European ancestry (Bustamante et al. 2011; Popejoy and Fullerton 2016). Ironically, what should be one important goal of human genetics research: uncovering an increasingly clearer and more complete picture of human genetic variation worldwide, portray a fairer representation of different human populations and advancing current knowledge on genetic diseases using diverse sets of populations (Zeggini 2014), has often been disregarded in favor of a Eurocentric perspective (Need and Goldstein 2009; Sirugo et al. 2019). Numerous studies addressing population isolates, indeed, contributed significantly to identify the loci underlying complex diseases: bipolar disorder and schizophrenia in Finland and Basque populations (Palo et al. 2007; Parsons et al. 2007), studies on Iceland individuals highlighting variants associated to atrial fibrillation, myocardial infarction, type 2 diabetes and glaucoma (Manolescu et al. 2004; Gudbjartsson et al. 2007; Helgadottir et al. 2007; Steinthorsdottir et al. 2007; Thorleifsson et al. 2007) and also traits as height and pigmentation in Finland, Iceland, Sardinia and Amish populations (Sulem et al. 2007, 2008; Gudbjartsson et al. 2008; Sanna et al. 2008). It has been suggested that addressing isolated populations for studying diseases can help in reducing the variance of environmental variables on pathogenic conditions, as homogeneity in phenotype and environment within isolates would facilitate the disease–gene recognition (Kristiansson et al. 2008), thus favoring the inclusion of underrepresented populations to advance our understating of health-related traits.
Data availability
Upon acceptance of this manuscript, a vcf file containing the CNV calls for the samples analyzed will be deposited in the European Genome-Phenome Archive (EGA). In-house scripts used in this work can be retrieved from https://github.com/marcoantinucci/CNVs_scripts.
References
Abyzov A, Urban AE, Snyder M, Gerstein M (2011) CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21:974–984. https://doi.org/10.1101/gr.114876.110
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664. https://doi.org/10.1101/gr.094052.109
Almarri MA, Bergström A, Prado-Martinez J et al (2020) Population structure, stratification, and introgression of human structural variation. Cell 182:189-199.e15. https://doi.org/10.1016/j.cell.2020.05.024
Angelicheva D, Turnev I, Dye D et al (1999) Congenital cataracts facial dysmorphism neuropathy (CCFDN) syndrome: a novel developmental disorder in Gypsies maps to 18qter. Eur J Hum Genet 7:560–566. https://doi.org/10.1038/sj.ejhg.5200319
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. https://doi.org/10.1038/75556
Audano PA, Sulovari A, Graves-Lindsay TA et al (2019) Characterizing the major structural variant alleles of the human genome. Cell 176:663-675.e19. https://doi.org/10.1016/j.cell.2018.12.019
Ayub Q, Tyler-Smith C (2009) Genetic variation in South Asia: Assessing the influences of geography, language and ethnicity for understanding history and disease risk. Brief Funct Genomic Proteomic 8:395–404. https://doi.org/10.1093/bfgp/elp015
Balick DJ, Do R, Cassa CA et al (2015) Dominance of deleterious alleles controls the response to a population bottleneck. PLoS Genet 11:1–23. https://doi.org/10.1371/journal.pgen.1005436
Behr AA, Liu KZ, Liu-Fang G et al (2016) Pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32:2817–2823. https://doi.org/10.1093/bioinformatics/btw327
Bergström A, McCarthy SA, Hui R et al (2020) Insights into human genetic variation and population history from 929 diverse genomes. Science 367:eaa5012. https://doi.org/10.1126/science.aay5012
Bianco E, Laval G, Font-Porterias N et al (2020) Recent common origin, reduced population size, and marked admixture have shaped European roma genomes. Mol Biol Evol 37:3175–3187. https://doi.org/10.1093/molbev/msaa156
Boerger BH (1984) Proto-Romanes phonology. Dissertation, University of Texas, Austin, USA, vol 195, pp 138–141
Buniello A, Macarthur JAL, Cerezo M et al (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47:D1005–D1012. https://doi.org/10.1093/nar/gky1120
Bustamante CD, de La Vega FM, Burchard EG (2011) Genomics for the world. Nature 475:163–165. https://doi.org/10.1038/475163a
Cabrera-Serrano M, Mavillard F, Biancalana V et al (2018) A Roma founder BIN1 mutation causes a novel phenotype of centronuclear myopathy with rigid spine. Neurology 91:e339–e348. https://doi.org/10.1212/WNL.0000000000005862
Casals F, Hodgkinson A, Hussin J et al (2013) Whole-exome sequencing reveals a rapid change in the frequency of rare functional variants in a founding population of humans. PLoS Genet 9:e1003815. https://doi.org/10.1371/journal.pgen.1003815
Chang CC, Chow CC, Tellier LCAM et al (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. https://doi.org/10.1186/s13742-015-0047-8
Chen K, Wallis JW, McLellan MD et al (2009) BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6:677–681. https://doi.org/10.1038/nmeth.1363
Chen W, Hayward C, Wright AF et al (2011) Copy number variation across European populations. PLoS ONE. https://doi.org/10.1371/journal.pone.0023087
Collins RL, Brand H, Karczewski KJ et al (2020) A structural variation reference for medical and population genetics. Nature 581:444–451. https://doi.org/10.1038/s41586-020-2287-8
Conrad DF, Pinto D, Redon R et al (2010) Origins and functional impact of copy number variation in the human genome. Nature 464:704–712. https://doi.org/10.1038/nature08516
Cunningham F, Allen JE, Allen J et al (2022) Ensembl 2022. Nucleic Acids Res 50:D988–D995. https://doi.org/10.1093/nar/gkab1049
De Cid R, Riveira-Munoz E, Zeeuwen PLJM et al (2009) Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet 41:211–215. https://doi.org/10.1038/ng.313
Delaneau O, Zagury JF, Robinson MR et al (2019) Accurate, scalable and integrative haplotype estimation. Nat Commun 10:5436. https://doi.org/10.1038/s41467-019-13225-y
Dennis MY, Harshman L, Nelson BJ et al (2017) The evolution and population diversity of human-specific segmental duplications. Nat Ecol Evol 1:1–10. https://doi.org/10.1038/s41559-016-0069
Dentro SC, Leshchiner I, Haase K et al (2021) Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 184:2239-2254.e39. https://doi.org/10.1016/j.cell.2021.03.009
Eggertsson HP, Kristmundsdottir S, Beyter D et al (2019) GraphTyper2 enables population-scale genotyping of structural variation using pangenome graphs. Nat Commun 10:1–8. https://doi.org/10.1038/s41467-019-13341-9
Ena GF, Aizpurua-Iraola J, Font-Porterias N et al (2022) Population genetics of the European Roma—a review. Genes (basel) 13:2068. https://doi.org/10.3390/genes13112068
Farrell JJ, Sherva RM, Chen Z, et al (2011) A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. 117:4935–4945. https://doi.org/10.1182/blood-2010-11-317081.HMIP
Firth HV, Wright CF (2011) The Deciphering Developmental Disorders (DDD) study. Dev Med Child Neurol 53:702–703. https://doi.org/10.1111/j.1469-8749.2011.04032.x
Font-Porterias N, Arauna LR, Poveda A et al (2019) European Roma groups show complex West Eurasian admixture footprints and a common South Asian genetic origin. PLoS Genet 15:e1008417. https://doi.org/10.1371/journal.pgen.1008417
Font-Porterias N, Giménez A, Carballo-Mesa A et al (2021) Admixture has shaped Romani genetic diversity in clinically relevant variants. Front Genet 12:1–12. https://doi.org/10.3389/fgene.2021.683880
Fox J, Weisberg S (2011) An R companion to applied regression. Sage publications
Fraser A (1992) The gypsies. Wiley-Blackwell, Oxford
Fu W, Gittelman RM, Bamshad MJ, Akey JM (2014) Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am J Hum Genet 95:421–436. https://doi.org/10.1016/j.ajhg.2014.09.006
García-Fernández C, Font-Porterias N, Kučinskas V et al (2020) Sex-biased patterns shaped the genetic history of Roma. Sci Rep 10:1–10
Gautam P, Jha P, Kumar D et al (2012) Spectrum of large copy number variations in 26 diverse Indian populations: potential involvement in phenotypic diversity. Hum Genet 131:131–143. https://doi.org/10.1007/s00439-011-1050-5
Gazave E, Ma L, Chang D et al (2014) Neutral genomic regions refine models of recent rapidhuman population growth. Proc Natl Acad Sci USA 111:757–762. https://doi.org/10.1073/pnas.1310398110
Gel B, Díez-Villanueva A, Serra E et al (2016) RegioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32:289–291. https://doi.org/10.1093/bioinformatics/btv562
Geoffroy V, Herenger Y, Kress A et al (2018) AnnotSV: an integrated tool for structural variations annotation. Bioinformatics 34:3572–3574. https://doi.org/10.1093/bioinformatics/bty304
Geoffroy V, Guignard T, Kress A et al (2021) AnnotSV and knotAnnotSV: a web server for human structural variations annotations, ranking and analysis. Nucleic Acids Res 49:W21–W28. https://doi.org/10.1093/nar/gkab402
Girirajan S, Dennis MY, Baker C et al (2013) Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet 92:221–237. https://doi.org/10.1016/j.ajhg.2012.12.016
Graffelman J (2015) Exploring diallelic genetic markers: The HardyWeinberg package. J Stat Softw 64:1–23. https://doi.org/10.18637/jss.v064.i03
Granot Y, Tal O, Rosset S, Skorecki K (2016) On the apportionment of population structure. PLoS ONE 11:e0160413. https://doi.org/10.1371/journal.pone.0160413
Gravel S (2016) When is selection effective? Genetics 203:451–462. https://doi.org/10.1534/genetics.115.184630
Gresham D, Morar B, Underhill PA et al (2001) Origins and divergence of the Roma (gypsies). Am J Hum Genet 69:1314–1331. https://doi.org/10.1086/324681
Gudbjartsson DF, Arnar DO, Helgadottir A et al (2007) Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 448:353–357. https://doi.org/10.1038/nature06007
Gudbjartsson DF, Walters GB, Thorleifsson G et al (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40:609–615. https://doi.org/10.1038/ng.122
Hamdan A, Ewing A (2022) Unravelling the tumour genome: the evolutionary and clinical impacts of structural variants in tumourigenesis. J Pathol 257:479–493
Hamosh A, Scott AF, Amberger JS et al (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33:D514–D517. https://doi.org/10.1093/nar/gki033
Handsaker RE, Korn JM, Nemesh J, McCarroll SA (2011) Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet 43:269–276. https://doi.org/10.1038/ng.768
Handsaker RE, Van Doren V, Berman JR et al (2015) Large multiallelic copy number variations in humans. Nat Genet 47:296–303. https://doi.org/10.1038/ng.3200
Hao W, Storey JD (2019) Extending tests of hardy-weinberg equilibrium to structured populations. Genetics 213:759–770. https://doi.org/10.1534/genetics.119.302370
Hehir-Kwa JY, Marschall T, Kloosterman WP et al (2016) A high-quality human reference panel reveals the complexity and distribution of genomic structural variants. Nat Commun 7:1–10. https://doi.org/10.1038/ncomms12989
Helgadottir A, Thorleifsson G, Manolescu A et al (2007) A common variant on chromosome 9p21 affects the risk of myocardial infarction. Obstet Gynecol Surv 62:585–587. https://doi.org/10.1097/01.ogx.0000279313.65556.85
Henn BM, Botigué LR, Peischl S et al (2016) Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci USA 113:E440–E449. https://doi.org/10.1073/pnas.1510805112
Hollox EJ, Zuccherato LW, Tucci S (2022) Genome structural variation in human evolution. Trends Genet 38:45–58. https://doi.org/10.1016/j.tig.2021.06.015
Hurles ME, Dermitzakis ET, Tyler-Smith C (2008) The functional impact of structural variation in humans. Trends Genet 24:238–245. https://doi.org/10.1016/j.tig.2008.03.001
Itsara A, Cooper GM, Baker C et al (2008) Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 84:148–161. https://doi.org/10.1016/j.ajhg.2008.12.014
Kaklamani VG, Wisinski KB, Sadim M et al (2008) Variants of the adiponectin (ADIPOQ) and adiponectin receptor 1 (ADIPOR1) genes and colorectal cancer risk. J Am Med Assoc 300:1523–1531. https://doi.org/10.1001/jama.300.13.1523
Kalaydjieva L, Hallmayer J, Chandler D et al (1996) Gene mapping in Gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet 14:214–217. https://doi.org/10.1038/ng1096-214
Kalaydjieva L, Perez-Lezaun A, Angelicheva D et al (1999) A founder mutation in the GK1 gene is responsible for galactokinase deficiency in Roma (Gypsies). Am J Hum Genet 65:1299–1307. https://doi.org/10.1086/302611
Kalaydjieva L, Gresham D, Calafell F (2001) Genetic studies of the Roma (Gypsies): a review. BMC Med Genet 2:5. https://doi.org/10.1186/1471-2350-2-5
Kalaydjieva L, Morar B, Chaix R, Tang H (2005) A newly discovered founder population: the Roma/Gypsies. BioEssays 27:1084–1094. https://doi.org/10.1002/bies.20287
Kanduri C, Ukkola-Vuoti L, Oikkonen J et al (2013) The genome-wide landscape of copy number variations in the MUSGEN study provides evidence for a founder effect in the isolated Finnish population. Eur J Hum Genetics 21:1411–1416. https://doi.org/10.1038/ejhg.2013.60
Karczewski KJ, Francioli LC, Tiao G et al (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. https://doi.org/10.1038/s41586-020-2308-7
Kato H, Kimura H, Kushima I et al (2022) The genetic architecture of schizophrenia: review of large-scale genetic studies. J Hum Genet 68:175–182. https://doi.org/10.1038/s10038-022-01059-4
Keegan NP, Wilton SD, Fletcher S (2022) Analysis of pathogenic pseudoexons reveals novel mechanisms driving cryptic splicing. Front Genet 12:2711. https://doi.org/10.3389/fgene.2021.806946
Khayat MM, Mohammad S, Sahraeian E et al (2021) Hidden biases in germline structural variant detection. Genome Biol 22:347
Kimura M, Ohta T (1969) The average number of generations until fixation of a mutant gene in a finite population. Genetics 61:763–771. https://doi.org/10.1093/genetics/61.3.763
Kimura M, Maruiama T, Crow JF (1963) The mutation load in small populations. Genetics 48:1303–1312. https://doi.org/10.1093/genetics/48.10.1303
Kosugi S, Momozawa Y, Liu X et al (2019) Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol 20:8–11. https://doi.org/10.1186/s13059-019-1720-5
Kristiansson K, Naukkarinen J, Peltonen L (2008) Isolated populations and complex disease gene identification. Genome Biol 9:109. https://doi.org/10.1186/gb-2008-9-8-109
Lawrence M, Huber W, Pagès H et al (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. https://doi.org/10.1371/journal.pcbi.1003118
Layer RM, Chiang C, Quinlan AR, Hall IM (2014) LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15:1–19. https://doi.org/10.1186/gb-2014-15-6-r84
Liao Y, Wang J, Jaehnig EJ et al (2019) WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47:W199–W205. https://doi.org/10.1093/nar/gkz401
Liégeois J-P (1994) Roma, gypsies, travellers. Council of Europe Press, Strasbourg, France
Lim ET, Würtz P, Havulinna AS et al (2014) Distribution and medical impact of loss-of-function variants in the finnish founder population. PLoS Genet 10:e1004494. https://doi.org/10.1371/journal.pgen.1004494
Linck E, Battey CJ (2019) Minor allele frequency thresholds strongly affect population structure inference with genomic data sets. Mol Ecol Resour 19:639–647. https://doi.org/10.1111/1755-0998.12995
Lohmueller KE (2014) The distribution of deleterious genetic variation in human populations. Curr Opin Genet Dev 29:139–146. https://doi.org/10.1016/j.gde.2014.09.005
Lohmueller KE, Indap AR, Schmidt S et al (2008) Proportionally more deleterious genetic variation in European than in African populations. Nature 451:994–997. https://doi.org/10.1038/nature06611
Loots GG, Kneissel M, Keller H et al (2005) Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res 15:928–935. https://doi.org/10.1101/gr.3437105
Lou H, Li S, Jin W et al (2015) Copy number variations and genetic admixtures in three Xinjiang ethnic minority groups. Eur J Hum Genet 23:536–542. https://doi.org/10.1038/ejhg.2014.134
Lutz BD (1995) Gypsies as victims of the holocaust. Holocaust Genocide Stud 9:346–359. https://doi.org/10.1093/hgs/9.3.346
Mallick S, Li H, Lipson M et al (2016) The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538:201–206. https://doi.org/10.1038/nature18964
Manolescu A, Helgadottir A, Kong A et al (2004) The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet 36:233–239
McCarroll SA, Huett A, Kuballa P et al (2008) Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet 40:1107–1112. https://doi.org/10.1038/ng.215
McInnes L, Healy J, Melville J (2018) Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426
Melegh BI, Banfai Z, Hadzsiev K et al (2017) Refining the South Asian Origin of the Romani people. BMC Genet 18:1–13. https://doi.org/10.1186/s12863-017-0547-x
Mendizabal I, Lao O, Marigorta UM et al (2013) Implications of population history of European Romani on genetic susceptibility to disease. Hum Hered 76:194–200. https://doi.org/10.1159/000360762
Mills RE, Walter K, Stewart C et al (2011) Mapping copy number variation by population-scale genome sequencing. Nature 470:59–65. https://doi.org/10.1038/nature09708
Milton S (1991) Gypsies and the Holocaust. Hist Teacher 24:375. https://doi.org/10.2307/494697
Mohamad Isa II, Jamaluddin J, Achim NH, Abubakar S (2020) Population-specific profiling of CCL3L1 copy number of the three major ethnic groups in Malaysia and the implication on HIV susceptibility. Gene 754:144821. https://doi.org/10.1016/j.gene.2020.144821
Mondal M, Casals F, Xu T et al (2016) Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat Genet 48:1066–1070. https://doi.org/10.1038/ng.3621
Moorjani P, Patterson N, Loh PR et al (2013) Reconstructing Roma history from genome-wide data. PLoS ONE 8:e58633. https://doi.org/10.1371/journal.pone.0058633
Morar B, Gresham D, Angelicheva D et al (2004) Mutation history of the roma/gypsies. Am J Hum Genet 75:596–609. https://doi.org/10.1086/424759
Moreno-Cabrera JM, del Valle J, Castellanos E et al (2021) CNVfilteR: an R/Bioconductor package to identify false positives produced by germline NGS CNV detection tools. Bioinformatics 37:4227–4229. https://doi.org/10.1093/bioinformatics/btab356
Morris-Rosendahl DJ, Crocq M-A (2020) Neurodevelopmental disorders—the history and future of a diagnostic concept. Dialogues Clin Neurosci 22:65–72. https://doi.org/10.31887/DCNS.2020.22.1/macrocq
Nakatsuka N, Moorjani P, Rai N et al (2017) The promise of discovering population-specific disease-associated genes in South Asia. Nat Genet 49:1403–1407. https://doi.org/10.1038/ng.3917
Narang A, Jha P, Kumar D et al (2014) Extensive copy number variations in admixed Indian population of African ancestry: potential involvement in adaptation. Genome Biol Evol 6:3171–3181. https://doi.org/10.1093/gbe/evu250
Need AC, Goldstein DB (2009) Next generation disparities in human genomics: concerns and remedies. Trends Genet 25:489–494. https://doi.org/10.1016/j.tig.2009.09.012
Palo OM, Antila M, Silander K et al (2007) Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum Mol Genet 16:2517–2528. https://doi.org/10.1093/hmg/ddm207
Parsons MJ, Mata I, Beperet M et al (2007) A dopamine D2 receptor gene-related polymorphism is associated with schizophrenia in a Spanish population isolate. Psychiatr Genet 17:159–163. https://doi.org/10.1097/YPG.0b013e328017f8a4
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:2074–2093. https://doi.org/10.1371/journal.pgen.0020190
Patterson MD, Marschall T, Pisanti N et al (2015) WhatsHap: weighted haplotype assembly for future-generation sequencing reads. J Comput Biol 22:498–509. https://doi.org/10.1089/cmb.2014.0157
Pedersen CET, Lohmueller KE, Grarup N et al (2017) The effect of an extreme and prolonged population bottleneck on patterns of deleterious variation: Insights from the Greenlandic Inuit. Genetics 205:787–801. https://doi.org/10.1534/genetics.116.193821
Pembleton LW, Cogan NOI, Forster JW (2013) StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol Ecol Resour 13:946–952. https://doi.org/10.1111/1755-0998.12129
Perry GH, Yang F, Marques-Bonet T et al (2008) Copy number variation and evolution in humans and chimpanzees. Genome Res 18:1698–1710. https://doi.org/10.1101/gr.082016.108
Petersen USS, Doktor TK, Andresen BS (2022) Pseudoexon activation in disease by non-splice site deep intronic sequence variation—wild type pseudoexons constitute high-risk sites in the human genome. Hum Mutat 43:103–127. https://doi.org/10.1002/humu.24306
Piccolo F, Jeanpierre M, Leturcq F et al (1996) A founder mutation in the γ-sarcoglycan gene of Gypsies possibly predating their migration out of India. Hum Mol Genet 5:2019–2022. https://doi.org/10.1093/hmg/5.12.2019
Popejoy AB, Fullerton SM (2016) Genomics is failing on diversity. Nature 538:161–164. https://doi.org/10.1038/538161a
R Core Team (2003) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Rainer J (2017) EnsDb.Hsapiens.v86: Ensembl based annotation package. R package version 2.99.0
Rainer J, Gatto L, Weichenberger CX (2019) Ensembldb: an R package to create and use ensembl-based annotation resources. Bioinformatics 35:3151–3153. https://doi.org/10.1093/bioinformatics/btz031
Redon R, Ishikawa S, Fitch KR et al (2006) Global variation in copy number in the human genome. Nature 444:444–454. https://doi.org/10.1038/nature05329
Reyniers A (1995) Gypsy populations and their movements within Central and Eastern Europe and towards some OECD countries. Organisation for Economic Co-Operation and Development Publications, Paris
Rigau M, Juan D, Valencia A, Rico D (2019) Intronic CNVs and gene expression variation in human populations. PLoS Genet 15:1–23. https://doi.org/10.1371/journal.pgen.1007902
Romdhane L, Mezzi N, Dallali H et al (2021) A map of copy number variations in the Tunisian population: a valuable tool for medical genomics in North Africa. NPJ Genom Med. https://doi.org/10.1038/s41525-020-00166-5
Sanna S, Jackson AU, Nagaraja R et al (2008) Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet 40:198–203. https://doi.org/10.1038/ng.74
Sebat J, Lakshmi B, Malhotra D et al (2007) Strong association of de novo copy number mutations with autism. Science 316:445–449. https://doi.org/10.1126/science.1138659
Sekiguchi M, Sobue A, Kushima I et al (2020) ARHGAP10, which encodes Rho GTPase-activating protein 10, is a novel gene for schizophrenia risk. Transl Psychiatry 10:247. https://doi.org/10.1038/s41398-020-00917-z
Serres-Armero A, Davis BW, Povolotskaya IS et al (2021) Copy number variation underlies complex phenotypes in domestic dog breeds and other canids. Genome Res 31:762–774. https://doi.org/10.1101/GR.266049.120
Singh T, Walters JTR, Johnstone M et al (2017) The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet 49:1167–1173. https://doi.org/10.1038/ng.3903
Sirugo G, Williams SM, Tishkoff SA (2019) The missing diversity in human genetic studies. Cell 177:26–31. https://doi.org/10.1016/j.cell.2019.02.048
Soylev A, Kockan C, Hormozdiari F, Alkan C (2017) Toolkit for automated and rapid discovery of structural variants. Methods 129:3–7. https://doi.org/10.1016/j.ymeth.2017.05.030
Sridhar CR (2006) Historical amnesia: the Romani holocaust. Econ Polit Wkly 41:3569–3571
Staehling-Hampton K, Proll S, Paeper BW et al (2002) A 52-kb deletion in the SOST-MEOX1 intergenic region on 17q12-q21 is associated with van Buchem disease in the Dutch population. Am J Med Genet 110:144–152. https://doi.org/10.1002/ajmg.10401
Stefansson H, Rujescu D, Cichon S et al (2008) Large recurrent microdeletions associated with schizophrenia. Nature 455:232–236. https://doi.org/10.1038/nature07229
Steinthorsdottir V, Thorleifsson G, Reynisdottir I et al (2007) A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 39:770–775. https://doi.org/10.1038/ng2043
Stranger BE, Forrest MS, Dunning M et al (2007) Relative impact of nucleotide and copy number variation on gene phenotypes. Science 315:848–853. https://doi.org/10.1126/science.1136678
Sudmant PH, Mallick S, Nelson BJ et al (2015a) Global diversity, population stratification, and selection of human copy-number variation. Science 349:aab3761. https://doi.org/10.1126/science.aab3761
Sudmant PH, Rausch T, Gardner EJ et al (2015b) An integrated map of structural variation in 2,504 human genomes. Nature 526:75–81. https://doi.org/10.1038/nature15394
Sulem P, Gudbjartsson DF, Stacey SN et al (2007) Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet 39:1443–1452. https://doi.org/10.1038/ng.2007.13
Sulem P, Gudbjartsson DF, Stacey SN et al (2008) Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet 40:835–837. https://doi.org/10.1038/ng.160
Telonis AG, Rigoutsos I (2021) The transcriptional trajectories of pluripotency and differentiation comprise genes with antithetical architecture and repetitive-element content. BMC Biol 19:1–19. https://doi.org/10.1186/s12915-020-00928-8
Thaler A, Ash E, Gan-Or Z et al (2009) The LRRK2 G2019S mutation as the cause of Parkinson’s disease in Ashkenazi Jews. J Neural Transm 116:1473–1482. https://doi.org/10.1007/s00702-009-0303-0
Thorleifsson G, Magnusson KP, Sulem P et al (2007) Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science 317:1397–1400. https://doi.org/10.1126/science.1146554
Urnikyte A, Domarkiene I, Stoma S et al (2016) CNV analysis in the Lithuanian population. BMC Genet 17:1–8. https://doi.org/10.1186/s12863-016-0373-6
Uyan Ö, Ömür Ö, Aǧim ZS et al (2013) Genome-wide copy number variation in sporadic amyotrophic lateral sclerosis in the Turkish population: deletion of EPHA3 Is a possible protective factor. PLoS ONE 8:e72381. https://doi.org/10.1371/journal.pone.0072381
Valls-Margarit J, Galván-Femenía I, Matías-Sánchez D et al (2022) GCAT|Panel, a comprehensive structural variant haplotype map of the Iberian population from high-coverage whole-genome sequencing. Nucleic Acids Res 50:2464–2479. https://doi.org/10.1093/nar/gkac076
Vaz-Drago R, Custódio N, Carmo-Fonseca M (2017) Deep intronic mutations and human disease. Hum Genet 136:1093–1111. https://doi.org/10.1007/s00439-017-1809-4
Weischenfeldt J, Symmons O, Spitz F, Korbel JO (2013) Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat Rev Genet 14:125–138. https://doi.org/10.1038/nrg3373
Willer CJ, Speliotes EK, Loos RJF et al (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 41:25–34. https://doi.org/10.1038/ng.287
Yang TL, Chen XD, Guo Y et al (2008) Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet 83:663–674. https://doi.org/10.1016/j.ajhg.2008.10.006
Ye K, Schulz MH, Long Q et al (2009) Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25:2865–2871. https://doi.org/10.1093/bioinformatics/btp394
Zeggini E (2014) Using genetically isolated populations to understand the genomic basis of disease. Genome Med 6:12–14. https://doi.org/10.1186/s13073-014-0083-5
Zhang B, Kirov S, Snoddy J (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33:W741–W748. https://doi.org/10.1093/nar/gki475
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by the Spanish Ministry of Economy and Competitiveness and Agencia Estatal de Investigación (grant numbers CGL2016-75389-P (MINEICO/FEDER, UE), PID2019-106485 GB-I00/AEI/10. 13039/501100011033 (MINEICO), and “Unidad María de Maeztu” (CEX2018-000792-M) to FC and DC; and Agència de Gestió d’Ajuts Universitaris i de la Recerca (Generalitat de Catalunya, grant 2017SGR00702).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by MA. The first draft of the manuscript was written by MA and FC and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Antinucci, M., Comas, D. & Calafell, F. Population history modulates the fitness effects of Copy Number Variation in the Roma. Hum. Genet. 142, 1327–1343 (2023). https://doi.org/10.1007/s00439-023-02579-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-023-02579-5