Abstract
Both safety and efficacy of medical treatment can vary depending on the ethnogeographic background of the patient. One of the reasons underlying this variability is differences in pharmacogenetic polymorphisms in genes involved in drug disposition, as well as in drug targets. Knowledge and appreciation of these differences is thus essential to optimize population-stratified care. Here, we provide an extensive updated analysis of population pharmacogenomics in ten pharmacokinetic genes (CYP2D6, CYP2C19, DPYD, TPMT, NUDT15 and SLC22A1), drug targets (CFTR) and genes involved in drug hypersensitivity (HLA-A, HLA-B) or drug-induced acute hemolytic anemia (G6PD). Combined, polymorphisms in the analyzed genes affect the pharmacology, efficacy or safety of 141 different drugs and therapeutic regimens. The data reveal pronounced differences in the genetic landscape, complexity and variant frequencies between ethnogeographic groups. Reduced function alleles of CYP2D6, SLC22A1 and CFTR were most prevalent in individuals of European descent, whereas DPYD and TPMT deficiencies were most common in Sub-Saharan Africa. Oceanian populations showed the highest frequencies of CYP2C19 loss-of-function alleles while their inferred CYP2D6 activity was among the highest worldwide. Frequencies of HLA-B*15:02 and HLA-B*58:01 were highest across Asia, which has important implications for the risk of severe cutaneous adverse reactions upon treatment with carbamazepine and allopurinol. G6PD deficiencies were most frequent in Africa, the Middle East and Southeast Asia with pronounced differences in variant composition. These variability data provide an important resource to inform cost-effectiveness modeling and guide population-specific genotyping strategies with the goal of optimizing the implementation of precision public health.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interindividual differences in drug response are a common phenomenon in pharmacological therapy. While some patients respond appropriately to a given treatment, in others, it can result in lack of efficacy, which affects an estimated 10–45% of patients (Salvà Lacombe et al. 1996; Trivedi et al. 2006). Furthermore, interindividual differences can give rise to sometimes severe adverse drug reactions (ADRs) in a subset of patients that overall account for approximately 7% of all hospitalizations and 0.3% of death among all hospitalized patients (Lazarou et al. 1998; Pirmohamed et al. 2004). Among the factors causing interindividual differences, genetic germline variations in genes that are involved in pharmacokinetics and pharmacodynamics are estimated to explain 20–30% of drug response variability.
Notably, many of these pharmacogenes are among the most polymorphic genes in the human genome and harbor thousands of genetic variants, which can change enzyme activity or disrupt drug-target interactions, thereby eventually altering drug effects (Lauschke et al. 2017; Zhou et al. 2021a). Much effort has been made to identify actionable associations between genetic variants and differential drug response. As of 2021, > 310 drugs have received pharmacogenomic information in their labels or have received guidelines by pharmacogenomic expert working groups, such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG), that can guide drug selection or posology (Lauschke et al. 2019; Shekhani et al. 2020).
Nevertheless, only a fraction of these established pharmacogenomic biomarkers is implemented in routine clinical care and the only preemptive tests that are mandated are for HLA-B*57:01 and DPYD variants to inform abacavir and fluoropyrimidine therapy, respectively. While the underlying reasons are complex and multifaceted, prevalence of the variants in question constitutes one of the factors that impacts the clinical utility of genetic testing (Lauschke and Ingelman-Sundberg 2016; Russell et al. 2021). Thus, mapping variant frequencies in different ethnogeographic groups can provide important information to inform cost-effectiveness modeling and guide population-specific genotyping strategies. Here, we provide an updated overview of population pharmacogenomics of ten important pharmacokinetic genes (CYP2D6, CYP2C19, DPYD, TPMT, NUDT15 and SLC22A1), drug targets (CFTR) and genes involved in adverse event risk independent of drug pharmacokinetics or target (HLA-A, HLA-B and G6PD). We provide a detailed overview of ethnogeographic differences in allele frequencies, infer functional consequences and discuss implications and relevance for the implementation of population-specific precision public health. For other clinically relevant pharmacogenes, such as CYP2B6 (Langmia et al. 2021), UGT1A1 (Hall et al. 1999) or NAT2 (Sabbagh et al. 2011), we refer the interested reader to excellent reviews on the topic.
CYP2D6
CYP2D6 is one of the most pleiotropic drug-metabolizing enzymes and is involved in the hepatic clearance of approximately 25% of all clinically used drugs, including tricyclic antidepressants, opioids, antiemetics and antiarrhythmics (Zanger and Schwab 2013). Importantly, at least in part due to the lack of important endogenous substrates and low evolutionary constraints, CYP2D6 constitutes one of the most polymorphic genes in the cytochrome P450 (CYP) gene family, resulting in drastic functional diversity of CYP2D6 (Fujikura et al. 2015; Ingelman-Sundberg 2005). Of the more than 100 different CYP2D6 alleles that have been described to date, the loss-of-function (LOF) alleles CYP2D6*3, *4, *5 and *6, the decreased function alleles *9, *10, *17, *29 and *41 as well as the CYP2D6 duplications *1xN and *2xN are functionally most relevant and are common with minor allele frequencies (MAF) > 1% in at least one population (Tables 1 and 2). Over the past decades, substantial interethnic differences have been revealed for these alleles, which translate into substantial variability in metabolic phenotypes across populations (Gaedigk et al. 2017; Zhou et al. 2017).
The splice variant CYP2D6*4 (rs3892097) constitutes the globally most common CYP2D6 variant allele of functional importance. In Europe, CYP2D6*4 is prevalent across North and Central Europe with frequencies around 20–25%. The highest frequency of this allele was observed on the Faroe Islands (33.4%), whereas it is substantially less prevalent in Southern Europe in Italy (16.4%), Greece (17.7%) and Turkey (13.2%), resulting in a European North-to-South gradient (Petrović et al. 2020). In addition, high CYP2D6*4 frequencies were observed in Ashkenazi Jews (22.6%), a genetic isolate historically living in Europe that harbors a genetic repertoire that is distinctly different from other populations (Scott et al. 2007; Zhou et al. 2018). CYP2D6*4 is also abundant in American populations, particularly in Columbia (19.4%) (Isaza et al. 2000), Costa Rica (15.8%) (Céspedes-Garro et al. 2014), Panama (15.4%) (Jorge et al. 1999) and Nicaragua (15.1%) (Agúndez et al. 1997). CYP2D6*4 frequencies are slightly lower in West Asia (7.8%) and Central-South Asia (8.5%), whereas the variant is almost absent among East Asians (0.6%) (Gaedigk et al. 2017). Similarly, CYP2D6*4 frequencies are lower in Africa, ranging from 2% in Zimbabwe (Dandara et al. 2001) to 7% in Ghana (Griese et al. 1999). Interestingly, CYP2D6*4 frequency in African Americans (6.3%) is higher than in most African populations that reside in Africa (3.3%), possibly at least in part because of genetic admixture.
Besides CYP2D6*4, also other CYP2D6 LOF alleles such as CYP2D6*3 (rs35742686) and *6 (rs5030655) are most abundant in European populations. The frequencies of CYP2D6*3 are above 1% in most European countries with the highest values found in Finland (3.5%) and the United Kingdom (3.3%) (Auton et al. 2015). In contrast, the allele is rare or absent in Portugal (0.7%) (Albuquerque et al. 2013; Correia et al. 2009), Turkey (0.7%) (Aydin et al. 2005; Aynacioglu et al. 1999; Mizzi et al. 2016; Serin et al. 2012), Austria (0.5%) (Beer et al. 2011) and Norway (0%) (Molden et al. 2002). Notably, while CYP2D6*3 is overall less abundant outside Europe, it is also found in countries with admixed populations, such as Brazil (Friedrich et al. 2014; Kohlrausch et al. 2009). Similarly, CYP2D6*6 is only common in some European populations with frequencies up to 2.1% in Finland.
In contrast to the LOF alleles CYP2D6*3, *4 and *6, deletion of CYP2D6 (CYP2D6*5) is most common in Africa, East Asia and Oceania with frequencies pivoting around 5–6% (Gaedigk et al. 2017). In Europe, CYP2D6*5 prevalence is overall lower with a South-East to North-West gradient, ranging from 1% in Croatia (Ganoci et al. 2017) to 7.2% in Netherlands (Poulussen et al. 2019). Frequencies of CYP2D6*5 are similarly low in the Americas (2.1%), as well as in South Asian populations (3.2%) with national frequencies up to 5.1% in Malaysia (Teh et al. 2001).
The reduced function variant CYP2D6*10 (rs1065852, rs1135840) is the most common allele in East Asians with frequencies up to 64.1% (Qin et al. 2008). Frequencies are high in Han Chinese (43.5%) and Hui (51%), but substantially lower in Mongolians (25.2%) and Tibetans (28.1%) (Yin et al. 2012). In contrast, CYP2D6*10 is substantially less prevalent in African (6.6%), Ashkenazim (6.2%), European (2.8%) and American populations (2.6%) (Gaedigk et al. 2017). The inframe deletion variant CYP2D6*9 (rs5030656) is globally rare but relatively common in European and American populations with highest frequencies in Denmark (3.4%) (Pedersen et al. 2005; Rasmussen et al. 2006) and Nicaragua (4.4%) (Agúndez et al. 1997).
CYP2D6*17 (rs16947, rs28371706) and CYP2D6*29 (rs16947, rs1135840, rs61736512, rs59421388) are both African-specific alleles with frequencies of 9–34% (Aklillu et al. 1996; Dandara et al. 2001; Masimirembwa et al. 1996) and 4–20% (Dodgen et al. 2016; Wennerholm et al. 2001), respectively. Although considered extremely rare in other populations, they have been also identified in admixed populations. In the Americas, both alleles were prevalent in the Afro–Trinidadian population (CYP2D6*17, 16.5%; CYP2D6*29, 8.7%) (Montané Jaime et al. 2013), as well as in Cuba (CYP2D6*17, 6.4%) (Llerena et al. 2012), Brazil (CYP2D6*17, 4.8%; CYP2D6*29, 2.8%) (Antunes et al. 2012; Friedrich et al. 2014; Kohlrausch et al. 2009) and Costa Rica (CYP2D6*17, 4.1%; CYP2D6*29, 2.2%) (Céspedes-Garro et al. 2014). In addition, both CYP2D6*17 and CYP2D6*29 were observed in the Middle East with frequencies pivoting around 2.5% and 1.6%, respectively (Khalaj et al. 2019; Luo et al. 2004; McLellan et al. 1997; Qumsieh et al. 2011; Sistonen et al. 2007).
The splicing variant CYP2D6*41 (rs28371725) is globally common with highest frequencies being reported in Bedouins (29%) (Luo et al. 2004) and Indians (12.5%) (Sistonen et al. 2009). CYP2D6*41 is also prevalent in African (9.7%), European (7.4%), American (3.7%) and East Asian (2.2%) populations albeit at overall lower frequencies. Notably, however, CYP2D6*41 frequencies can be substantially higher than the respective superpopulation averages, as observed in Ethiopia (22.9%) (Aklillu et al. 2002), Italy (15.2%) (Carano et al. 2018) and the Netherlands (15%) (Poulussen et al. 2019).
In contrast to the aforementioned decreased function and LOF CYP2D6 alleles, the gain-of-function (GOF) duplication allele CYP2D6*1xN is most prevalent in Oceanian Aborigines (11.5%) (Sistonen et al. 2007), particularly in Papua New Guinea (12%) (von Ahsen et al. 2010), whereas the genetically distinct GOF allele CYP2D6*2xN is most common in the Mozabite population in North Africa (28.3%) (Sistonen et al. 2007). In contrast, in Sub-Saharan Africa, the frequencies of CYP2D6 duplications are overall low (2.4% and 0.8% for CYP2D6*1xN and CYP2D6*2xN, respectively) (Sistonen et al. 2007). CYP2D6 duplications are moreover common in Ashkenazim and Middle Eastern populations with combined frequencies of 8% and 3.9% (Fuselli et al. 2004; Scott et al. 2007). CYP2D6 gene duplications are rare in Central European populations, such as Germans (1.3%), Austrians (1.6%) and Hungarians (1.8%), but considerably higher in both Northern and Southern European groups, such as Finnish (4.3%), Spanish (3.5%), Greek (6%) and Turkish (5.6%) (Petrović et al. 2020). In Asian populations, both CYP2D6*1xN and CYP2D6*2xN are rare with frequencies below 1% (Sistonen et al. 2007).
The country-specific CYP2D6 allele frequency data can be aggregated to infer CYP2D6 phenotypes (Gaedigk et al. 2017; Koopmans et al. 2021). The frequency of CYP2D6 poor metabolizers (PM), defined as individuals carrying two LOF alleles, is highest in Ashkenazi Jews (6%) and European population (5.4–11.4%) and lowest in populations from the Middle East (0.9%), East Asia (0.4%) and Oceania (0.4%; Fig. 1). In contrast, the prevalence of intermediate metabolizers (IM) that exhibit reduced but measurable CYP2D6 metabolism was found to be highest in African populations (10–60%) and Ashkenazim (10–40%), and lowest in South Asia (3.8%) and the Americas (2.8%). Ultrarapid metabolizers (UM) that carry at least one functional gene duplication, are most common in indigenous Oceanian populations (21.2%) and North Africa (up to 39%), whereas they are lowest in East Asia (1.4%). These functional extrapolations can provide important information for population-specific drug selection and the posology of CYP2D6 substrates.
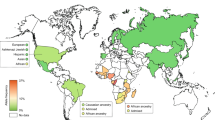
Global distribution of inferred CYP2D6 phenotypes. Frequencies of CYP2D6 poor metabolizer (A), intermediate metabolizer (B) and ultrarapid metabolizer (C) phenotypes were calculated based on the frequencies of loss-of-function alleles (*3, *4, *5 and *6), decreased function alleles (*9, *10, *17, *29 and *41) and increased function alleles (*1xN and *2xN) from 53 countries/populations (Tables 1 and 2; Supplementary Table 1). Countries are color-coded with the highest frequency in red, the average frequency across all populations (\(\overline{f }\)) in yellow, and the lowest frequency in green. In case of missing population frequencies, averaged continent frequency data from the literature (Gaedigk et al. 2017) were used to infer metabolizer phenotypes
CYP2C19
CYP2C19 is a key enzyme involved in the metabolism of the antiplatelet drug clopidogrel, selective serotonin reuptake inhibitors (SSRIs) as well as proton pump inhibitors, and genetic variability in CYP2C19 contributes to the differential response to these substrates. The clinically most relevant variant alleles are CYP2C19*2 (rs4244285) and CYP2C19*3 (rs4986893) that abolish enzyme activity and the regulatory CYP2C19*17 variant (rs12248560) that results in increased gene activity (Table 3).
CYP2C19*2 is globally common with highest frequencies found in Oceanian (61%) (Scott et al. 2013) and Asian populations (28.4% in East Asian and 31.8% in South Asian)(Ionova et al. 2020). On a per-country level, CYP2C19*2 was most prevalent in the Vanuatu atoll with a reported frequency of 71% (Kaneko et al. 1997, 1999). In Africa, the Americas and Europe, the frequencies of this allele pivot around 12–15% (Scott et al. 2013) with South African Xhosa (21%), Cypriots (21%), Romani (20.8%) and Maltese (20%) constituting the ethnogeographic hotspots (Drögemöller et al. 2010; Pimenoff et al. 2012; Sipeky et al. 2013; Mizzi et al. 2016).
Like CYP2C19*2, also the CYP2C19*3 allele is common in Oceania (15%) (Scott et al. 2013) and across East Asia (6%) (Ionova et al. 2020). Notably, the frequency of CYP2C19*3 in East Asia exhibits an East-to-West gradient with highest frequencies in Japanese (11.3%), followed by South Koreans (8.6%) and Chinese (4.4%) (Dorji et al. 2019). Interestingly, while CYP2C19*2 and CYP2C19*3 are both common across Oceania, their frequencies in Polynesian populations, including Samoan, Tongan, Fijian, Cook Islander and Maori, are substantially lower than in Melanesians, including Papua New Guinean, Vanuatuan and Aboriginal Australian (CYP2C19*2: 22% in Polynesians vs. 51% in Melanesians; CYP2C19*3: 4% in Polynesians vs. 19% in Melanesians) (Helsby 2016).
CYP2C19*17 is prevalent worldwide with frequencies above 15% except for East Asian populations (3.7%) (Ionova et al. 2020). In Europe, the highest prevalence was reported in Slovakia (33%), Poland (29.8%) and the Czech Republic (29%), whereas frequencies are lower in South and East Europe (Cyprus, 11%; Span, 17%; Russia, 15%) (Gawrońska-Szklarz et al. 2012; Mizzi et al. 2016; Vicente et al. 2014).
The functional allele frequency data has been used to predict CYP2C19 phenotypes across ethnicities (Koopmans et al. 2021). CYP2C19 PM status was most common in Oceania where around 58% of individuals are homozygous or compound heterozygous for CYP2C19 LOF alleles (Fig. 2). Considerable numbers of CYP2C19 PMs were also reported in East Asian (14.2%) and Central/South Asian (11.8%) populations, whereas their numbers are lower in Latin America (1.1%), Europe (2.7%) and Africa (3.3%). CYP2C19 UMs are most common in European, African and Latin American populations with frequencies pivoting around 20–30%, whereas only 2.1% of the East Asians are UMs (Koopmans et al. 2021).
Global distribution of inferred CYP2C19 phenotypes. Frequencies of CYP2C19 poor metabolizers (A), intermediate metabolizers (B) and ultrarapid metabolizers (C) were calculated based on frequencies of the loss-of-function alleles CYP2C19*2 and *3, as well as the increased function allele CYP2C19*17 for 52 countries/populations (Table 3; Supplementary Table 2). Countries are color-coded with the highest frequency in red, the average frequency across all populations (\(\overline{f }\)) in yellow, and the lowest frequency in green. In case of missing population frequencies, averaged continent frequency data from the literature (Ionova et al. 2020; Scott et al. 2013) were used to infer metabolizer phenotypes
DPYD
Fluoropyrimidines, including 5-fluorouracil and its prodrugs capecitabine and tegafur, are important chemotherapeutics for the treatment of various solid tumors. They are among the most prescribed anticancer drugs worldwide with more than two million patients estimated to use fluoropyrimidines each year (Ezzeldin and Diasio 2004). However, up to 40% of patients experience fluoropyrimidine-induced toxicity that is severe enough to require discontinuation of therapy, and in 0.5–1% of patients these ADRs are fatal (Hoff et al. 2001; Van Cutsem et al. 2001). The toxicity of fluoropyrimidines is strongly associated with the metabolic activity of dihydropyrimidine dehydrogenase (DPD), the enzyme catalyzing the rate-limiting step in the biotransformation of fluoropyrimidines into non-toxic metabolites. As such, reduced activity of DPD increases fluoropyrimidine exposure, resulting in increased cytotoxicity.
Interindividual variation in DPD activity is strongly associated with genetic variability of the respective gene, DPYD. The most well-studied DPYD variant is DPYD*2A (rs3918290; c.1059 + 1G > A; IVS14 + 1G > A), a splicing variant that results in exon skipping and gives rise to a truncated gene product with no catalytic activity (Vreken et al. 1996). The highest frequency of DPYD*2A is found in the Finnish population (2.4%) (Zhou et al. 2020), whereas frequencies in Central, South and East Europe are > twofold lower, pivoting around 1%, 0.5% and 0.3%, respectively (Raida et al. 2001; Salgueiro et al. 2004; Sulzyc-Bielicka et al. 2008; Uzunkoy et al. 2007; van Kuilenburg et al. 2001) (Table 4). DPYD*2A is extremely rare in Asian, African and indigenous American populations (Elraiyah et al. 2017; Hariprakash et al. 2018; Zhou et al. 2020).
In addition to DPYD*2A, the DPYD haplotype HapB3, comprising three intronic variants (c.483 + 18 G > A/rs56276561, c.680 + 139 G > A/rs6668296 and c.959-51T > C/rs115349832) and one synonymous variant (E412E; c.1236 G > A; rs56038477), has been associated with severe fluoropyrimidine toxicity (Amstutz et al. 2009). This association is likely due to c.1129–5923C > G (rs75017182), a deep intronic variant that is in strong linkage with HapB3 and that impairs DPD function by affecting pre-mRNA splicing (van Kuilenburg et al. 2010). Importantly, c.1129–5923C > G/HapB3 is common in many populations. In Europe, it is considered the most common reduced function DPYD variant with an averaged frequency of 2.1% and highest prevalence in the Netherlands (2.6%) and Germany (3.3%) (van Kuilenburg et al. 2010; Zhou et al. 2020). In contrast, HapB3 is less frequent in Africa (0.2%), East Asia (0.2%), Latinos (0.8%) and Ashkenazim (0.7%) (Zhou et al. 2020). Another well-established decreased function DPYD variant is p.Y186C (rs115232898), a variant that is prevalent with frequencies up to 3.3% among individuals of African ancestry but is almost absent in other populations (Offer et al. 2013). Other functionally relevant DPYD variants, such as p.D949V (rs67376798), are rare with frequencies below 1% in all populations.
Previous estimates for the global prevalence of partial and full DPD deficiency are 3–8% and 0.02–0.2%, respectively, with highest frequencies in Africans and Finnish and lowest in Ashkenazi Jews and East Asians (Caudle et al. 2013; Zhou et al. 2020). As frequencies of DPD deficiency differ by up to tenfold between populations, these data thus emphasize the importance of population-adjusted strategies for the optimization of fluoropyrimidine dosing and solid cancer therapy.
TPMT and NUDT15
Thiopurine methyltransferase (encoded by TPMT) and nudix hydrolase 15 (encoded by NUDT15) play important roles in the metabolism of the thiopurines mercaptopurine and thioguanine, which are widely used in the treatment of acute lymphoblastic leukemia, inflammatory bowel diseases and autoimmune disorders. Thiopurines are metabolized intracellularly into thioguanosine monophosphate (TGMP), which is further converted into the active thioguanine di- and triphosphates that exert their cytotoxic and antiproliferative effects by blocking purine synthesis and by causing direct damage to DNA and RNA (Bökkerink et al. 1993; Inamochi et al. 1999; Karim et al. 2013). Furthermore, they have anti-inflammatory effects by inducing T cell apoptosis via inhibition of the GTPase RAC1 (Poppe et al. 2006). TPMT plays a central role in the metabolism of thiopurines into inactive methyl-metabolites thereby shunting TGMP away from further metabolic activation. Similarly, NUDT15 dephosphorylates thioguanine di- and triphosphates back into its monophosphate form, counteracting its incorporation into DNA and RNA.
Genetic variations can cause TMPT and NUDT15 deficiency, resulting in excessive formation of thioguanine di- and triphosphates and an increased risk of severe myelosuppression. The most common and well-characterized TPMT alleles are TPMT*3A (rs1142345 and rs1800460), *3C (rs1142345) and *2 (rs1800462), which together explain more than 90% of decreased TPMT activity phenotypes (Schaeffeler et al. 2004; Zhou et al. 2020). TPMT*3A is most common in European and Latin American populations with frequencies pivoting around 2–4%. The highest TPMT*3A frequencies in Europe were observed in the UK (4.5%) (Ameyaw et al. 1999) and Greenland (8.1%) (Toft et al. 2006), whereas frequencies in Croatia were substantially lower (1.9%) (Ladić et al. 2016). No TPMT*3A alleles were found in 194 indigenous Saami in Norway (Loennechen et al. 2001). In Latin America, frequencies were highest in Brazil (up to 3.9%) (Ferreira et al. 2020), Colombia (3.6%) (Isaza et al. 2003) and Argentina (3.1%) (Laróvere et al. 2003).
In Asian and African populations TPMT*3A is very rare and instead TPMT*3C is the predominant allele underlying TPMT deficiency (Chang et al. 2002; Hon et al. 1999). In Asia, frequencies of TPMT*3C range between 0.8% in Japanese, 0.9% in Koreans (Lee et al. 2008), 1.3–3% in Chinese populations and 0.8–2.8% across South Asia (Hiratsuka et al. 2000; Kham et al. 2002; Lee et al. 2008; Zhang et al. 2003). These allele-specific interethnic differences are even more striking in Sub-Saharan Africa where TPMT*3C is highly abundant in Ghana (7.6%) (Ameyaw et al. 1999), Kenya (5.4%) (McLeod et al. 1999) and Nigeria (5.3%) (Adehin et al. 2017), but relatively rare in North African populations, such as Tunisians (1.4%) (Melaouhia et al. 2012), Egyptians (1.3%) (Hamdy et al. 2003) and Libyans (1%) (Zeglam et al. 2015). The other reduced function variant, TPMT*2, is globally rare with MAF < 1% with few reported exceptions, such as in Iran (2.2%)(Bahari et al. 2010) and Sardinia (1.7%) (Rossino et al. 2006).
Based on frequencies of TPMT*3A, *3C, *2, it is estimated that the frequency of patients harboring intermediate TPMT activity is around 3–14%, and approximately 1 in 178 to 1 in 3,736 patients are fully TPMT deficient (Relling et al. 2011). When extending these analyses using Next Generation Sequencing to also include other functional variations, recent studies suggested highest prevalence of intermediate and full TMPT deficiency in Africa with frequencies of 11% and 0.3%, respectively, whereas the corresponding frequencies were lowest in Asian populations (0.03–0.04% full deficiency; 3.3–3.9% intermediate activity) and Ashkenazim (0.02% full deficiency; 2.9% intermediate activity) (Zhou et al. 2020).
While polymorphisms in TPMT alone explain around 40% of thiopurine-induced ADRs (Schaeffeler et al. 2019), predictions can be further improved by including the missense variant p.R139C in NUDT15 (c.415C > T; rs116855232) (Yang et al. 2015b, 2014). Mechanistically, this variant destabilizes the protein structure, thereby resulting in lower enzymatic activity (Rehling et al. 2021). p.R139C defines NUDT15*3 and is moreover part of NUDT15*2 in combination with the inframe deletion variant (rs746071566), in both cases resulting in a loss of gene product function. The frequency of p.R139C differs > 20-fold across populations. It is most abundant in Asian populations, including Japanese (16%) (Tanaka et al. 2015), Koreans (11.3%) (Kim et al. 2017), Chinese (12.7%) (Chao et al. 2017) and Indians (10.7%) (Shah et al. 2018), as well as Amerindian groups (5–32%) (Suarez-Kurtz et al. 2019). In contrast, frequencies are considerably lower in admixed Brazilian populations (6.8%) (Rodrigues et al. 2020) and across Europe (0.4%) with the exception of Nordic populations, such as Finns (2.3%) and Swedes (2%) (Wahlund et al. 2020). Similarly, p.R139C is almost absent in Africa and the Middle East (Jarrar and Ghishan 2019).
Due to the high frequency of p.R139C, NUDT15 deficiency is common in East Asian (22.6%), South Asian (13.6%) and Latin American (12.5–21.2%) populations (Moriyama et al. 2016), surpassing the prevalence of TPMT deficiency and suggesting that variations in NUDT15 rather than in TPMT are the major drivers of thiopurine-induced toxicity across Asia and Latin America. In contrast, TMPT reduced function alleles explain the majority of thiopurine toxicity in Europe and Africa.
SLC22A1 (OCT1)
The SLC22A1 gene encodes the organic cation transporter OCT1 that is highly expressed in hepatocytes, immune cells and most epithelial barriers. OCT1 partakes in the disposition of a large number of structurally diverse drugs (including metformin, tramadol, lamivudine, oxaliplatin, sorafenib and morphine), endogenous substrates (choline, acetylcholine and agmatine), vitamins (vitamin B1) and toxins (1-methyl-4-phenylpyridinium), and genetic variants in SLC22A1 have been reproducibly associated with altered substrate pharmacokinetics (Arimany-Nardi et al. 2015; Chen et al. 2014; Herraez et al. 2013; Tzvetkov et al. 2013, 2011). Importantly, SLC22A1 is highly polymorphic with more than 1,000 genetic variants of which 450 alter the amino acid sequence of the transporter (Schaller and Lauschke 2019). While most of these variations are very rare and poorly characterized, at least 15 functionally relevant alleles have been identified that are common in at least one population (Seitz et al. 2015).
In European populations, the reduced function alleles SLC22A1*2 (p.M420del; rs202220802) and SLC22A1*3 (p.R61C; rs12208357) constitute the most abundant alleles with frequencies of 10–20% and 2–10%, respectively (Schaller and Lauschke 2019; Zazuli et al. 2020). In addition, the LOF alleles SLC22A1*4 (p.G401S; rs34130495), SLC22A1*5 (p.G465R; rs34059508) and SLC22A1*6 (p.C88R; rs55918055) occur in Europe with frequencies of 1–7%, 0–8% and 0–2%. Notably, SLC22A1*4 seems to be graded from 7.1% in Spain, 5.4% in Sardinia and 4.2% among French Basques in the South of Europe to 1.6% in Finland, 2% in Britain and 0% on the Orkney islands in Northern Europe (Seitz et al. 2015). SLC22A1*7 to *15 are not found across Europe. In aggregate, these data indicate that around 44% of individuals of European descent carry at least one SLC22A1 reduced function allele.
The patterns of genetic SLC22A1 variability are substantially different in African populations. In Sub-Saharan Africa, SLC22A1*8 (p.R488M; rs35270274), a variant allele with slightly increased activity towards morphine and metformin, constitutes the most common allele with frequencies between 2 and 18% (Seitz et al. 2015). Furthermore, SLC22A1*7 (p.S14F; rs34447885) is common with frequencies up to 9%. Effects of this allele are substrate-specific, entailing reduced transport of metformin, tropisetron and tyramine, whereas no differences are observed for morphine, debrisoquine and tramadol. SLC22A1*2 is found across Sub-Saharan Africa albeit with lower prevalence than in Europe (0–11% compared to 10–20%). In aggregate, only around 15% of individuals in Africa harbor reduced function variants, whereas around 12% carry the African increased activity allele SLC22A1*8. In contrast to Sub-Saharan Africa, Northern Africa and the Middle East recapitulates the variant pattern observed in European populations with SLC22A1*2 and SLC22A1*3 being most common, while SLC22A1*7 and SLC22A1*8 are only rare with frequencies around 1%.
Compared to European and African populations, the genetic complexity in East Asian and indigenous American populations is considerably lower. In Pima, Maya, Surui and Colombian populations, OCT1 deficiencies are highly common with frequencies up to 94%, which is almost exclusively allotted to SLC22A1*2. In contrast, in East Asia, 95–98% of alleles are normactive with only few ethnogeographic hotspots of Asian-specific reduced activity variants, such as SLC22A1*12 (p.S29L; rs375175439) in She (10%), as well as SLC22A1*9 (p.P117L; rs200684404), SLC22A1*11 (p.I449T; rs183240019) and SLC22A1*15 (p.E284K) with frequencies of 5–6% in Mongolians, Nashi and Monghour in China, respectively (Chen et al. 2010; Cheong et al. 2011).
Pharmacogenetically important HLA alleles
While around 80% of ADRs are consequences of excessive pharmacological actions, the remaining 20% are idiosyncratic events that are unrelated to the therapeutic effect of the drug (Uetrecht and Naisbitt 2013). Many but likely not all idiosyncratic ADRs are immunologically mediated and can affect virtually any tissue, either in isolation or in combination with systemic effects (Phillips 2016). Idiosyncratic ADRs are more often severe or life-threatening with specific manifestations, such as Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) resulting in mortality rates up to 13–60% (Schulz et al. 2000; Watanabe et al. 2021). The human leukocyte antigen (HLA) gene family encodes the major histocompatibility complex (MHC), which regulates T-cell mediated immunity. HLA genes have been strongly implicated in the etiology of immune-related adverse events caused by a multitude of drugs (Lauschke et al. 2019). The established models suggest that drugs (1) act as haptens, binding covalently to proteins and forming new antigens, (2) directly interact with the T cell receptor via non-covalent bonds or (3) bind non-covalently to the MHC, resulting in deformations of the peptide-binding groove and altered antigen presentation (Pavlos et al. 2015).
Notably, HLA genes are extremely polymorphic, but most idiosyncratic immunological ADRs are restricted to carriers of one or few specific HLA variant alleles. For instance, abacavir binds exclusively to the peptide-binding groove of HLA-B*5701, resulting in altered presentation of self-peptides, which in turn triggers polyclonal alloreactive autoimmunity and gives rise to abacavir hypersensitivity syndrome (AHS)(Illing et al. 2012; Ostrov et al. 2012). Further prominent and clinically well-established associations are associations of allopurinol-induced cutaneous adverse events with HLA-B*58:01 and links between carbamazepine-induced SJS/TEN and HLA-B*15:02 and HLA-A*31:01.
Abacavir is a nucleoside analog reverse transcriptase inhibitor that is used in combination with other antiretrovirals for the treatment of HIV/AIDS. In historic studies before the identification of HLA-B*5701 as a genetic risk factor, AHS occurs in around 5% of patients treated with abacavir with a mortality rate of around 3 per 1000 (Bannister et al. 2008; Hetherington et al. 2001). Importantly, while almost half of all HLA-B*57:01 carriers develop AHS after abacavir exposure, AHS was not observed in any of the patients without HLA-B*57:01 (Mallal et al. 2008). Based on these unambiguous data, preemptive testing of HLA-B*57:01 has become mandatory across the US and Europe before the initiation of abacavir therapy. HLA-B*57:01 allele frequency is a key factor to assess AHS risk in a population-scale. We recently evaluated the ethnogeographic distribution of pharmacogenetically relevant HLA alleles based on genetic information from 6.5 million individuals across 74 countries (Zhou et al. 2021b). The results showed that HLA-B*57:01 is generally rare in Africa, the Middle East and East Asia, whereas in Europe frequencies are reported between 1% in Sweden to 5.8% in Ireland (Fig. 3A). Globally, HLA-B*57:01 is most frequent in India (6.2%) and Sri Lanka (9.3%), whereas it is much less abundant in other South Asian countries such as Malaysia (1.1%), Thailand (2.1%) and Vietnam (2.6%).
Global distribution of clinically important human leukocyte antigen (HLA) alleles. Allele frequencies of HLA-B*57:01 (A), HLA-B*15:02 (B), HLA-A*31:01 (C), and HLA-B*58:01 (D) across up to 74 countries are shown. Countries are color-coded with the highest frequency in red, the average frequency across all populations (\(\overline{f}\)) in yellow, and the lowest frequency in blue. Countries for which no HLA frequency information was available are colored white. Figure modified with permission from (Zhou et al. 2021b)
Carbamazepine-induced severe cutaneous adverse reactions (SCAR) are associated with two alleles, HLA-B*15:02 and HLA-A*31:01, and odds ratios up to 2,504 (Chung et al. 2004) and 58 (Genin et al. 2014) have been reported, respectively. HLA-B*15:02 is exclusively found in Southeast Asian populations where allele frequencies are particularly high in the Philippines (22%), Vietnam (13.8%), Indonesia (11.6%) and Malaysia (11.5%), with the notable exception of Japan (< 0.1%; Fig. 3B). Consequently, genetic testing of HLA-B*15:02 is recommended in individuals of Asian ancestry but not for other populations. In contrast to the region-specific HLA-B*15:02, HLA-A*31:01 is common worldwide (Fig. 3C). It is most prevalent in indigenous populations in the Americas, such as in Argentina (28.8%), Mexico (10.1%), the United States (7.8%), Nicaragua (6.7%) and Chile (6.6%), whereas frequencies in Africa and Oceania seem to be lower (approximately 1%). However, frequency estimates of the latter are only based on small cohorts and further information in these populations is needed to corroborate these observations.
The xanthine oxidase inhibitor allopurinol is used for the treatment for hyperuricemia, but its utility is limited by the development of SCAR in up to 0.5% of patients (Yang et al. 2015a). HLA-B*58:01 is the predominant risk allele in Asian populations (Hung et al. 2005; Lonjou et al. 2008) where it is very common in Mongolia (8.8%), China (7.8%), Thailand (7.8%) and Singapore (7.2%; Fig. 3D). In addition, it is prevalent in several African countries, including Kenya (8.2%), Guinea Bissau (7.8%) and Senegal (6.9%). In contrast, HLA-B*58:01 frequencies are overall low across Europe and the Americas with frequencies ranging from 0.5 to 3.5%. Combined, these data provide the molecular basis for ethnogeographic differences in idiosyncratic ADR risk and suggest that preemptive testing can reduce idiosyncratic toxicity particularly in at-risk populations where the frequency of the respective HLA alleles are high.
CFTR
The CFTR gene encodes a chloride channel that is part of the ATP-binding cassette (ABC) transporter superfamily (ABCC7). The gene product plays essential roles in ion and water secretion and absorption in epithelial tissues. Genetic variations that impact CFTR function constitute the cause of cystic fibrosis (CF), an autosomal recessive disorder most commonly observed in populations of European descent. CF manifests primarily as lung disease with symptoms that resemble pneumonia, bronchiectasis and asthma. Further non-pulmonary symptoms include pancreatic dysfunction, intestinal obstructions and elevated sweat electrolytes. Notably however, phenotypes, ages of onset and clinical manifestations differ considerably between patients.
By now, more than 2,100 genetic variants in CFTR have been described of which more than 400 are assumed to be pathogenic (Kounelis et al. 2020; Xiao and Lauschke 2021). Pathogenic variants are classified into five categories: variants that cause defective protein production, mostly due to premature stop codons or frameshift mutations or large insertions (class I); variants that result in defective protein trafficking (class II); variants causing defects in protein gating (class III) or dysfunctional protein conductance (class IV); and variants that cause reduced amounts of functional proteins, mostly due to splicing defects (class V).
Overall, the class II variant p.F508del (rs1801178) is most common, accounting for 70–75% of CF cases in individuals of European descent (Watson et al. 2004). In contrast, p.F508del is less common in ethnogeographic groups from Africa and Asia. Further misfolding variants include p.N1303K (rs80034486) and p.I507del (rs1490508086) that explain up to 2.8% of CF cases in Ashkenazim and up to 1.9% in Africans, respectively (Table 5). Splicing defect variants (class V) that substantially reduce the amount of functional CFTR at the plasma membrane include c.2988G > A (3120 + 1G > A), c.3717 + 12191C > T (3849 + 10kbC > T) as well as various other rare CFTR rearrangements and are of substantial relevance in Africa, where they constitute a frequent, in some groups even the most common, variant class associated with CF (Goldman et al. 2001; Macek et al. 1997; Schrijver et al. 2016; Owusu et al. 2020).
The major variant that causes the generation of correctly trafficked but dysfunctional channel proteins (class III) is p.G551D (rs75527207). While this variant only contributes minorly (< 1%) to cystic fibrosis risk in Hispanics and Ashkenazim, it explains between 2 and 3.5% of cases in non-Hispanic Caucasians and Asian Americans (Watson et al. 2004). Other variants resulting in CFTR dysfunction include p.R347P (rs77932196) and the Asian-specific variant p.S549N (rs121908755).
There is substantial heterogeneity within the larger populations. For instance, on average only 3–5% of European CF patients carry class III, IV or V variants; however, up to 14% of CF patients in Ireland have at least one class III variant, while more than 12% of patients in Moldova carry at least one class V mutation (De Boeck et al. 2014). Importantly, which genetic factors underlie the disease in a given patient determines the choice of pertinent therapy. Activity of reduced function CFTR proteins that have been correctly trafficked to the plasma membrane can be stimulated using “CFTR potentiators” (ivacaftor), while “CFTR correctors” (lumacaftor, tezacaftor and elexacaftor) can act as molecular chaperones to support channel folding and correct delivery of the transporter to the plasma membrane. Read-through agents (ataluren and ELX-02) have been suggested for carriers of premature termination codons that account for up to 12% of pathogenic CF alleles. However, ataluren failed to show improvement in clinical outcomes in a phase III trial and further development was hence halted (Aslam et al. 2017). ELX-02 showed promising results in vitro and phase II trials are currently ongoing (Kerem 2020). Combined, these data indicate that around 80% of CF patients in European populations carry at least one allele that renders them susceptible to treatment with currently available CFTR potentiators and CFTR correctors (p.F508del, p.G551D, p.S549N and c.3717 + 12191C > T). In contrast, the fraction of patients with suitable genotypes is considerably lower in African (∼ 60%), Hispanic (∼ 55%), Asian (∼ 45%) and Ashkenazi Jewish individuals (∼ 40%).
G6PD
G6PD encodes glucose-6-phosphate dehydrogenase, a key enzyme in the pentose phosphate pathway that regulates NADPH levels, which is essential for redox homeostasis. Importantly, G6PD is highly polymorphic, and more than 200 variants have been shown to cause reduced G6PD activity (Beutler and Vulliamy 2002). While mostly asymptomatic, G6PD deficiency can be of importance upon exposure to certain triggers of oxidative stress, particularly in erythrocytes that lack mitochondria and are thus reliant on G6PD for the synthesis of redox equivalents. Triggers can be dietary components, such as fava beans or legumes, different bacterial or viral infections, or various chemically diverse drugs, such as primaquine, dapsone, sulfonamide antibiotics and rasburicase. Under these circumstances G6PD deficiency strongly increases the risk of sometimes life-threatening acute hemolytic anemia. Notably, G6PD is located on the X-chromosome and thus primarily impacts hemizygous males and homozygous females, whereas among heterozygous females only around 8–20% exhibit clinically relevant levels of reduced G6PD activity (Chu et al. 2018; Dechyotin et al. 2021; Johnson et al. 2009; Satyagraha et al. 2021).
G6PD deficiency is most common in Africa, followed by Southeast Asia and the Middle East (Koromina et al. 2021; Nkhoma et al. 2009). While overall disease prevalence might be similar between these regions, the genetic basis of G6PD deficiency differs drastically (Table 6). Of note, G6PD variant alleles are commonly referred to by their trivial names, which is a convention we will also follow in this review. In Sub-Saharan Africa, the A-202A/376G allele is most common with frequencies around 10% and local peaks up to 24%, followed by A-968C/376G with frequencies around 1% (Awandu et al. 2018; May et al. 2000; Pernaute-Lau et al. 2021). However, frequency profiles can be reversed in specific ethnogeographic groups, as demonstrated for West African populations in Senegal and Guinea where the A-968C/376G is predominant (approximately 7–11% for A-968C/376G vs. 1–3% for A-202A/376G) (De Araujo et al. 2006; Howes et al. 2013). Further West African alleles include the Sierra Leone (or A-311A/376G) variant, which however has not been extensively characterized with high geographic resolution (Jalloh et al. 2008). In contrast to Sub-Saharan Africa, the different A- alleles are very rare in East African populations (Assefa et al. 2018; Hamid et al. 2019). These results have potentially important implications for public health decisions, particularly for the treatment and prevention of malaria, as they support the roll out of primaquine, a drug associated with major anemia risk in G6PD deficient individuals, as radical cure for Plasmodium vivax and as transmission interruption for Plasmodium falciparum in East Africa, whereas G6PD genotyping before the initiation of 8-aminoquinolone therapy is warranted in South and West Africa. However, the status of other deficient variants beyond A- should be evaluated in East Africa to further corroborate this conclusion.
In Middle Eastern populations G6PD deficiency is primarily attributed to the Mediterranean allele (Doss et al. 2016), accounting, for instance, for 88% and 74% of G6PD deficiency among the Kurdish population in Northern Iraq and in Kuwaiti Arabs (MAF in the general population = 1–4%), respectively (Al-Allawi et al. 2010; Alfadhli et al. 2005). Further relevant G6PD deficient variants in the Middle East are A-968C/376G, Cairo and Chatham, with overall MAFs of 0.4–0.8%. The Mediterranean variant is furthermore common in Southcentral Asia with frequencies up to 8.9% in Afghani Pashtun (Jamornthanyawat et al. 2014). While it also constitutes a relevant factor in India, explaining around 24% of G6PD deficiencies in a country-wide survey, the overall most prevalent allele was Orissa, which accounted for 57% of all deficiencies (Devendra et al. 2020). Further rare variants of relevance in specific South Asian subpopulations and tribal groups are Kalyan–Kerala and Namoru (Chalvam et al. 2007). In Southeast Asia, the predominant allele is Mahidol, which explains 38–96% in of G6PD deficiencies in Burma, Thailand and Myanmar (Matsuoka et al. 2004; Phompradit et al. 2011). In contrast, G6PD deficiency in Cambodia was almost exclusively caused by the Viangchan allele (Matsuoka et al. 2005). Furthermore, specific subpopulations feature unique molecular G6PD patterns; for instance, the otherwise rare Aures allele constitutes the most common G6PD deficient variant in the Lao Theung population, the second largest ethnic group in Laos (Sanephonasa et al. 2021).
Compared to the variant profile in South and Southeast Asian populations, G6PD variability in China is distinctly different. In Han Chinese, Kaiping (MAF = 0.3%) and Canton (MAF = 0.3%) were the most common G6PD deficient alleles and showed a clear South-to-North national gradient (He et al. 2020). In other Chinese ethnic groups, such as Dai, Miao, Tibetans and Yi, variant signatures showed pronounced differences with the G6PD Gaohe, Baise, Fushan and Union alleles explaining > 10% of population-specific deficiencies (Zheng et al. 2020). In contrast to China where the country-wide prevalence of G6PD deficiency is around 1.9% among males, G6PD deficiency is a rare disorder in Japan with an overall frequency of < 0.1%. Notably, despite this low frequency, a multitude of distinct very rare Japanese deficient alleles have been described, including Fukushima, Morioka, Yamaguchi and Musashino. Combined, these results demonstrate the conspicuous differences in G6PD molecular genetics even across ethnic groups in close geographical proximity and indicate that it is essential to employ genotyping strategies that are tailored to the specific population or ethnic background of a given patient.
Opportunities for precision public health
Population pharmacogenomic profiling can reveal genetic differences that predispose to differences in drug response. In Europeans, reduced function alleles of CYP2D6 are considerably more frequent than in other populations. Thus, genetic testing is particularly beneficial in these populations for identifying outlier patients, such as CYP2D6 poor metabolizers. The respective information can be utilized clinically, e.g. for prescribing alternatives to tramadol and codeine analgesics for pain relief (Crews et al. 2021) and for recommending aromatase inhibitors, such as anastrozole instead of tamoxifen for the prevention of breast cancer recurrence (Goetz et al. 2018)). Furthermore, European populations harbour the highest frequencies of CFTR trafficking mutations, suggesting that the rate of cystic fibrosis patients responding to CFTR correctors is overall higher in Europe compared to other populations.
Reduced function variants of DPYD and TPMT are most prevalent in Sub-Saharan Africa and, thus, preemptive genetic testing and genotype-guided dose adjustments of fluoropyrimidines and thiopurines are particularly beneficial in those populations. Similarly, African populations have the highest frequencies of genetic G6PD deficiency, which constitutes a contraindication to treatment with the 8-aminoquinoline antimalarials primaquine and tafenoquine, the only curative treatments for Plasmodium vivax malaria, due to drastically elevated risk of severe acute haemolytic anaemia (Watson et al. 2018). Furthermore, G6PD deficiency status is useful to guide treatment with various other drugs, including pegloticase, rasburicase, flutamide, as well as sulfonamide antibiotics.
Southeast Asia constitutes the main hotspot of the HLA-B*15:02 and HLA-B*58:01 alleles, entailing that testing for carbamazepine and allopurinol induced severe cutaneous adverse reactions is most important in these groups. Notably, country-specific frequency information can refine pharmacogenomic decision making at the national level. For example, while HLA-B*15:02 is generally common in Asian populations with average minor allele frequencies of 5–10%, rates are much higher in the Philippines where about half of the population are carriers, whereas frequencies in Japan are < 0.1%. With increasing availability of genotype information, genetic differences between ethnic groups are revealed with higher and higher resolution and the resulting data shows that pronounced genetic differences can exist even across relatively small geographic regions. However, we want to emphasize that both high resolution studies with well-defined cohorts as well as population-scale aggregated information should be considered to allow for an integration of information about ethnogeographic differences with modern human migration and admixture.
Conclusions
Interindividual differences in drug response are in part caused by genetic variants with differential ethnogeographic prevalence and information about their distribution can be important for population-stratified therapy (Mette et al. 2012; Roberts et al. 2021; Yasuda et al. 2008). In this review, we provide a current update of population differences in the genetic variability of ten different genes that are included in the labels of 141 different drugs or therapeutic regimens as warnings or as factors impacting the clinical pharmacology of the agents in question (Supplementary Table 5). The aggregated data suggest strong differences in variant distribution and gene functionality between major ethnogeographic groups. We hope that the overview provided herein can serve as a useful resource for pharmacologists, clinical geneticists and public health researchers to evaluate treatment risks and inform population-adjusted genotyping strategies.
References
Adehin A, Bolaji OO, Kennedy MA, Adeagbo BA (2017) Allele frequencies of thiopurine S-methyltransferase (TPMT) variants in the Nigerian population. Pol Ann Med 24:144–147. https://doi.org/10.1016/j.poamed.2016.06.007
Agúndez JA, Ramirez R, Hernandez M, Llerena A, Benítez J (1997) Molecular heterogeneity at the CYP2D gene locus in Nicaraguans: impact of gene-flow from Europe. Pharmacogenetics 7:337–340. https://doi.org/10.1097/00008571-199708000-00010
Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M (1996) Frequent distribution of ultrarapid metabolizers of debrisoquine in an Ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther 278:441–446
Aklillu E, Herrlin K, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M (2002) Evidence for environmental influence on CYP2D6-catalysed debrisoquine hydroxylation as demonstrated by phenotyping and genotyping of Ethiopians living in Ethiopia or in Sweden. Pharmacogenetics 12:375–383. https://doi.org/10.1097/00008571-200207000-00005
Al-Allawi N, Eissa AA, Jubrael JM, Jamal SA, Hamamy H (2010) Prevalence and molecular characterization of Glucose-6-Phosphate dehydrogenase deficient variants among the Kurdish population of Northern Iraq. BMC Blood Disord 10:6. https://doi.org/10.1186/1471-2326-10-6
Albuquerque J, Ribeiro C, Naranjo MEG, Llerena A, Grazina M (2013) Characterization of CYP2D6 genotypes and metabolic profiles in the Portuguese population: pharmacogenetic implications. Per Med 10:709–718. https://doi.org/10.2217/pme.13.56
Alfadhli S, Kaaba S, Elshafey A, Salim M, AlAwadi A, Bastaki L (2005) Molecular characterization of glucose-6-phosphate dehydrogenase gene defect in the Kuwaiti population. Arch Pathol Lab Med 129:1144–1147. https://doi.org/10.5858/2005-129-1144-mcogdg
Ameyaw MM, Collie-Duguid ES, Powrie RH, Ofori-Adjei D, McLeod HL (1999) Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum Mol Genet 8:367–370. https://doi.org/10.1093/hmg/8.2.367
Amstutz U, Farese S, Aebi S, Largiadèr CR (2009) Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: a haplotype assessment. Pharmacogenomics 10:931–944. https://doi.org/10.2217/pgs.09.28
Antunes MV, Linden R, Santos TV, Wallemacq P, Haufroid V, Classen JF, Andreolla H, Costa N, Fontanive TO, Rosa DD (2012) Endoxifen levels and its association with CYP2D6 genotype and phenotype: evaluation of a southern Brazilian population under tamoxifen pharmacotherapy. Ther Drug Monit 34:422–431. https://doi.org/10.1097/FTD.0b013e318260b46e
Arimany-Nardi C, Koepsell H, Pastor-Anglada M (2015) Role of SLC22A1 polymorphic variants in drug disposition, therapeutic responses, and drug-drug interactions. Pharmacogenomics J 15:473–487. https://doi.org/10.1038/tpj.2015.78
Aslam AA, Higgins C, Sinha IP, Southern KW (2017) Ataluren and similar compounds (specific therapies for premature termination codon class I mutations) for cystic fibrosis. Cochrane Database Syst Rev 1:CD012040. https://doi.org/10.1002/14651858.CD012040.pub2
Assefa A, Ali A, Deressa W, Tsegaye W, Abebe G, Sime H, Kebede A, Jima D, Kassa M, Abreha T, Teka H, Solomon H, Malone J, Shi YP, Zhou Z, Reithinger R, Hwang J (2018) Glucose-6-phosphate dehydrogenase (G6PD) deficiency in Ethiopia: absence of common African and Mediterranean allelic variants in a nationwide study. Malar J 17:388. https://doi.org/10.1186/s12936-018-2538-4
Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526:68–74. https://doi.org/10.1038/nature15393
Awandu SS, Raman J, Makhanthisa TI, Kruger P, Frean J, Bousema T, Niemand J, Birkholtz LM (2018) Understanding human genetic factors influencing primaquine safety and efficacy to guide primaquine roll-out in a pre-elimination setting in southern Africa. Malar J 17:120. https://doi.org/10.1186/s12936-018-2271-z
Aydin M, Hatirnaz O, Erensoy N, Ozbek U (2005) CYP2D6 and CYP1A1 mutations in the Turkish population. Cell Biochem Funct 23:133–135. https://doi.org/10.1002/cbf.1222
Aynacioglu AS, Sachse C, Bozkurt A, Kortunay S, Nacak M, Schröder T, Kayaalp SO, Roots I, Brockmöller J (1999) Low frequency of defective alleles of cytochrome P450 enzymes 2C19 and 2D6 in the Turkish population. Clin Pharmacol Ther 66:185–192. https://doi.org/10.1053/cp.1999.v66.100072001
Bahari A, Hashemi M, Bari Z, Moazeni-Roodi A, Kaykhaei MA, Narouie B (2010) Frequency of thiopurine S-methyltransferase (TPMT) alleles in southeast Iranian population. Nucleosides Nucleotides Nucleic Acids 29:237–244. https://doi.org/10.1080/15257771003720418
Bannister WP, Friis-Møller N, Mocroft A, Viard JP, van Lunzen J, Kirk O, Gargalianos P, Bánhegyi D, Chiesi A, Lundgren JD (2008) Incidence of abacavir hypersensitivity reactions in euroSIDA. Antivir Ther 13:687–696
Beer B, Erb R, Pitterl F, Niederstätter H, Maroñas O, Gesteira A, Carracedo A, Piatkov I, Oberacher H (2011) CYP2D6 genotyping by liquid chromatography-electrospray ionization mass spectrometry. Anal Bioanal Chem 400:2361–2370. https://doi.org/10.1007/s00216-010-4597-4
Beutler E, Vulliamy TJ (2002) Hematologically important mutations: glucose-6-phosphate dehydrogenase. Blood Cells Mol Dis 28:93–103. https://doi.org/10.1006/bcmd.2002.0490
Bökkerink JP, Stet EH, De Abreu RA, Damen FJ, Hulscher TW, Bakker MA, van Baal JA (1993) 6-Mercaptopurine: cytotoxicity and biochemical pharmacology in human malignant T lymphoblasts. Biochem Pharmacol 45:1455–1463. https://doi.org/10.1016/0006-2952(93)90045-x
Carano F, Sarno S, De Fanti S, Serventi P, Bini C, Luiselli D, Pelotti S (2018) Genetic variability of CYP2D6, CYP2B6, CYP2C9 and CYP2C19 genes across the Italian Peninsula. Ann Hum Biol 45:66–71. https://doi.org/10.1080/03014460.2017.1378368
Caudle KE, Thorn CF, Klein TE, Swen JJ, McLeod HL, Diasio RB, Schwab M (2013) Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing. Clin Pharmacol Ther 94:640–645. https://doi.org/10.1038/clpt.2013.172
Céspedes-Garro C, Jiménez-Arce G, Naranjo ME, Barrantes R, Llerena A (2014) Ethnic background and CYP2D6 genetic polymorphisms in Costa Ricans. Rev Biol Trop 62:1659–1671
Chalvam R, Mukherjee MB, Colah RB, Mohanty D, Ghosh K (2007) G6PD Namoru (208 T–> C) is the major polymorphic variant in the tribal populations in southern India. Br J Haematol 136:512–513. https://doi.org/10.1111/j.1365-2141.2006.06463.x
Chang JG, Lee LS, Chen CM, Shih MC, Wu MC, Tsai FJ, Liang DC (2002) Molecular analysis of thiopurine S-methyltransferase alleles in South-east Asian populations. Pharmacogenetics 12:191–195. https://doi.org/10.1097/00008571-200204000-00003
Chao K, Wang X, Cao Q, Qian J, Wu K, Zhu X, Yang H, Liang J, Lin L, Huang Z, Zhang Y, Huang Y, Sun Y, Xue X, Huang M, Hu P, Lan P, Gao X (2017) Combined detection of NUDT15 variants could highly predict thiopurine-induced leukopenia in chinese patients with inflammatory bowel disease: a multicenter analysis. Inflamm Bowel Dis 23:1592–1599. https://doi.org/10.1097/mib.0000000000001148
Chen L, Takizawa M, Chen E, Schlessinger A, Segenthelar J, Choi JH, Sali A, Kubo M, Nakamura S, Iwamoto Y, Iwasaki N, Giacomini KM (2010) Genetic polymorphisms in organic cation transporter 1 (OCT1) in Chinese and Japanese populations exhibit altered function. J Pharmacol Exp Ther 335:42–50. https://doi.org/10.1124/jpet.110.170159
Chen L, Shu Y, Liang X, Chen EC, Yee SW, Zur AA, Li S, Xu L, Keshari KR, Lin MJ, Chien HC, Zhang Y, Morrissey KM, Liu J, Ostrem J, Younger NS, Kurhanewicz J, Shokat KM, Ashrafi K, Giacomini KM (2014) OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. Proc Natl Acad Sci U S A 111:9983–9988. https://doi.org/10.1073/pnas.1314939111
Cheong HS, Kim HD, Na HS, Kim JO, Kim LH, Kim SH, Bae JS, Chung MW, Shin HD (2011) Screening of genetic variations of SLC15A2, SLC22A1, SLC22A2 and SLC22A6 genes. J Hum Genet 56:666–670. https://doi.org/10.1038/jhg.2011.77
Chu CS, Bancone G, Nosten F, White NJ, Luzzatto L (2018) Primaquine-induced haemolysis in females heterozygous for G6PD deficiency. Malar J 17:101. https://doi.org/10.1186/s12936-018-2248-y
Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, Wu JY, Chen YT (2004) Medical genetics: a marker for Stevens–Johnson syndrome. Nature 428:486. https://doi.org/10.1038/428486a
Correia C, Santos P, Coutinho AM, Vicente AM (2009) Characterization of pharmacogenetically relevant CYP2D6 and ABCB1 gene polymorphisms in a Portuguese population sample. Cell Biochem Funct 27:251–255. https://doi.org/10.1002/cbf.1561
Crews KR, Monte AA, Huddart R, Caudle KE, Kharasch ED, Gaedigk A, Dunnenberger HM, Leeder JS, Callaghan JT, Samer CF, Klein TE, Haidar CE, van Driest SL, Ruano G, Sangkuhl K, Cavallari LH, Müller DJ, Prows CA, Nagy M, Somogyi AA, Skaar TC (2021) Clinical pharmacogenetics implementation consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin Pharmacol Ther 110:888–896. https://doi.org/10.1002/cpt.2149
Dandara C, Masimirembwa CM, Magimba A, Sayi J, Kaaya S, Sommers DK, Snyman JR, Hasler JA (2001) Genetic polymorphism of CYP2D6 and CYP2C19 in east- and southern African populations including psychiatric patients. Eur J Clin Pharmacol 57:11–17. https://doi.org/10.1007/s002280100282
De Araujo C, Migot-Nabias F, Guitard J, Pelleau S, Vulliamy T, Ducrocq R (2006) The role of the G6PD AEth376G/968C allele in glucose-6-phosphate dehydrogenase deficiency in the seerer population of Senegal. Haematologica 91:262–263
De Boeck K, Zolin A, Cuppens H, Olesen HV, Viviani L (2014) The relative frequency of CFTR mutation classes in European patients with cystic fibrosis. J Cyst Fibros 13:403–409. https://doi.org/10.1016/j.jcf.2013.12.003
Dechyotin S, Sakunthai K, Khemtonglang N, Yamsri S, Sanchaisuriya K, Kitcharoen K, Kitcharoen S (2021) Prevalence and molecular characterization of glucose-6-phosphate dehydrogenase (G6PD) deficiency in females from previously malaria endemic regions in Northeastern Thailand and identification of a novel G6PD variant. Mediterr J Hematol Infect Dis 13:e2021029. https://doi.org/10.4084/MJHID.2021.029
Devendra R, Gupta V, Shanmugam R, Singh M, Patel P, Valecha N, Mishra N, Ahmed N, Hoti SL, Hegde HV, Warang P, Chiddarwar A, Kedar P, Mayekar P, Mukherjee MB (2020) Prevalence and spectrum of mutations causing G6PD deficiency in Indian populations. Infect Genet Evol 86:104597. https://doi.org/10.1016/j.meegid.2020.104597
Dodgen TM, Labuschagne CJ, van Schalkwyk A, Steffens FE, Gaedigk A, Cromarty AD, Alessandrini M, Pepper MS (2016) Pharmacogenetic comparison of CYP2D6 predictive and measured phenotypes in a South African cohort. Pharmacogenom J 16:566–572. https://doi.org/10.1038/tpj.2015.76
Dorji PW, Tshering G, Na-Bangchang K (2019) CYP2C9, CYP2C19, CYP2D6 and CYP3A5 polymorphisms in south-east and east Asian populations: a systematic review. J Clin Pharm Ther 44:508–524. https://doi.org/10.1111/jcpt.12835
Doss CG, Alasmar DR, Bux RI, Sneha P, Bakhsh FD, Al-Azwani I, Bekay RE, Zayed H (2016) Genetic epidemiology of glucose-6-phosphate dehydrogenase deficiency in the arab world. Sci Rep 6:37284. https://doi.org/10.1038/srep37284
Drögemöller BI, Wright GE, Niehaus DJ, Koen L, Malan S, Da Silva DM, Hillermann-Rebello R, La Grange AM, Venter M, Warnich L (2010) Characterization of the genetic profile of CYP2C19 in two South African populations. Pharmacogenomics 11:1095–1103. https://doi.org/10.2217/pgs.10.90
Elraiyah T, Jerde CR, Shrestha S, Wu R, Nie Q, Giama NH, Sarangi V, Roberts LR, Offer SM, Diasio RB (2017) Novel deleterious dihydropyrimidine dehydrogenase variants may contribute to 5-fluorouracil sensitivity in an east African population. Clin Pharmacol Ther 101:382–390. https://doi.org/10.1002/cpt.531
Ezzeldin H, Diasio R (2004) Dihydropyrimidine dehydrogenase deficiency, a pharmacogenetic syndrome associated with potentially life-threatening toxicity following 5-fluorouracil administration. Clin Colorectal Cancer 4:181–189. https://doi.org/10.3816/ccc.2004.n.018
Ferreira GMA, Ribeiro Elias AB, Nascimento J, Monteiro WM, Melo GC, Baia-da-Silva DC, Lacerda MVG, Suarez-Kurtz G (2020) Pharmacogenomics of thiopurines: distribution of TPMT and NUDT15 polymorphisms in the Brazilian Amazon. Pharmacogenet Genomics 30:184–189. https://doi.org/10.1097/fpc.0000000000000411
Friedrich DC, Genro JP, Sortica VA, Suarez-Kurtz G, de Moraes ME, Pena SD, dos Santos AK, Romano-Silva MA, Hutz MH (2014) Distribution of CYP2D6 alleles and phenotypes in the Brazilian population. PLoS ONE 9:e110691. https://doi.org/10.1371/journal.pone.0110691
Fujikura K, Ingelman-Sundberg M, Lauschke VM (2015) Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet Genomics 25:584–594. https://doi.org/10.1097/fpc.0000000000000172
Fuselli S, Dupanloup I, Frigato E, Cruciani F, Scozzari R, Moral P, Sistonen J, Sajantila A, Barbujani G (2004) Molecular diversity at the CYP2D6 locus in the Mediterranean region. Eur J Hum Genet 12:916–924. https://doi.org/10.1038/sj.ejhg.5201243
Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS (2017) Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 19:69–76. https://doi.org/10.1038/gim.2016.80
Ganoci L, Božina T, Mirošević Skvrce N, Lovrić M, Mas P, Božina N (2017) Genetic polymorphisms of cytochrome P450 enzymes: CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5 in the Croatian population. Drug Metab Pers Ther 32:11–21. https://doi.org/10.1515/dmpt-2016-0024
Ganzcakowski M, Town M, Bowden DK, Vulliamy TJ, Kaneko A, Clegg JB, Weatherall DJ, Luzzatto L (1995) Multiple glucose 6-phosphate dehydrogenase-deficient variants correlate with malaria endemicity in the Vanuatu archipelago (southwestern Pacific). Am J Hum Genet 56:294–301
Gawrońska-Szklarz B, Adamiak-Giera U, Wyska E, Kurzawski M, Gornik W, Kaldonska M, Drozdzik M (2012) CYP2C19 polymorphism affects single-dose pharmacokinetics of oral pantoprazole in healthy volunteers. Eur J Clin Pharmacol 68:1267–1274. https://doi.org/10.1007/s00228-012-1252-3
Genin E, Chen DP, Hung SI, Sekula P, Schumacher M, Chang PY, Tsai SH, Wu TL, Bellón T, Tamouza R, Fortier C, Toubert A, Charron D, Hovnanian A, Wolkenstein P, Chung WH, Mockenhaupt M, Roujeau JC (2014) HLA-A*31:01 and different types of carbamazepine-induced severe cutaneous adverse reactions: an international study and meta-analysis. Pharmacogenom J 14:281–288. https://doi.org/10.1038/tpj.2013.40
Goetz MP, Sangkuhl K, Guchelaar H-J, Schwab M, Province M, Whirl-Carrillo M, Symmans WF, McLeod HL, Ratain MJ, Zembutsu H, Gaedigk A, van Schaik RH, Ingle JN, Caudle KE, Klein TE (2018) Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy. Clin Phamracol Ther 103:770–777. https://doi.org/10.1002/cpt.1007
Goldman A, Labrum R, Claustres M, Desgeorges M, Guittard C, Wallace A, Ramsay M (2001) The molecular basis of cystic fibrosis in South Africa. Clin Genet 59:37–41. https://doi.org/10.1034/j.1399-0004.2001.590106.x
Griese EU, Asante-Poku S, Ofori-Adjei D, Mikus G, Eichelbaum M (1999) Analysis of the CYP2D6 gene mutations and their consequences for enzyme function in a West African population. Pharmacogenetics 9:715–723
Hall D, Ybazeta G, Destro-Bisol G, Petzl-Erler ML, Di Rienzo A (1999) Variability at the uridine diphosphate glucuronosyltransferase 1A1 promoter in human populations and primates. Pharmacogenetics 9:591–599
Hamdy SI, Hiratsuka M, Narahara K, Endo N, El-Enany M, Moursi N, Ahmed MS, Mizugaki M (2003) Genotype and allele frequencies of TPMT, NAT2, GST, SULT1A1 and MDR-1 in the Egyptian population. Br J Clin Pharmacol 55:560–569. https://doi.org/10.1046/j.1365-2125.2003.01786.x
Hamid MMA, Albsheer M, Muneer M, Altinae L, Lover AA (2019) PO 8609 Prevalence and risk factors for glucose-6-phosphate dehydrogenase (G6PD) deficiency in two P. vivax malaria-endemic areas in Sudan. BMJ Glob Health 4:A62. https://doi.org/10.1136/bmjgh-2019-EDC.164
Hariprakash JM, Vellarikkal SK, Keechilat P, Verma A, Jayarajan R, Dixit V, Ravi R, Senthivel V, Kumar A, Sehgal P, Sonakar AK, Ambawat S, Giri AK, Philip A, Sivadas A, Faruq M, Bharadwaj D, Sivasubbu S, Scaria V (2018) Pharmacogenetic landscape of DPYD variants in south Asian populations by integration of genome-scale data. Pharmacogenomics 19:227–241. https://doi.org/10.2217/pgs-2017-0101
He Y, Zhang Y, Chen X, Wang Q, Ling L, Xu Y (2020) Glucose-6-phosphate dehydrogenase deficiency in the Han Chinese population: molecular characterization and genotype-phenotype association throughout an activity distribution. Sci Rep 10:17106. https://doi.org/10.1038/s41598-020-74200-y
Helsby NA (2016) CYP2C19 and CYP2D6 genotypes in Pacific peoples. Br J Clin Pharmacol 82:1303–1307. https://doi.org/10.1111/bcp.13045
Herraez E, Lozano E, Macias RI, Vaquero J, Bujanda L, Banales JM, Marin JJ, Briz O (2013) Expression of SLC22A1 variants may affect the response of hepatocellular carcinoma and cholangiocarcinoma to sorafenib. Hepatology 58:1065–1073. https://doi.org/10.1002/hep.26425
Hetherington S, McGuirk S, Powell G, Cutrell A, Naderer O, Spreen B, Lafon S, Pearce G, Steel H (2001) Hypersensitivity reactions during therapy with the nucleoside reverse transcriptase inhibitor abacavir. Clin Ther 23:1603–1614. https://doi.org/10.1016/s0149-2918(01)80132-6
Hiratsuka M, Inoue T, Omori F, Agatsuma Y, Mizugaki M (2000) Genetic analysis of thiopurine methyltransferase polymorphism in a Japanese population. Mutat Res 448:91–95. https://doi.org/10.1016/s0027-5107(00)00004-x
Hoff PM, Ansari R, Batist G, Cox J, Kocha W, Kuperminc M, Maroun J, Walde D, Weaver C, Harrison E, Burger HU, Osterwalder B, Wong AO, Wong R (2001) Comparison of oral capecitabine versus intravenous fluorouracil plus leucovorin as first-line treatment in 605 patients with metastatic colorectal cancer: results of a randomized phase III study. J Clin Oncol 19:2282–2292. https://doi.org/10.1200/jco.2001.19.8.2282
Hon YY, Fessing MY, Pui CH, Relling MV, Krynetski EY, Evans WE (1999) Polymorphism of the thiopurine S-methyltransferase gene in African–Americans. Hum Mol Genet 8:371–376. https://doi.org/10.1093/hmg/8.2.371
Howes RE, Dewi M, Piel FB, Monteiro WM, Battle KE, Messina JP, Sakuntabhai A, Satyagraha AW, Williams TN, Baird JK, Hay SI (2013) Spatial distribution of G6PD deficiency variants across malaria-endemic regions. Malar J 12:418. https://doi.org/10.1186/1475-2875-12-418
Hung SI, Chung WH, Liou LB, Chu CC, Lin M, Huang HP, Lin YL, Lan JL, Yang LC, Hong HS, Chen MJ, Lai PC, Wu MS, Chu CY, Wang KH, Chen CH, Fann CS, Wu JY, Chen YT (2005) HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci U S A 102:4134–4139. https://doi.org/10.1073/pnas.0409500102
Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, Miles JJ, Kjer-Nielsen L, Gras S, Williamson NA, Burrows SR, Purcell AW, Rossjohn J, McCluskey J (2012) Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486:554–558. https://doi.org/10.1038/nature11147
Inamochi H, Higashigawa M, Shimono Y, Nagata T, Cao DC, Mao XY, M’Soka T, Hori H, Kawasaki H, Sakurai M (1999) Delayed cytotoxicity of 6-mercaptopurine is compatible with mitotic death caused by DNA damage due to incorporation of 6-thioguanine into DNA as 6-thioguanine nucleotide. J Exp Clin Cancer Res 18:417–424
Ingelman-Sundberg M (2005) Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenom J 5:6–13. https://doi.org/10.1038/sj.tpj.6500285
Ionova Y, Ashenhurst J, Zhan J, Nhan H, Kosinski C, Tamraz B, Chubb A (2020) CYP2C19 allele frequencies in over 2.2 million direct-to-consumer genetics research participants and the potential implication for prescriptions in a large health system. Clin Transl Sci 13:1298–1306. https://doi.org/10.1111/cts.12830
Isaza CA, Henao J, López AM, Cacabelos R (2000) Isolation, sequence and genotyping of the drug metabolizer CYP2D6 gene in the Colombian population. Methods Find Exp Clin Pharmacol 22:695–705. https://doi.org/10.1358/mf.2000.22.9.802286
Isaza C, Henao J, López AM, Cacabelos R (2003) Allelic variants of the thiopurine methyltransferase (TPMT) gene in the Colombian population. Methods Find Exp Clin Pharmacol 25:423–429. https://doi.org/10.1358/mf.2003.25.6.769646
Jalloh A, Jalloh M, Gamanga I, Baion D, Sahr F, Gbakima A, Willoughby VR, Matsuoka H (2008) G6PD deficiency assessment in Freetown, Sierra Leone, reveals further insight into the molecular heterogeneity of G6PD A. J Hum Genet 53:675–679. https://doi.org/10.1007/s10038-008-0294-y
Jamornthanyawat N, Awab GR, Tanomsing N, Pukrittayakamee S, Yamin F, Dondorp AM, Day NP, White NJ, Woodrow CJ, Imwong M (2014) A population survey of the glucose-6-phosphate dehydrogenase (G6PD) 563C>T (Mediterranean) mutation in Afghanistan. PLoS ONE 9:e88605. https://doi.org/10.1371/journal.pone.0088605
Jarrar YB, Ghishan M (2019) The nudix hydrolase 15 (nudt15) gene variants among Jordanian Arab population. Asian Pac J Cancer Prev 20:801–808. https://doi.org/10.31557/apjcp.2019.20.3.801
Johnson MK, Clark TD, Njama-Meya D, Rosenthal PJ, Parikh S (2009) Impact of the method of G6PD deficiency assessment on genetic association studies of malaria susceptibility. PLoS ONE 4:e7246. https://doi.org/10.1371/journal.pone.0007246
Jorge LF, Eichelbaum M, Griese EU, Inaba T, Arias TD (1999) Comparative evolutionary pharmacogenetics of CYP2D6 in Ngawbe and Embera Amerindians of Panama and Colombia: role of selection versus drift in world populations. Pharmacogenetics 9:217–228
Kaneko A, Kaneko O, Taleo G, Björkman A, Kobayakawa T (1997) High frequencies of CYP2C19 mutations and poor metabolism of proguanil in Vanuatu. Lancet 349:921–922. https://doi.org/10.1016/s0140-6736(05)62696-7
Kaneko A, Lum JK, Yaviong L, Takahashi N, Ishizaki T, Bertilsson L, Kobayakawa T, Björkman A (1999) High and variable frequencies of CYP2C19 mutations: medical consequences of poor drug metabolism in Vanuatu and other Pacific islands. Pharmacogenetics 9:581–590
Karim H, Ghalali A, Lafolie P, Vitols S, Fotoohi AK (2013) Differential role of thiopurine methyltransferase in the cytotoxic effects of 6-mercaptopurine and 6-thioguanine on human leukemia cells. Biochem Biophys Res Commun 437:280–286. https://doi.org/10.1016/j.bbrc.2013.06.067
Karimi M, Martinez di Montemuros F, Danielli MG, Farjadian S, Afrasiabi A, Fiorelli G, Cappellini MD (2003) Molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Fars province of Iran. Haematologica 88:346–347
Kerem E (2020) ELX-02: an investigational read-through agent for the treatment of nonsense mutation-related genetic disease. Expert Opin Investig Drugs 29:1347–1354. https://doi.org/10.1080/13543784.2020.1828862
Khalaj Z, Baratieh Z, Nikpour P, Khanahmad H, Mokarian F, Salehi R, Salehi M (2019) Distribution of CYP2D6 polymorphism in the Middle Eastern region. J Res Med Sci 24:61. https://doi.org/10.4103/jrms.JRMS_1076_18
Kham SK, Tan PL, Tay AH, Heng CK, Yeoh AE, Quah TC (2002) Thiopurine methyltransferase polymorphisms in a multiracial Asian population and children with acute lymphoblastic leukemia. J Pediatr Hematol Oncol 24:353–359. https://doi.org/10.1097/00043426-200206000-00006
Kim HT, Choi R, Won HH, Choe YH, Kang B, Lee K, Koo HH, Yoo KH, Kim YH, Lee SY (2017) NUDT15 genotype distributions in the Korean population. Pharmacogenet Genomics 27:197–200. https://doi.org/10.1097/fpc.0000000000000274
Kohlrausch FB, Gama CS, Lobato MI, Belmonte-de-Abreu P, Gesteira A, Barros F, Carracedo A, Hutz MH (2009) Molecular diversity at the CYP2D6 locus in healthy and schizophrenic southern Brazilians. Pharmacogenomics 10:1457–1466. https://doi.org/10.2217/pgs.09.76
Koopmans AB, Braakman MH, Vinkers DJ, Hoek HW, van Harten PN (2021) Meta-analysis of probability estimates of worldwide variation of CYP2D6 and CYP2C19. Transl Psychiatry 11:141. https://doi.org/10.1038/s41398-020-01129-1
Koromina M, Pandi MT, van der Spek PJ, Patrinos GP, Lauschke VM (2021) The ethnogeographic variability of genetic factors underlying G6PD deficiency. Pharmacol Res 173:105904. https://doi.org/10.1016/j.phrs.2021.105904
Kounelis F, Kanterakis A, Kanavos A, Pandi MT, Kordou Z, Manusama O, Vonitsanos G, Katsila T, Tsermpini EE, Lauschke VM, Koromina M, van der Spek PJ, Patrinos GP (2020) Documentation of clinically relevant genomic biomarker allele frequencies in the next-generation FINDbase worldwide database. Hum Mutat 41:1112–1122. https://doi.org/10.1002/humu.24018
Ladić A, Božina N, Borzan V, Brinar M, Vucelić B, Cuković-Cavka S (2016) An epidemiological study of thiopurine-methyltransferase variants in a Croatian inflammatory bowel disease patient cohort. Acta Clin Croat 55:16–22. https://doi.org/10.20471/ACC.2016.55.01.3
Langmia IM, Just KS, Yamoune S, Brockmöller J, Masimirembwa C, Stingl JC (2021) CYP2B6 functional variability in drug metabolism and exposure across populations-implication for drug safety, dosing, and individualized therapy. Front Gen 12:692234. https://doi.org/10.3389/fgene.2021.692234
Laróvere LE, de Kremer RD, Lambooy LH, De Abreu RA (2003) Genetic polymorphism of thiopurine S-methyltransferase in Argentina. Ann Clin Biochem 40:388–393. https://doi.org/10.1258/000456303766477039
Lauschke VM, Ingelman-Sundberg M (2016) Requirements for comprehensive pharmacogenetic genotyping platforms. Pharmacogenomics 17:917–924. https://doi.org/10.2217/pgs-2016-0023
Lauschke VM, Milani L, Ingelman-Sundberg M (2017) Pharmacogenomic biomarkers for improved drug therapy-recent progress and future developments. Aaps j 20:4. https://doi.org/10.1208/s12248-017-0161-x
Lauschke VM, Zhou Y, Ingelman-Sundberg M (2019) Novel genetic and epigenetic factors of importance for inter-individual differences in drug disposition, response and toxicity. Pharmacol Ther 197:122–152. https://doi.org/10.1016/j.pharmthera.2019.01.002
Lazarou J, Pomeranz BH, Corey PN (1998) Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 279:1200–1205. https://doi.org/10.1001/jama.279.15.1200
Lee SS, Kim WY, Jang YJ, Shin JG (2008) Duplex pyrosequencing of the TPMT*3C and TPMT*6 alleles in Korean and Vietnamese populations. Clin Chim Acta 398:82–85. https://doi.org/10.1016/j.cca.2008.08.014
Llerena A, Dorado P, Ramírez R, González I, Alvarez M, Peñas-Lledó EM, Pérez B, Calzadilla LR (2012) CYP2D6 genotype and debrisoquine hydroxylation phenotype in Cubans and Nicaraguans. Pharmacogenom J 12:176–183. https://doi.org/10.1038/tpj.2010.85
Loennechen T, Utsi E, Hartz I, Lysaa R, Kildalsen H, Aarbakke J (2001) Detection of one single mutation predicts thiopurine S-methyltransferase activity in a population of Saami in northern Norway. Clin Pharmacol Ther 70:183–188. https://doi.org/10.1067/mcp.2001.117445
Lonjou C, Borot N, Sekula P, Ledger N, Thomas L, Halevy S, Naldi L, Bouwes-Bavinck JN, Sidoroff A, de Toma C, Schumacher M, Roujeau JC, Hovnanian A, Mockenhaupt M (2008) A European study of HLA-B in Stevens–Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet Genom 18:99–107. https://doi.org/10.1097/FPC.0b013e3282f3ef9c
Luo HR, Aloumanis V, Lin KM, Gurwitz D, Wan YJ (2004) Polymorphisms of CYP2C19 and CYP2D6 in Israeli ethnic groups. Am J Pharmacogenom 4:395–401. https://doi.org/10.2165/00129785-200404060-00006
Macek M Jr, Mackova A, Hamosh A, Hilman BC, Selden RF, Lucotte G, Friedman KJ, Knowles MR, Rosenstein BJ, Cutting GR (1997) Identification of common cystic fibrosis mutations in African–Americans with cystic fibrosis increases the detection rate to 75%. Am J Hum Genet 60:1122–1127
Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, Jägel-Guedes E, Rugina S, Kozyrev O, Cid JF, Hay P, Nolan D, Hughes S, Hughes A, Ryan S, Fitch N, Thorborn D, Benbow A (2008) HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 358:568–579. https://doi.org/10.1056/NEJMoa0706135
Masimirembwa C, Persson I, Bertilsson L, Hasler J, Ingelman-Sundberg M (1996) A novel mutant variant of the CYP2D6 gene (CYP2D6*17) common in a black African population: association with diminished debrisoquine hydroxylase activity. Br J Clin Pharmacol 42:713–719. https://doi.org/10.1046/j.1365-2125.1996.00489.x
Matsuoka H, Wang J, Hirai M, Arai M, Yoshida S, Kobayashi T, Jalloh A, Lin K, Kawamoto F (2004) Glucose-6-phosphate dehydrogenase (G6PD) mutations in Myanmar: G6PD Mahidol (487G>A) is the most common variant in the Myanmar population. J Hum Genet 49:544–547. https://doi.org/10.1007/s10038-004-0187-7
Matsuoka H, Nguon C, Kanbe T, Jalloh A, Sato H, Yoshida S, Hirai M, Arai M, Socheat D, Kawamoto F (2005) Glucose-6-phosphate dehydrogenase (G6PD) mutations in Cambodia: G6PD Viangchan (871G>A) is the most common variant in the Cambodian population. J Hum Genet 50:468–472. https://doi.org/10.1007/s10038-005-0279-z
May J, Meyer CG, Grossterlinden L, Ademowo OG, Mockenhaupt FP, Olumese PE, Falusi AG, Luzzatto L, Bienzle U (2000) Red cell glucose-6-phosphate dehydrogenase status and pyruvate kinase activity in a Nigerian population. Trop Med Int Health 5:119–123. https://doi.org/10.1046/j.1365-3156.2000.00529.x
McLellan RA, Oscarson M, Seidegård J, Evans DA, Ingelman-Sundberg M (1997) Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics 7:187–191. https://doi.org/10.1097/00008571-199706000-00003
McLeod HL, Pritchard SC, Githang’a J, Indalo A, Ameyaw MM, Powrie RH, Booth L, Collie-Duguid ES (1999) Ethnic differences in thiopurine methyltransferase pharmacogenetics: evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics 9:773–776. https://doi.org/10.1097/00008571-199912000-00012
Melaouhia S, Fékih M, Garat A, Allorge D, Ferchichi H, Klouz A, Boubaker J, Broly F, Lakhal M (2012) Allele frequency of inosine triphosphate pyrophosphatase (ITPA) and thiopurine-S-methyl transferase (TPMT) genes in the Tunisian population. Clin Res Hepatol Gastroenterol 36:178–184. https://doi.org/10.1016/j.clinre.2011.12.001
Mesbah-Namin SA, Sanati MH, Mowjoodi A, Mason PJ, Vulliamy TJ, Noori-Saloii MR (2002) Three major glucose-6-phosphate dehydrogenase-deficient polymorphic variants identified in Mazandaran state of Iran. Br J Haematol 117:763–764. https://doi.org/10.1046/j.1365-2141.2002.03483.x
Mette L, Mitropoulos K, Vozikis A, Patrinos GP (2012) Pharmacogenomics and public health: implementing “populationalized” medicine. Pharmacogenomics 13:803–813. https://doi.org/10.2217/pgs.12.52
Mizzi C, Dalabira E, Kumuthini J, Dzimiri N, Balogh I, Başak N, Böhm R, Borg J, Borgiani P, Bozina N, Bruckmueller H, Burzynska B, Carracedo A, Cascorbi I, Deltas C, Dolzan V, Fenech A, Grech G, Kasiulevicius V, Kádaši Ľ, Kučinskas V, Khusnutdinova E, Loukas YL, Macek M Jr, Makukh H, Mathijssen R, Mitropoulos K, Mitropoulou C, Novelli G, Papantoni I, Pavlovic S, Saglio G, Setric J, Stojiljkovic M, Stubbs AP, Squassina A, Torres M, Turnovec M, van Schaik RH, Voskarides K, Wakil SM, Werk A, Del Zompo M, Zukic B, Katsila T, Lee MT, Motsinger-Rief A, Mc Leod HL, van der Spek PJ, Patrinos GP (2016) A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS ONE 11:e0162866. https://doi.org/10.1371/journal.pone.0162866
Molden E, Johansen PW, Bøe GH, Bergan S, Christensen H, Rugstad HE, Rootwelt H, Reubsaet L, Lehne G (2002) Pharmacokinetics of diltiazem and its metabolites in relation to CYP2D6 genotype. Clin Pharmacol Ther 72:333–342. https://doi.org/10.1067/mcp.2002.127396
Montané Jaime LK, Lalla A, Steimer W, Gaedigk A (2013) Characterization of the CYP2D6 gene locus and metabolic activity in Indo- and Afro-Trinidadians: discovery of novel allelic variants. Pharmacogenomics 14:261–276. https://doi.org/10.2217/pgs.12.207
Moriyama T, Nishii R, Perez-Andreu V, Yang W, Klussmann FA, Zhao X, Lin TN, Hoshitsuki K, Nersting J, Kihira K, Hofmann U, Komada Y, Kato M, McCorkle R, Li L, Koh K, Najera CR, Kham SK, Isobe T, Chen Z, Chiew EK, Bhojwani D, Jeffries C, Lu Y, Schwab M, Inaba H, Pui CH, Relling MV, Manabe A, Hori H, Schmiegelow K, Yeoh AE, Evans WE, Yang JJ (2016) NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet 48:367–373. https://doi.org/10.1038/ng.3508
Nkhoma ET, Poole C, Vannappagari V, Hall SA, Beutler E (2009) The global prevalence of glucose-6-phosphate dehydrogenase deficiency: a systematic review and meta-analysis. Blood Cells Mol Dis 42:267–278. https://doi.org/10.1016/j.bcmd.2008.12.005
Offer SM, Lee AM, Mattison LK, Fossum C, Wegner NJ, Diasio RB (2013) A DPYD variant (Y186C) in individuals of African ancestry is associated with reduced DPD enzyme activity. Clin Pharmacol Ther 94:158–166. https://doi.org/10.1038/clpt.2013.69
Ostrov DA, Grant BJ, Pompeu YA, Sidney J, Harndahl M, Southwood S, Oseroff C, Lu S, Jakoncic J, de Oliveira CA, Yang L, Mei H, Shi L, Shabanowitz J, English AM, Wriston A, Lucas A, Phillips E, Mallal S, Grey HM, Sette A, Hunt DF, Buus S, Peters B (2012) Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc Natl Acad Sci U S A 109:9959–9964. https://doi.org/10.1073/pnas.1207934109
Owusu SK, Morrow BM, White D, Klugman S, Vanker A, Gray D, Zampoli M (2020) Cystic fibrosis in black African children in South Africa: a case control study. J Cyst Fibros 19:540–545. https://doi.org/10.1016/j.jcf.2019.09.007
Pavlos R, Mallal S, Ostrov D, Buus S, Metushi I, Peters B, Phillips E (2015) T cell-mediated hypersensitivity reactions to drugs. Annu Rev Med 66:439–454. https://doi.org/10.1146/annurev-med-050913-022745
Pedersen RS, Damkier P, Brosen K (2005) Tramadol as a new probe for cytochrome P450 2D6 phenotyping: a population study. Clin Pharmacol Ther 77:458–467. https://doi.org/10.1016/j.clpt.2005.01.014
Pernaute-Lau L, Adegnika AA, Zhou Y, Zinsou JF, Gil JP, Krishna S, Kremsner PG, Lauschke VM, Velavan TP (2021) Pharmacogene sequencing of a gabonese population with severe plasmodium falciparum malaria reveals multiple novel variants with putative relevance for antimalarial treatment. Antimicrob Agents Chemother 65:e0027521. https://doi.org/10.1128/aac.00275-21
Petrović J, Pešić V, Lauschke VM (2020) Frequencies of clinically important CYP2C19 and CYP2D6 alleles are graded across Europe. Eur J Hum Genet 28:88–94. https://doi.org/10.1038/s41431-019-0480-8
Phillips EJ (2016) Classifying ADRs–does dose matter? Br J Clin Pharmacol 81:10–12. https://doi.org/10.1111/bcp.12749
Phompradit P, Kuesap J, Chaijaroenkul W, Rueangweerayut R, Hongkaew Y, Yamnuan R, Na-Bangchang K (2011) Prevalence and distribution of glucose-6-phosphate dehydrogenase (G6PD) variants in Thai and Burmese populations in malaria endemic areas of Thailand. Malar J 10:368. https://doi.org/10.1186/1475-2875-10-368
Pimenoff VN, Laval G, Comas D, Palo JU, Gut I, Cann H, Excoffier L, Sajantila A (2012) Similarity in recombination rate and linkage disequilibrium at CYP2C and CYP2D cytochrome P450 gene regions among Europeans indicates signs of selection and no advantage of using tagSNPs in population isolates. Pharmacogenet Genomics 22:846–857. https://doi.org/10.1097/FPC.0b013e32835a3a6d
Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, Farrar K, Park BK, Breckenridge AM (2004) Adverse drug reactions as cause of admission to hospital: prospective analysis of 18,820 patients. BMJ 329:15–19. https://doi.org/10.1136/bmj.329.7456.15
Poppe D, Tiede I, Fritz G, Becker C, Bartsch B, Wirtz S, Strand D, Tanaka S, Galle PR, Bustelo XR, Neurath MF (2006) Azathioprine suppresses ezrin-radixin-moesin-dependent T cell-APC conjugation through inhibition of Vav guanosine exchange activity on Rac proteins. J Immunol 176:640–651. https://doi.org/10.4049/jimmunol.176.1.640
Poulussen FCP, Peters BJ, Hua KH, Houthuizen P, Grouls RJ, Deenen MJ (2019) The effect of the CYP2D6 genotype on the maintenance dose of metoprolol in a chronic Dutch patient population. Pharmacogenet Genomics 29:179–182. https://doi.org/10.1097/fpc.0000000000000381
Qin S, Shen L, Zhang A, Xie J, Shen W, Chen L, Tang J, Xiong Y, Yang L, Shi Y, Feng G, He L, Xing Q (2008) Systematic polymorphism analysis of the CYP2D6 gene in four different geographical Han populations in mainland China. Genomics 92:152–158. https://doi.org/10.1016/j.ygeno.2008.05.004
Qumsieh RY, Ali BR, Abdulrazzaq YM, Osman O, Akawi NA, Bastaki SM (2011) Identification of new alleles and the determination of alleles and genotypes frequencies at the CYP2D6 gene in Emiratis. PLoS ONE 6:e28943. https://doi.org/10.1371/journal.pone.0028943
Raida M, Schwabe W, Häusler P, Van Kuilenburg AB, Van Gennip AH, Behnke D, Höffken K (2001) Prevalence of a common point mutation in the dihydropyrimidine dehydrogenase (DPD) gene within the 5’-splice donor site of intron 14 in patients with severe 5-fluorouracil (5-FU)- related toxicity compared with controls. Clin Cancer Res 7:2832–2839
Rasmussen JO, Christensen M, Svendsen JM, Skausig O, Hansen EL, Nielsen KA (2006) CYP2D6 gene test in psychiatric patients and healthy volunteers. Scand J Clin Lab Investig 66:129–136. https://doi.org/10.1080/00365510500469702
Rehling D, Zhang SM, Jemth AS, Koolmeister T, Throup A, Wallner O, Scaletti E, Moriyama T, Nishii R, Davies J, Desroses M, Rudd SG, Scobie M, Homan E, Berglund UW, Yang JJ, Helleday T, Stenmark P (2021) Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity. J Biol Chem 296:100568. https://doi.org/10.1016/j.jbc.2021.100568
Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE, Klein TE (2011) Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther 89:387–391. https://doi.org/10.1038/clpt.2010.320
Roberts MC, Fohner AE, Landry L, Olstad DL, Smit AK, Turbitt E, Allen CG (2021) Advancing precision public health using human genomics: examples from the field and future research opportunities. Genome Med 13:97. https://doi.org/10.1186/s13073-021-00911-0
Rodrigues JCG, Souza TP, Pastana LF, Ribeiro Dos Santos AM, Fernandes MR, Pinto P, Wanderley AV, Souza SJ, Kroll JE, Pereira AL, Magalhães L, Mercês LRD, Vidal AF, Vinasco-Sandoval T, Cavalcante GC, Guerreiro JF, Assumpção PP, Ribeiro-Dos-Santos Â, Santos S, Santos N (2020) Identification of NUDT15 gene variants in Amazonian Amerindians and admixed individuals from northern Brazil. PLoS ONE 15:e0231651. https://doi.org/10.1371/journal.pone.0231651
Rossino R, Vincis C, Alves S, Prata MJ, Macis MD, Nucaro AL, Schirru E, Congia M (2006) Frequency of the thiopurine S-methyltransferase alleles in the ancient genetic population isolate of Sardinia. J Clin Pharm Ther 31:283–287. https://doi.org/10.1111/j.1365-2710.2006.00736.x
Russell LE, Zhou Y, Almousa AA, Sodhi JK, Nwabufo CK, Lauschke VM (2021) Pharmacogenomics in the era of next generation sequencing—from byte to bedside. Drug Metab Rev 53:253–278. https://doi.org/10.1080/03602532.2021.1909613
Sabbagh A, Darlu P, Crouau-Roy B, Poloni ES (2011) Arylamine N-acetyltransferase 2 (NAT2) genetic diversity and traditional subsistence: a worldwide population survey. PLoS ONE 6:e18507. https://doi.org/10.1371/journal.pone.0018507
Salgueiro N, Veiga I, Fragoso M, Sousa O, Costa N, Pellon ML, Sanches E, dos Santos JG, Teixeira MR, Castedo S (2004) Mutations in exon 14 of dihydropyrimidine dehydrogenase and 5-Fluorouracil toxicity in Portuguese colorectal cancer patients. Genet Med 6:102–107. https://doi.org/10.1097/01.gim.0000118061.66602.a5
Salvà Lacombe P, García Vicente JA, Costa Pagès J, Lucio Morselli P (1996) Causes and problems of nonresponse or poor response to drugs. Drugs 51:552–570. https://doi.org/10.2165/00003495-199651040-00004
Sanephonasa A, Cheepsunthorn CL, Khaminsou N, Savongsy O, Nuchprayoon I, Leecharoenkiat K (2021) Molecular characterization of G6PD mutations reveals the high frequency of G6PD Aures in the Lao Theung population. Malar J 20:30. https://doi.org/10.1186/s12936-020-03560-7
SathupakS LK, Kampuansai J (2021) Prevalence and molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Lue ethnic group of northern Thailand. Sci Rep 11:2956. https://doi.org/10.1038/s41598-021-82477-w
Satyagraha AW, Sadhewa A, Baramuli V, Elvira R, Ridenour C, Elyazar I, Noviyanti R, Coutrier FN, Harahap AR, Baird JK (2015) PLoS Negl Trop Dis 9:e0003602. https://doi.org/10.1371/journal.pntd.0003602
Satyagraha AW, Sadhewa A, Panggalo LV, Subekti D, Elyazar I, Soebianto S, Mahpud N, Harahap AR, Baird JK (2021) Genotypes and phenotypes of G6PD deficiency among Indonesian females across diagnostic thresholds of G6PD activity guiding safe primaquine therapy of latent malaria. PLoS Negl Trop Dis 15:e0009610. https://doi.org/10.1371/journal.pntd.0009610
Schaeffeler E, Fischer C, Brockmeier D, Wernet D, Moerike K, Eichelbaum M, Zanger UM, Schwab M (2004) Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German–Caucasians and identification of novel TPMT variants. Pharmacogenetics 14:407–417. https://doi.org/10.1097/01.fpc.0000114745.08559.db
Schaeffeler E, Jaeger SU, Klumpp V, Yang JJ, Igel S, Hinze L, Stanulla M, Schwab M (2019) Impact of NUDT15 genetics on severe thiopurine-related hematotoxicity in patients with European ancestry. Genet Med 21:2145–2150. https://doi.org/10.1038/s41436-019-0448-7
Schaller L, Lauschke VM (2019) The genetic landscape of the human solute carrier (SLC) transporter superfamily. Hum Genet 138:1359–1377. https://doi.org/10.1007/s00439-019-02081-x
Schrijver I, Pique L, Graham S, Pearl M, Cherry A, Kharrazi M (2016) The spectrum of CFTR variants in nonwhite cystic fibrosis patients: implications for molecular diagnostic testing. J Mol Diagn 18:39–50. https://doi.org/10.1016/j.jmoldx.2015.07.005
Schulz JT, Sheridan RL, Ryan CM, MacKool B, Tompkins RG (2000) A 10-year experience with toxic epidermal necrolysis. J Burn Care Rehabil 21:199–204. https://doi.org/10.1097/00004630-200021030-00004
Scott SA, Edelmann L, Kornreich R, Erazo M, Desnick RJ (2007) CYP2C9, CYP2C19 and CYP2D6 allele frequencies in the Ashkenazi Jewish population. Pharmacogenomics 8:721–730. https://doi.org/10.2217/14622416.8.7.721
Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR (2013) Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94:317–323. https://doi.org/10.1038/clpt.2013.105
Seitz T, Stalmann R, Dalila N, Chen J, Pojar S, Dos Santos Pereira JN, Krätzner R, Brockmöller J, Tzvetkov MV (2015) Global genetic analyses reveal strong inter-ethnic variability in the loss of activity of the organic cation transporter OCT1. Genome Med 7:56. https://doi.org/10.1186/s13073-015-0172-0
Serin A, Canan H, Alper B, Gulmen M (2012) The frequencies of mutated alleles of CYP2D6 gene in a Turkish population. Forensic Sci Int 222:332–334. https://doi.org/10.1016/j.forsciint.2012.07.012
Shah SAV, Paradkar MU, Desai DC, Ashavaid TF (2018) Preemptive NUDT15 genotyping: redefining the management of patients with thiopurine-induced toxicity. Drug Metab Pers Ther 33:57–60. https://doi.org/10.1515/dmpt-2017-0038
Shekhani R, Steinacher L, Swen JJ, Ingelman-Sundberg M (2020) Evaluation of current regulation and guidelines of pharmacogenomic drug labels: opportunities for improvements. Clin Pharmacol Ther 107:1240–1255. https://doi.org/10.1002/cpt.1720
Sipeky C, Weber A, Szabo M, Melegh BI, Janicsek I, Tarlos G, Szabo I, Sumegi K, Melegh B (2013) High prevalence of CYP2C19*2 allele in Roma samples: study on Roma and Hungarian population samples with review of the literature. Mol Biol Rep 40:4727–4735. https://doi.org/10.1007/s11033-013-2569-4
Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S (2007) CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genom 17:93–101. https://doi.org/10.1097/01.fpc.0000239974.69464.f2
Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A (2009) Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genom 19:170–179. https://doi.org/10.1097/FPC.0b013e32831ebb30
Suarez-Kurtz G, Brisson GD, Hutz MH, Petzl-Erler ML, Salzano FM (2019) NUDT15 polymorphism in native American populations of Brazil. Clin Pharmacol Ther 105:1321–1322. https://doi.org/10.1002/cpt.1379
Sulzyc-Bielicka V, Bińczak-Kuleta A, Pioch W, Kładny J, Gziut K, Bielicki D, Ciechanowicz A (2008) 5-Fluorouracil toxicity-attributable IVS14 + 1G > A mutation of the dihydropyrimidine dehydrogenase gene in Polish colorectal cancer patients. Pharmacol Rep 60:238–242
Tanaka Y, Kato M, Hasegawa D, Urayama KY, Nakadate H, Kondoh K, Nakamura K, Koh K, Komiyama T, Manabe A (2015) Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br J Haematol 171:109–115. https://doi.org/10.1111/bjh.13518
Teh LK, Ismail R, Yusoff R, Hussein A, Isa MN, Rahman AR (2001) Heterogeneity of the CYP2D6 gene among Malays in Malaysia. J Clin Pharm Ther 26:205–211. https://doi.org/10.1046/j.1365-2710.2001.00347.x
Toft N, Nygaard U, Gregers J, Schmiegelow K (2006) Genetic analyses of thiopurine methyltransferase polymorphisms in Greenlandic and Danish populations. Acta Paediatr 95:1665–1667. https://doi.org/10.1080/08035250600743788
Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, Norquist G, Howland RH, Lebowitz B, McGrath PJ, Shores-Wilson K, Biggs MM, Balasubramani GK, Fava M (2006) Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 163:28–40. https://doi.org/10.1176/appi.ajp.163.1.28
Tzvetkov MV, Saadatmand AR, Lötsch J, Tegeder I, Stingl JC, Brockmöller J (2011) Genetically polymorphic OCT1: another piece in the puzzle of the variable pharmacokinetics and pharmacodynamics of the opioidergic drug tramadol. Clin Pharmacol Ther 90:143–150. https://doi.org/10.1038/clpt.2011.56
Tzvetkov MV, dos Santos Pereira JN, Meineke I, Saadatmand AR, Stingl JC, Brockmöller J (2013) Morphine is a substrate of the organic cation transporter OCT1 and polymorphisms in OCT1 gene affect morphine pharmacokinetics after codeine administration. Biochem Pharmacol 86:666–678. https://doi.org/10.1016/j.bcp.2013.06.019
Uetrecht J, Naisbitt DJ (2013) Idiosyncratic adverse drug reactions: current concepts. Pharmacol Rev 65:779–808. https://doi.org/10.1124/pr.113.007450
Uzunkoy A, Dilmec F, Ozgonul A, van Kuilenburg AB, Akkafa F (2007) Investigation of IVS14+ 1G > A polymorphism of DPYD gene in a group of Turkish patients with colorectal cancer. Anticancer Res 27:3899–3902
Van Cutsem E, Twelves C, Cassidy J, Allman D, Bajetta E, Boyer M, Bugat R, Findlay M, Frings S, Jahn M, McKendrick J, Osterwalder B, Perez-Manga G, Rosso R, Rougier P, Schmiegel WH, Seitz JF, Thompson P, Vieitez JM, Weitzel C, Harper P (2001) Oral capecitabine compared with intravenous fluorouracil plus leucovorin in patients with metastatic colorectal cancer: results of a large phase III study. J Clin Oncol 19:4097–4106. https://doi.org/10.1200/jco.2001.19.21.4097
van Kuilenburg AB, Muller EW, Haasjes J, Meinsma R, Zoetekouw L, Waterham HR, Baas F, Richel DJ, van Gennip AH (2001) Lethal outcome of a patient with a complete dihydropyrimidine dehydrogenase (DPD) deficiency after administration of 5-fluorouracil: frequency of the common IVS14+1G>A mutation causing DPD deficiency. Clin Cancer Res 7:1149–1153
van Kuilenburg AB, Meijer J, Mul AN, Meinsma R, Schmid V, Dobritzsch D, Hennekam RC, Mannens MM, Kiechle M, Etienne-Grimaldi MC, Klümpen HJ, Maring JG, Derleyn VA, Maartense E, Milano G, Vijzelaar R, Gross E (2010) Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity. Hum Genet 128:529–538. https://doi.org/10.1007/s00439-010-0879-3
Vicente J, González-Andrade F, Soriano A, Fanlo A, Martínez-Jarreta B, Sinués B (2014) Genetic polymorphisms of CYP2C8, CYP2C9 and CYP2C19 in Ecuadorian Mestizo and Spaniard populations: a comparative study. Mol Biol Rep 41:1267–1272. https://doi.org/10.1007/s11033-013-2971-y
von Ahsen N, Tzvetkov M, Karunajeewa HA, Gomorrai S, Ura A, Brockmöller J, Davis TM, Mueller I, Ilett KF, Oellerich M (2010) CYP2D6 and CYP2C19 in Papua New Guinea: high frequency of previously uncharacterized CYP2D6 alleles and heterozygote excess. Int J Mol Epidemiol Genet 1:310–319
Vreken P, Van Kuilenburg AB, Meinsma R, Smit GP, Bakker HD, De Abreu RA, van Gennip AH (1996) A point mutation in an invariant splice donor site leads to exon skipping in two unrelated Dutch patients with dihydropyrimidine dehydrogenase deficiency. J Inherit Metab Dis 19:645–654. https://doi.org/10.1007/bf01799841
Wahlund M, Nilsson A, Kahlin AZ, Broliden K, Myrberg IH, Appell ML, Berggren A (2020) The role of TPMT, ITPA, and NUDT15 variants during mercaptopurine treatment of Swedish pediatric patients with acute lymphoblastic leukemia. J Pediatr 216:150-157.e1. https://doi.org/10.1016/j.jpeds.2019.09.024
Watanabe T, Go H, Saigusa Y, Takamura N, Watanabe Y, Yamane Y, Totsuka M, Ishikawa H, Nakamura K, Matsukura S, Kambara T, Takaki S, Yamaguchi Y, Aihara M (2021) Mortality and risk factors on admission in toxic epidermal necrolysis: a cohort study of 59 patients. Allergol Int 70:229–234. https://doi.org/10.1016/j.alit.2020.11.004
Watson MS, Cutting GR, Desnick RJ, Driscoll DA, Klinger K, Mennuti M, Palomaki GE, Popovich BW, Pratt VM, Rohlfs EM, Strom CM, Richards CS, Witt DR, Grody WW (2004) Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet Med 6:387–391. https://doi.org/10.1097/01.gim.0000139506.11694.7c
Watson J, Taylor WR, Bancone G, Chu CS, Jittamala P, White NJ (2018) Implications of current therapeutic restrictions for primaquine and tafenoquine in the radical cure of vivax malaria. PLoS Negl Trop Dis 12:e0006440. https://doi.org/10.1371/journal.pntd.0006440
Wennerholm A, Johansson I, Hidestrand M, Bertilsson L, Gustafsson LL, Ingelman-Sundberg M (2001) Characterization of the CYP2D6*29 allele commonly present in a black Tanzanian population causing reduced catalytic activity. Pharmacogenetics 11:417–427. https://doi.org/10.1097/00008571-200107000-00005
Xiao Q, Lauschke VM (2021) The prevalence, genetic complexity and population-specific founder effects of human autosomal recessive disorders. NPJ Genom Med 6:41. https://doi.org/10.1038/s41525-021-00203-x
Yang SK, Hong M, Baek J, Choi H, Zhao W, Jung Y, Haritunians T, Ye BD, Kim KJ, Park SH, Park SK, Yang DH, Dubinsky M, Lee I, McGovern DP, Liu J, Song K (2014) A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet 46:1017–1020. https://doi.org/10.1038/ng.3060
Yang CY, Chen CH, Deng ST, Huang CS, Lin YJ, Chen YJ, Wu CY, Hung SI, Chung WH (2015a) Allopurinol use and risk of fatal hypersensitivity reactions: a nationwide population-based study in Taiwan. JAMA Intern Med 175:1550–1557. https://doi.org/10.1001/jamainternmed.2015.3536
Yang JJ, Landier W, Yang W, Liu C, Hageman L, Cheng C, Pei D, Chen Y, Crews KR, Kornegay N, Wong FL, Evans WE, Pui CH, Bhatia S, Relling MV (2015b) Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol 33:1235–1242. https://doi.org/10.1200/jco.2014.59.4671
Yasuda SU, Zhang L, Huang SM (2008) The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther 84:417–423. https://doi.org/10.1038/clpt.2008.141
Yin SJ, Ni YB, Wang SM, Wang X, Lou YQ, Zhang GL (2012) Differences in genotype and allele frequency distributions of polymorphic drug metabolizing enzymes CYP2C19 and CYP2D6 in mainland Chinese Mongolian, Hui and Han populations. J Clin Pharm Ther 37:364–369. https://doi.org/10.1111/j.1365-2710.2011.01298.x
Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. https://doi.org/10.1016/j.pharmthera.2012.12.007
Zazuli Z, Duin N, Jansen K, Vijverberg SJH, Maitland-van der Zee AH, Masereeuw R (2020) The impact of genetic polymorphisms in organic cation transporters on renal drug disposition. Int J Mol Sci. https://doi.org/10.3390/ijms21186627
Zeglam HB, Benhamer A, Aboud A, Rtemi H, Mattardi M, Saleh SS, Bashein A, Enattah N (2015) Polymorphisms of the thiopurine S-methyltransferase gene among the Libyan population. Libyan J Med 10:27053. https://doi.org/10.3402/ljm.v10.27053
Zhang JP, Guan YY, Wu JH, Jiang WQ, Huang M (2003) Genetic polymorphism of the thiopurine S-methyltransferase of healthy Han Chinese. Ai Zheng 22:385–388
Zheng Y, Wang J, Liang X, Huang H, Ma Y, Lin L, Wang C, Zhan X, Yang L, Zha G, Yang P, Zou X, Chen Z, Chen X, Chen W, Liu X, Lin M (2020) Epidemiology, evolutionary origin, and malaria-induced positive selection effects of G6PD-deficient alleles in Chinese populations. Mol Genet Genomic Med 8:e1540. https://doi.org/10.1002/mgg3.1540
Zhou Y, Lauschke VM (2018) Comprehensive overview of the pharmacogenetic diversity in Ashkenazi Jews. J Med Genet 55:617–627. https://doi.org/10.1136/jmedgenet-2018-105429
Zhou Y, Ingelman-Sundberg M, Lauschke VM (2017) Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 102:688–700. https://doi.org/10.1002/cpt.690
Zhou Y, Dagli Hernandez C, Lauschke VM (2020) Population-scale predictions of DPD and TPMT phenotypes using a quantitative pharmacogene-specific ensemble classifier. Br J Cancer 123:1782–1789. https://doi.org/10.1038/s41416-020-01084-0
Zhou Y, Herras Arribas G, Turku A, Jürgenson T, Mkrtchian S, Krebs K, Wang Y, Svobodova B, Milani L, Schulte G, Korabecny J, Gastaldello S, Lauschke VM (2021a) Rare genetic variability in human drug target genes modulates drug response and can guide precision medicine. Sci Adv
Zhou Y, Krebs K, Milani L, Lauschke VM (2021b) Global frequencies of clinically important HLA alleles and their implications for the cost-effectiveness of preemptive pharmacogenetic testing. Clin Pharmacol Ther 109:160–174. https://doi.org/10.1002/cpt.1944
Acknowledgements
V.M.L. receives support from the Swedish Research Council (Grant Agreement Nos. 2016-01153, 2016-01154 and 2019-01837), by the EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant number 875510), by the Swedish Strategic Research Programmes in Diabetes (SFO Diabetes) and Stem Cells and Regenerative Medicine (SFO StratRegen), as well as by the European Union’s Horizon 2020 research and innovation program U-PGx (Grant Agreement Nos. 668353). Furthermore, the lab acknowledges support from Merck KGaA and Eli Lilly and Company. The authors thank the team behind The Clinical and Functional Translation of CFTR (CFTR2) database (available at http://cftr2.org) for providing data.
Funding
Open access funding provided by Karolinska Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
YZ and VML are co-founders and shareholders of PersoMedix AB. In addition, VML is CEO and shareholder of HepaPredict AB and discloses consultancy work for Enginzyme AB.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
439_2021_2385_MOESM1_ESM.xlsx
Supplementary Table 1: References for population-specific frequencies of CYP2D6 alleles. PMIDs or, if the paper was not indexed in Pubmed, DOIs are provided (XLSX 13 KB)
439_2021_2385_MOESM2_ESM.xlsx
Supplementary Table 2: References for population-specific frequencies of CYP2C19 alleles. PMIDs or, if the paper was not indexed in Pubmed, DOIs are provided (XLSX 11 KB)
439_2021_2385_MOESM3_ESM.xlsx
Supplementary Table 3: References for population-specific frequencies of DPYD alleles. PMIDs or database sources are provided (XLSX 9 KB)
439_2021_2385_MOESM4_ESM.xlsx
Supplementary Table 4: Population-specific frequencies of the most common functionally deficient CFTR alleles in the general population. Data is extracted from 141,456 individuals provided by gnomAD v2.1.2 (https://gnomad.broadinstitute.org/) (XLSX 10 KB)
439_2021_2385_MOESM5_ESM.xlsx
Supplementary Table 5: Drugs and regimens for which the 10 analyzed genes impact the clinical pharmacology, efficacy or safety. Information obtained from https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling [Accessed 08.08.2021] (XLSX 13 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, Y., Lauschke, V.M. Population pharmacogenomics: an update on ethnogeographic differences and opportunities for precision public health. Hum Genet 141, 1113–1136 (2022). https://doi.org/10.1007/s00439-021-02385-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02385-x