Abstract
Modern wheat (Triticum aestivum L.) cultivars have a free-threshing habit, which allows for easy manual or mechanical threshing. However, when harvesting is delayed or extreme weather events occur at harvest time, grain shattering can cause severe loss of harvestable yield. In the past, grain size was considered a predisposing factor as large, plump kernels can lead to buckling and breaking of the outer glume, but the correlation between glume strength and shattering is not strong in modern wheat, and it is hypothesised that there may be other genetic mechanisms. Data from two bi-parent populations and a wheat diversity panel were analyzed to explore the underlying genetic basis for grain shattering observed in multiple field experiments through quantitative trait loci (QTL) analysis. Grain shattering had a significant and negative association with grain yield, irrespective of populations and environments. The correlation with plant height was positive in all populations, but correlations with phenology were population specific, being negative in the diversity panel and the Drysdale × Waagan population, and positive in the Crusader × RT812 population. In the wheat diversity panel, allelic variations at well-known major genes (Rht-B1, Rht-D1 and Ppd-D1) showed minimal association with grain shattering. Instead, the genome-wide analysis identified a single locus on chromosome 2DS, which explained 50% of the phenotypic variation, and mapping to ~ 10 Mb from Tenacious glume (Tg) gene. In the Drysdale × Waagan cross, however, the reduced height (Rht) genes showed major effects on grain shattering. At the Rht-B1 locus, the Rht-B1b allele was associated with 10.4 cm shorter plant height, and 18% decreased grain shattering, whereas Rht-D1b reduced plant height by 11.4 cm and reduced grain shattering by 20%. Ten QTL were detected in the Crusader × RT812, including a major locus detected on the long arm of chromosome 5A. All the QTL identified in this population were non-pleiotropic, as they were still significant even after removing the influence of plant height. In conclusion, these results indicated a complex genetic system for grain shattering in modern wheat, which varied with genetic background, involved pleiotropic as well as independent gene actions, and which might be different from shattering in wild wheat species caused by major domestication genes. The influence of Rht genes was confirmed, and this provides valuable information in breeding crops of the future. Further, the SNP marker close to Tg on chromosome 2DS should be considered for utility in marker-assisted selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern wheat (Triticum aestivum L.) cultivars have a free-threshing habit, with soft glumes that allow for manual or mechanical harvesting. However, when harvesting is delayed or extreme weather events occur at harvest time, grain shattering can account for up to 17% loss of harvestable yield. The propensity to shatter was eliminated in early forms of the domesticated crops (Dubcovsky and Dvorak 2007), but not completely (Li et al. 2006). In the wild type, the head fractures easily at the junction of the rachilla with the rachis but modern wheat have a tough rachis governed by major genes located on homoeologous regions on the short arms of chromosomes 3A (Br2), 3B (Br3) and 3D (Br1) (Nalam et al. 2006; Watanabe et al. 2006). Independent recessive mutations in each of the brittle rachis genes cause the cell wall at specific ‘‘constriction grooves’’ or fracture zones to thicken, converting the wild-type brittle rachis into a tough, non-brittle form that promotes head retention (Pourkheirandish et al. 2015). The brittle rachis, like other domestication traits, is easily distinguishable due to its high penetrance and is heavily selected against during the early generations of breeding programs. Despite heavy selection, however, grain loss due to shattering still occurs in the field in modern wheat (Chang 1943; Clarke and De Pauw 1983). Grain loss in this context is defined as the dropping of individual grains from the rachis at apical end of the spikes (Beck 1951; Porter 1959; Clarke 1981) (Fig. 1a), or the loss of a spike segment and the grains (Kandemir et al. 2000), which is different from the loss of whole spikes, as in barley (Clarke 1981). There are suggestions in the literature that the size of individual grains might be a predisposing factor (Platt and Wells 1949; Clarke 1981) as large, plump kernels and more grains per head can lead to buckling and breaking of the outer glume (bracts enclosing the grain), making the grain more easily removable from the spike (Vogel 1938). Genes that control soft glume (sog) and glume tenacity (Tg) have been identified and localised to chromosomes 2AS and 2DS (Sood et al. 2009), but according to Zhang et al. (2009), the correlation between glume strength and shattering is not strong in modern wheat. It was hypothesised, therefore, that there may be other genetic mechanisms controlling differences in grain shattering between varieties (Doebley et al. 2006).
Wheat is the second most-widely cultivated crop grown in the world, driven by the plethora of products that can be made from the grain, each of which has a near-limitless number of variations (Kiszonas and Morris 2018). It is pivotal to global food security, with demand now outstripping supply, as global production volume in 2020–2021 of over 700 million metric tonnes was outstripped by the consumption volume of over 710 million metric tonnes (Shahbandeh 2022). It is expected that climate change will exacerbate food security in areas that already currently have a high prevalence of hunger and malnutrition (Wheeler and Braun 2013). Globally, annual wheat yield ranges from < 1 t/ha/year when water or nutrients are limiting to > 10 t/ha/year in cooler, well-watered environments (Asseng et al. 2020). Causes of the yield gap are well documented (see Hochman and Horan 2018) but a significant component that is often overlooked is the loss of harvests in standing crops due to various reasons, including grain shattering.
Grain loss due to shattering (Fig. 1b) is a direct loss of income to the grower, as the more grains a grower can get into the machine at harvest, the greater the returns (Hofman and Kucera 1978). Clarke and De Pauw (1983) calculated the potential loss to range from 3 to 17% of harvestable yield, while Vogel (1938) reported it to be between 5 and 15%. Not only is the grain lost when shattering occurs, but a high amount of volunteer crop can also be expected during fallow, and if uncontrolled, can reduce water storage and contribute to the field seed bank. The amount of grain loss due to shattering depends on environmental conditions, such as high temperatures and wind speed, two weather factors that are predicted to increase in intensity due to climate change. Efforts to limit the loss are focused on cultivar choice and management, and a good understanding of the genetics would enable identifying susceptible genotypes before field trials and allow agronomic research to focus on managing the risk by varietal selection. The objectives of this study were to determine the underlying genetic basis for grain shattering in standing crop of modern wheat through quantitative trait loci (QTL) analysis and investigate the linkage with important agronomic characters.
Materials and methods
Diversity panel
The diversity panel comprised of 295 hexaploid and tetraploid genotypes, as well as advanced breeding lines from the International Maize and Wheat Improvement Center (CIMMYT), and landrace cultivars sourced from heat-prone environments. They formed part of a larger research on wheat heat tolerance reported by Collins et al. (2017). Molecular data for the diversity panel were derived from publicly available Illumina iSelect 90 K SNP array markers, made available by Garcia et al. (2019) for more than 500 diverse wheat accessions. The total marker set of 30,548 SNPs was filtered to remove markers with minor allele frequency of 5% or below. Then, the data were further reduced to retain 10,826 markers using PLINK’s function for SNP pruning based on linkage disequilibrium (Purcell et al. 2007). Missing data were imputed using the Random Forest regression method in R package (Stekhoven and Bühlmann 2011).
Drysdale × Waagan
This is a population of 142 doubled haploid (DH) lines derived from the F1 of a cross between the Australian wheat cultivars, ‘Drysdale’ and ‘Waagan’. Drysdale (named after the Australian artist, Russell Drysdale) was released in 2002 with the pedigree Hartog*3/Quarrion and was one of two varieties (the second was Rees) bred for increased water use efficiency by selecting for reduced carbon isotope discrimination (Richards 2006). Waagan was derived from a cross of Janz with a CIMMYT line, 24IBWSN-244. The point mutations responsible for the two major semi-dwarfing genes Rht-B1 (Syn. Rht1) and Rht-D1 (Syn. Rht2), segregate in the population, with Drysdale carrying the tall allele at Rht-B1 locus (Rht-B1a) and the dwarfing allele (Rht-D1b) at Rht-D1, and vice versa for Waagan (Shirdelmoghanloo et al. 2016).
Details of the Drysdale × Waagan DH population development and genotyping with the 9 k SNP array (Cavanagh et al. 2013) were described by (Shirdelmoghanloo et al. 2016). The total marker set consisted of 2,711 SNP markers, and these were assigned to physical positions on the wheat genome by searching against the T3/Wheat repository (https://wheat.triticeaetoolbox.org/). From these, a subset of 908 markers located on the wheat genome was selected for QTL analysis. They covered 13.7 Gigabase (Gb) of the 17-gigabase hexaploid bread wheat genome (80.6%), with an average inter-marker interval of 15.1 Mb.
Crusader × RT812
The Crusader × RT812 population is a DH population constructed from the F1 of a cross between the Australian cultivar, ‘Longreach-Crusader’ (Sunbrook/H45), and a breeding line, ‘RT812’, developed at CIMMYT. RT812 has the pedigree, Pastor//HXL7573/2*Bau/3/CMH82.575/CMH82.801. Crusader is extremely well adapted to main and later season sowings in New South Wales and Queensland, Australia. The population comprised of 243 lines, genotyped with 9,792 DArTseq single-nucleotide polymorphic markers assayed across the whole genome. DArTseq sequences (available at https://www.diversityarrays.com/) were used to query the IWGSC RefSeq v2.1 wheat reference genome for physical positions (Mb) of the markers. Missing values were imputed using the ‘fill.geno” in R/QTL (Broman et al. 2003).
Field experiments
Eleven field experiments were conducted between 2015 and 2018, and the locations for individual populations are indicated on Table 1. The soil at Wagga Wagga (latitude 35.05° S, longitude 147.35° E) is a sandy clay loam described as a Red Kandosol. It is moderately permeable, moderately well-drained with a greyish brown colour. At Condobolin (latitude 33.07° S, longitude 147.26° E), the soil is red brown in colour with near neutral pH and low inherent fertility and organic matter, and at Leeton (latitude 34.36° S, longitude 146.21° E), the soil is a vertosol, with shrink-swell properties that exhibit strong cracking when dry.
The experiments were sown with entries in a partial replication (p-rep), in which 75% of the entries were duplicated twice. Plot size was 7.5 m2 (6 rows with 30 cm spacing, 6 m long), and sown with 60 g seed. All experiments were fertilized at the time of sowing with mono-ammonium phosphate (MAP) at the rate of 100 kg ha−1. Weed and disease control, including irrigation, followed standard procedures described in Sissons et al. (2018).
Resistance to shattering was based on visual scoring using a scale of 1 to 9, in which 1 = no shattering and 9 = severe shattering (Haley et al. 2005). Plant height was measured in each plot, from soil surface to tip of the spike and defined as the average of three locations within each plot. Phenology was measured as the date of awn emergence, defined in days from sowing to when awns on 50% of the plants in a plot were approximately 1 cm above the flag leaf auricle (Sissons et al. 2018).
Plots were trimmed from 6 m long to 5 m prior to harvest, and grain yields were harvested using a plot machine, and converted to tonnes per hectare, based on the weight of uncleaned seed from each plot. A sub-sample of 300 g grains were taken from each plot and used to determine grain size, first by counting out 250 kernels randomly on a grain counter (Numigral, Rousseau, Paris, France), and then weighing the kernels, and the weight expressed as the weight of 1000 kernels.
Statistical analysis
Statistical analyses were performed in R (https://cran.r-project.org/) using mixed linear models to partition the phenotypic variance into genetic and non-genetic components. A baseline model was fitted within a multi-environment framework, which involved using information of the plot layouts (rows and columns) to model position of each plot. The analyses were performed using ASREML-R (version 4) package (Butler et al. 2017) according to the following form:
where y is a vector of trait observations, β is a vector of fixed effects to test for trends in the row and column directions, g is a vector of the underlying genetic variation of the trait among the progeny lines, X and Z are the associated design matrices, and ε is a vector of random residual errors. The residual variance also included a correlation structure parameterized as AR1 × AR1 (AR1 = autoregressive first order process) to model the correlation along the rows and columns due to neighbourliness of the experimental plots. These were fitted separately for each environment as fixed factors using the ASReml-R function at(), tested for significance (P < 0.05) using the wald statistics, and non-significant terms were removed from the final model. Normality checking was performed on the residuals as suggested in Kozak and Piepho (2018), and phenotypes with extreme values were replaced with ‘NA’ in multiple iterations until no residuals beyond ± 3 standard deviations were left.
Heritability (h2) was calculated as described by Cullis et al. (2006):
where mean.sed is the average pairwise prediction error of the genetic effects for the tested genotypes and σ2g is the genetic variance for the respective treatments.
Genetic correlations (rA) between grain yield, grain shattering and agronomic traits were calculated as:
where σ2x and σ2y are the genotypic variance components for traits x and y, and Covxy is the genetic covariance component, calculated after creating a dummy variable (z = x + y) as [(σ2z − (σ2x + σ2y)]/2. The statistical significance of rA was assessed as proposed by Schefler (1979), assuming a population correlation coefficient of zero, as:t = \(\frac{{r}_{A}}{\sqrt{(1-{r}_{A}^{2})/(n-2)}}\)where t is a student’s t value with (n-2) degrees of freedom, and n is the number of pairs of observations.
Genome-wide association analysis (GWA)
The population used for GWA comprised of 131 genotypes selected from the diversity panel for their ploidy (2n = 6x = 42) and availability of high-density molecular data, including KASP markers for the presence of well-known major semi-dwarf genes, Rht-B1 and Rht-D1, and photoperiod response gene, Ppd-D1. Population structure in the GWA panel was analyzed using discriminant analysis of principal components (DAPC) in the adegenet R package (Jombart et al. 2010). The final marker matrix after pre-processing contained 131 individuals and 3,711 markers. Genome-wide association (GWA) analysis was performed with the R package, GAPIT3 (Lipka et al. 2012), using the multi-locus mixed model approach (mlmm) described by Segura et al. (2012), which accounts for kinship. The method uses forward–backward stepwise regression to sequentially incorporate significant markers as covariates in the GWA model before scanning all other markers. Four principal component dimensions, fitted as fixed effects, were used to control for the confounding effect of population structure, along with a kinship matrix calculated according to VanRaden (2008) to define the variance and covariance structure of random individual effects (Yu et al. 2006). Markers exhibiting a genome-wide adjusted FDR P value < 0.05 were identified as significant.
Bi-parent QTL analysis
QTL analysis was performed using the Whole Genome Average Interval Mapping (RWGAIM) algorithm (Verbyla et al. 2012). The SNP molecular marker data were imported using the ‘read.cross’ command of R/QTL (Broman et al. 2003), and converted into an ‘interval’ object using the ‘cross2int’ command of R/WGAIM, with co-located markers placed in consensus bins. Then, the genetic ‘interval’ datum was merged with the baseline model defined in (1), and in the final step, R/WGAIM extended the model by incorporating all markers simultaneously as random covariates to detect main effect QTL. In the R script, the ‘fix.lines’ command was set to TRUE to fix the lines that do not exist in the genetic map. The gen.type = "marker" and the genome-wide Type I error for declaring a significant QTL was set at 0.01. Detailed summaries of detected QTL, including their position, effect size, and level of significance were provided by the package, and the QTL graphically displayed using linkMap() function.
Pleiotropic QTL analysis
In this step, we performed conditional genome scans using plant height as a fixed covariate in the R/WGAIM analysis of grain shattering. Comparison of unconditioned and conditioned scans would reveal changes in the LOD score for a QTL (Li et al. 2006), and if an unconditioned QTL is still detected after removing the influence of plant height, then this would likely represent an independent locus, but if the QTL is no longer significant, then that would suggest a pleiotropic QTL, causally linked to grain shattering through the effect of plant height. This approached yielded results that were identical to those from conditional genetic analysis proposed by Zhu (1995) and in some cases, revealed new QTL whose presence was made possible by removing the influence of plant height.
QTL × E analysis
Following the approach used by many authors (e.g., Lukens and Doebley 1999; Jermstad et al. 2003; Weinig et al. 2003; Geshnizjani et al. 2020), we used analysis of variance (ANOVA) to test the QTLs for environmental sensitivity. The approach also served to validate results obtain by RWGAIM algorithm.
Bioinformatic analysis
The physical positions of all SNP markers were determined by aligning the sequences harbouring each SNP to the updated reference sequence of the wheat genome (RefSeq v2.1) by BLAST search through the URGI portal (https://wheat-urgi.versailles.inra.fr/). The start position of each locus was extracted from the BLASTN output and used in the QTL analyses.
Results
Genotypic variation in landrace and elite wheat
Eleven field experiments were monitored for grain shattering in 2015, 2017 and 2018, and significant genotypic variations were observed in all cases (Fig. 2; Table 1). The variability in grain shattering was largest in the elite wheat cultivars grown under normal (June sowing) conditions, as opposed to those grown under late (August) sowing conditions (Fig. 2). Broad-sense heritability estimates obtained for the different population/year data (Table 1) showed higher genetic variances relative to error in eight of the eleven field experiments. This indicates high trait repeatability, which ranged from 0.30 to 0.87. Equally important, these values compared well with those for plant height (Table 1), suggesting either a strong, independent genetic basis for the varietal differences in grain shattering, or pleiotropic effect of genes controlling plant height. Using data for the elite wheat cultivars (because of the diverse genetic background), we found grain shattering in standing crop to show a high degree of repeatability across environments (Fig. 3), with highly significant positive correlation across sites (r = 0.77; P < 0.001) and time of sowing (r = 0.73; P < 0.001).
Genetic correlations
Grain shattering had a negative association with grain yield, irrespective of populations and environments (Table 2). The degree of association was small (− 0.07) in only 1 of the 11 population/year experiments we analyzed and was strong and consistently significant in majority of the experiments, ranging from – 0.20 to – 0.83 (Table 2). The correlations with grain size were positive, indicating that large grains increased the propensity to shattering, but data were not complete, particularly in the landraces, which were compromised by heavy lodging and had to be discarded.
The correlation with plant height was positive in all populations, and strongest in the Drysdale × Waagan doubled haploids, which segregated for the semi-dwarfing genes. The genetic correlation with plant height was also strong in the diversity panel, which comprised of genotypes with different genetic background, but was weak in the Crusader × RT812 population. Phenology had a significant effect on the propensity to grain shattering, but the influence was population-specific (Table 2), being negative in the diversity panel and the Drysdale × Waagan populations, and positive in the Crusader × RT812 population.
Genome-wide study of diversity panel
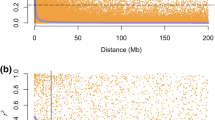
The GWA identified a marker (Fig. 4), BobWhite_c2949_1083 (Syn. IWB2281), which explained 50% of the phenotypic variability (GAPIT estimate). This could be due to the low statistical power associated with the relatively small population size, but to confirm the GWA result was not spurious, we analyzed data on plant height in the panel, which was genotyped with KASP markers that targeted major semi-dwarf genes, Rht-B1 and Rht-D1. The analysis identified the presence of the major genes in the panel, and their allelic effects conformed to expectations (Fig. 4).
For grain shattering, the quantile–quantile (QQ) plot (Fig. 4) showed further evidence that observed P values closely adhered to the expected values, with the genomic inflation factor less than 1.0 (λ = 0.90), indicating there were no systematic, spurious associations due to confounding factors. In the consensus wheat 90 K SNP array (Wang et al. 2014), the identified marker, BobWhite_c2949_1083 (Syn. IWB2281), was placed on chromosome 2BS, but physical mapping localised the SNP marker to the short arm of chromosome 2D, based on the recent Chinese Spring genome assembly (RefSeq v2.1).
QTL detection in Drysdale × Waagan
Both Drysdale and Waagan were present in the diversity panel, and they carried the alternate, non-shattering allele identified for grain shattering in the GWA analysis. In field experiments, however, Waagan ranked better than Drysdale for grain shattering, and their doubled haploid progenies were significantly different, with moderate-to-high heritability observed across the sowing times, and estimated at an average of 57.6% (Table 1). For plant height, the average heritability was 62.0%, and for phenology, it was 60.4%. The comparable heritability values showed grain shattering to be under strong genetic control, and further investigations were undertaken to unravel the genetic basis by scanning the wheat genome for allelic differences associated with the phenotype.
In the R/WGAIM analysis, 857 SNP markers representing unique, non-redundant marker bins were used for QTL analysis. The markers satisfied the expected ratio of 1:1 segregation, with 50.9% of ‘AA’ alleles, and 49.1% of the ‘BB’ alleles. Six genomic regions were found to be significantly linked to the variability in the grain shattering (Fig. 5a), and all QTL were verified to be significant (P < 0.01) by independent ANOVA tests (Table 3). Two of the QTL had major effects, collectively explaining almost 50% of the phenotypic variation. The two major QTL were located on chromosomes 4B and 4D, and directly linked to Rht-B1 and Rht-D1 semi-dwarfing genes. At the Rht-B1 locus, the Rht-B1b allele for reduced height carried by Waagan was associated with 10.4 cm shorter plant height, and 18% decreased grain shattering, whereas Rht-D1b carried by Drysdale reduced plant height by 11.4 cm and reduced grain shattering by 20% (Fig. 6). These QTL were still significant, even after adjusting for the effect of plant height, but the explained variation was substantially reduced from 27.4 to 13.9% in the case of the Rht-B1 locus, and from 18.9 to 6.7% for the Rht-D1 locus. These results indicated a pleiotropic influence of the plant height major genes on grain shattering in the population.
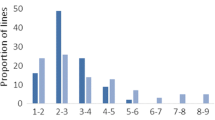
Chromosomal location of main-effect QTL identified for grain shattering and agronomic traits in two bi-parent populations of elite wheat cultivars. Chromosomes harbouring the QTL are represented by vertical lines, and each horizontal black line represents one of the unique SNP markers. Physical distances are reported on the scales to the left of the chromosomes
There were four minor QTL, located on chromosomes 3A, 6A, 6B and 7B (Fig. 5; Table 3). The minor QTL on 3A and 7B shared co-location with QTL associated with other agronomic traits, but the loci on chromosomes 6A and 6B were independent (Fig. 5a). Indeed, the locus on chromosome 3A was no longer significant, after adjusting for plant height, indicating it was a pleiotropic effect. However, the two loci detected on chromosome 6A and 6B, along with the QTL on chromosome 7B were still significant, even after conditioning on plant height, indicating they represent independent QTL. A QTL was detected for plant height on the short arm of chromosome 6A, which did not appear to affect shattering propensity. All identified QTL had some level of environmental sensitivity (Table 3), but this involved a change in magnitude rather than a change in direction.
QTL detected in Crusader × RT812
Amongst parent of this mapping population, RT812 was shatter resistant, while Crusader was susceptible. As shown in Table 1, the doubled haploid progeny from the cross exhibited significant variability for grain shattering, with heritability that ranged from 0.42 to 0.87, depending on the environment. To investigate the genetic basis, the DNA of the progeny lines were assayed for polymorphism at DArTseq markers, and from a total of 9792, a subset of 3948 non-redundant markers were used for QTL analysis. These covered 15.7 Gigabase (Gb) of the 17-Gb hexaploid bread wheat genome (92.4%), with a density of one marker per 4 Mb. The markers, on average, satisfied the expected ratio of 1:1 (AA = 48.9%; BB = 51.1%).
Ten QTL were detected for grain shattering using the WGAIM algorithm, and the QTL × environment ANOVA testing showed the effects were largely stable across environments (Table 3). Half of the identified QTL (located on chromosomes 2A, 2B, 5A and 7D) were closely linked to QTL affecting other agronomic traits, while the other half were largely isolated (Fig. 5b). The major locus for grain shattering was detected on the long arm of chromosome 5A, approximately 433 Mb from the centromere. Based on in silico mapping against the reference wheat genome, this QTL was located about 54.1 Mb downstream of the wheat domestication gene, Q, which determines spike morphology. Support interval for the QTL was small, spanning 374.3 Kb in length, and contained six genes, including genes that encodes for Cytochrome P450 and UDP-glucosyltransferase.
The QTL with minor effects explained between 3 and 7.8% of the phenotypic variability, and the ANOVA test for main effects confirmed that all were significantly associated with grain shattering, and stable across environments. Two loci located at the apical and centromeric ends of chromosome 4A were no longer significant after adjusting for the plant height, despite there being no QTL for plant height at these regions. Apart from these two, all other QTL identified in the population were non-pleiotropic, independent genetic factors, as they were still significant after removing the influence of plant height.
Discussion
In this study, a major locus was identified for grain shattering in the diversity panel, which mapped close to the Tenacious glume gene, Tg, on chromosome 2D. This locus was not evident in either of the bi-parent populations. This result is one of the benefits of using a combined linkage-based QTL mapping and LD-based association mapping to dissect the genetics of complex traits, as more genes can be identified by the combined use of these two methods (Lou et al. 2015). However, results obtained from such a combined approach are often not identical (Famoso et al. 2011; Lou et al. 2015; Altendorf et al. 2021), just as results from different bi-parent populations could be different due to heterogeneous genetic backgrounds (Holland 2007). In this case, one explanation is that the chromosome 2DS locus is probably fixed in the bi-parent populations. Parents of the doubled haploid populations were elite, high-yielding lines and not selected because of their contrasting phenotypes. QTL identified would be limited to the genetic loci segregating in the cross (Honsdorf et al. 2010; Brachi et al. 2010). The diversity panel, on the other hand, was comprised of old and current varieties, and the higher number of recombination events in such materials increase the range of natural variation that can be surveyed and would increase QTL detection power and resolution (Ewens and Spielman 2001).
Grain shattering is a difficult trait to phenotype accurately under field conditions (Zhang and Mergoum 2007a; Zhang et al. 2009), and in the past, breeding (Haley et al. 2005) and genetic studies (Marza et al. 2006; Bokore et al. 2022) have used visual scoring methods that are applied at harvest maturity. We followed the same approach here, and in a diverse panel of wheat cultivars, which comprised of elite lines and landrace varieties, this provided a good indication of a genotype’s propensity to shatter, with good stability across environments (Fig. 3). Substantial genetic variation was found to exist, with Spearman’s rank correlations ranging from 0.67 to 0.73, depending on the environment (Fig. 3). This meant that some wheat cultivars were more shatter resistant than others, and interestingly, more grain shattering occurred when crops were sown early (average score 4.3) than when sown late (average score 1.7). This has implications for early sowing systems to boost wheat yields (Hunt et al. 2019) because in bad years, a good portion of the harvest could be lost due to shattering.
Most of the shattering we observed in the field were at the apical end of the spike, that is, the top one-third of the spike (Fig. 1). This supports the findings made over 8 decades ago (Vogel 1941) that florets in the upper spikelets tend to shatter more easily than those towards the base of the spike. It could be that grains at the apical spikelets dry out faster during senescence and, therefore, more susceptible to be removable from the spike by buffeting wind and/or birds, but the physiological basis needs to be explored further.
We explored the genetic basis of grain shattering by QTL mapping in populations of different genetic backgrounds and to account for possible pleiotropy, we included other plant traits, such as phenology, plant height and grain size as covariates. The estimates of broad-sense heritability calculated for grain shattering in each population compared well with those calculated for plant height and phenology, suggesting it is either highly heritable on its own or it is the pleiotropic influence of genes affecting these traits. In the diversity panel, genome-wide analysis accurately identified the association of Rht-B1 and Rht-D1 with plant height, and these were the only markers above the threshold (Fig. 4), indicating presence of the Rht genes in the population. But there was no significant association of the Rht genes with grain shattering. Rather, the GWA identified a single locus, which explained 50% of the phenotypic variation in grain shattering. This was mapped to chromosome 2B (Wang et al. 2014), but the physical location was established to be the short arm of chromosome 2D, based on the updated Chinese Spring genome (RefSeq v2.1). Other previously reported genes on 2DS located in this vicinity include Ppd-D1 and Rht8, based on sequence matches of closely linked markers (Jantasuriyarat et al. 2004). However, the most likely candidate gene is the Tenacious glume, Tg1, which is located ~ 10.0 Mb on the distal side, based on sequence matches of closely linked markers reported by Sood et al. (2009). The Tg genes governs the free-threshing habit in wheat (Kerber and Rowland 1974) and was mapped to short arms of chromosomes 2B and 2D by both Jantasuriyarat et al. (2004) and Sood et al. (2009). Until recently, most of the molecular markers for the genes were based on simple sequence repeats (SSR), but Arif et al. (2021) reported a SNP marker, wsnp_Ra_c25656_3522705, close to Tg gene on chromosome 2D. Its physical position on the Chinese Spring genome suggests it is co-located (~ 7.7 Mb) with the marker identified in this study.
Grain shattering may be due to pleiotropic gene action
The genetic relationship of plant height with grain shattering is poorly understood, and some studies have suggested it might be entirely environmental (Zhang et al. 2009). Clarke and De Pauw (1983) found the amount of shattering was positively related with plant height, although not significant, and suggested that tall-strawed lines might tend to shatter more easily than short-strawed lines due to greater exposure of the spikes. In the Drysdale × Waagan cross, half of the QTL detected for grain shattering co-located with QTL associated with plant height and/or grain size (Fig. 5a). The two semi-dwarfing genes, Rht-B1 and Rht-D1, accounted for most of the observed variation in grain shattering, with alleles at the Rht-B1 locus contributing the most (R2 = 27.4%), and alleles at Rht-D1 accounting for a large proportion (18.9%) of the observed variability. The Rht-B1 locus explained 37.1% of the variation in plant height and 10.8% of the variability in grain size. On the other hand, the Rht-D1 locus explained 43.2% of variation in plant height and only a small proportion (3.4%) of the variability in grain size.
It is conceivable, therefore, that the effect of these regions on grain shattering might be a pleiotropic action of the reduced height genes. The Rht genes encode mutant DELLA proteins that are negative regulators of several gibberellic acid responses required for growth and have been associated with some undesirable agronomic characteristics, especially in water-limited environments (Jatayev et al. 2020). A linkage of grain shattering with semi-dwarfism was suggested by Oba et al. (1990), and later, Nakamura et al. (1995) showed that grain shattering caused by a single recessive gene, sh-2, is linked to the well-known semi-dwarf gene, sd-1 locus on chromosome 1. Zhang et al. (2013) reported the cloning of TaqSH1, a wheat ortholog of the rice grain shattering gene, qSH1, whose over-expression in Arabidopsis thaliana down-regulated known abscission genes and resulted in dwarfed plants, linking the reduced propensity to grain shattering with plant height.
The locus detected on chromosome 3A in the Drysdale × Waagan cross was no longer significant after adjusting for plant height. A significant QTL for plant height was detected within the interval, and this might be related to a gene for plant height identified by Martinez et al. (2021). QTL were also detected for grain shattering at the apical and centromeric ends of chromosome 4A in the Crusader × RT812 cross (Fig. 5b), but these became non-significant after adjusting for the plant height. This would indicate that these loci were associated with plant height and causally linked to grain shattering through the effect of plant height. Inability to detect a QTL for plant height at these regions might be related to allele frequency and small size of the effect (Würschum et al. 2017).
Independent QTL for grain shattering
In the two bi-parent populations, we found significant QTL on multiple chromosomal regions other than those harbouring known Rht genes. The Q gene (TRAESCS5A02G473800), located on the long arm of chromosome 5A, controls the ease with which grains can be separated from the chaff (Simons et al. 2006). We did not detect the presence of the Q gene in either of the two bi-parent populations used for this study. In fact, no QTLs were detected on chromosome 5A in the Drysdale × Waagan cross. In the Crusader × RT812 population, a QTL with strong phenotypic effect was identified on chromosome 5AL, with the peak signal at marker, 1107268 (Table 3), but it is unlikely to be linked to the Q gene locus, as it is physically located about 55.8 Mb distal of Q gene location. Other previously reported QTL in this vicinity include loci identified by Jantasuriyarat et al. (2004) and more recently by Bokore et al. (2022).
We found multiple QTLs on homoeologous group 2 chromosomes in the Crusader × RT812 population (Fig. 5b; Table 3), and this was intriguing because genes controlling classic domestication traits are located on these chromosomes. The spike-compacting gene, compactum (C), which affects rachilla morphology and grain size, is located on long arm of chromosome 2D, in a segment near the centromere (Johnson et al. 2008). The marker for QTL detected on chromosome 2D physically mapped to the long arm of the chromosome, closer to the telomere than the centromere, and is therefore unlinked. Peleg et al. (2011) identified a gene for brittle rachis in a durum wheat × wild emmer population, which mapped to the long arm of chromosome 2A, but the QTL detected in the current study on the long arm of chromosome 2A (Fig. 5b; Table 3) mapped to approximately 41.2 Mb from this gene, and they are therefore unlikely to be linked.
In conclusion, our results suggest that grain shattering in standing crop of modern wheat is not related to any lingering presence of classical domestication genes. Rather, in a population segregating for the semi-dwarf genes, strong genetic linkage to the known Rht genes, Rht-B1 and Rht-D1, was found, which supports suggestions in the literature of an association with plant height. The genetic correlation of grain shattering with plant height was strong and positive in the diversity panel, like in the Drysdale × Waagan bi-parent population (Table 2). Unlike the bi-parent population, however, there GWA did not find an association with Rht genes in the diversity panel. In fact, the landrace cultivars in the wheat diversity panel were taller than the elite cultivars by an average of 18.1 cm, but they were also the less likely to shatter. Single-marker analysis of variance showed that, to varying degrees of error, allelic variation at both Rht-B1 (P = 0.002) and Rht-D1 (P = 0.08) were associated with grain shattering in the diversity panel. Failure of GWA to detect an association might be due to stringency and can be regarded as a case of false negative.
QTL for grain shattering have been mapped to the Rht gene positions in previous studies (Marza et al. 2006; Bokore et al. 2022), but the current study is the first to use gene-based markers to confirm the association. The level of resolution is limited in these studies, and it would be worthwhile to further decipher the association of grain shattering with plant height and establish whether it is entirely pleiotropic or due to linkage disequilibrium. A SNP marker close to Tenacious glume gene, Tg, on chromosome 2DS was identified in the diversity panel and should be optimised for utility in marker-assisted selection.
Data availability
Data will be made available on request.
References
Altendorf KR, DeHaan LR, Larson SR, Anderson JA (2021) QTL for seed shattering and threshability in intermediate wheatgrass align closely with well-studied orthologs from wheat, barley, and rice. Plant Genome 14(3):e20145
Arif MAR, Shokat S, Plieske J, Ganal M, Lohwasser U, Chesnokov YV, Kocherina NV, Kulwal P, Kumar N, McGuire PE, Sorrells ME (2021) A SNP-based genetic dissection of versatile traits in bread wheat (Triticum aestivum L.). Plant J 108:960–976
Asseng S, Jose RG, Ra M, Oscar M, Gregory K, Dickson DD, Forrest MM, Gauthier PPG (2020) Wheat yield potential in controlled-environment vertical farms. PNAS 117:19131–19135
Beck TV (1951) A study of shattering and weathering in wheat and barley (Doctoral dissertation, University of Saskatchewan)
Bokore FE, Cuthbert RD, Knox RE, Campbell HL, Meyer B, N’Diaye A, Pozniak CJ, DePauw R (2022) Main effect and epistatic QTL affecting spike shattering and association with plant height revealed in two spring wheat (Triticum aestivum L.) populations. Theor Appl Genet 135:1143–1162
Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F (2010) Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet 6(5):e1000940
Broman KW, Wu H, Sen Ś, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Butler DG, Cullis, BR, Gilmour, AR, Gogel, BJ, Thompson R (2017) ASReml-R reference manual version 4. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. PNAS 110:8057–8062
Chang SC (1943) Morphological causes for varietal differences in shattering of wheat. Agron J 35:435–441
Clarke JM, Pauw RM (1983) The dynamics of shattering in maturing wheat. Euphytica 32:225–230
Clarke JM (1981) Effect of delayed harvest on shattering losses in oats, barley and wheat. Can J Plant Sci 61:25–28
Collins NC, Hildebrand S, Taylor K, Taylor H, Pleming D, Lohraseb I, Shirdelmoghanloo H, Erena M, Rahman M, Taylor J, Munoz-Santa S, Mather D, Heuer S, Sissons M, Emebiri L (2017) Understanding heat impacts on wheat to breed future tolerance. https://grdccomau/resources-and-publications/grdc-update-papers/tab-content/. Accessed 11 Feb 2022
Cullis BR, Smith AB, Coombes NE (2006) On the design of early generation variety trials with correlated data. J Agric Biol Environ Stat 11:381–393
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321
Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316:1862–1866
Ewens WJ, Spielman RS (2001) Locating genes by linkage and association. Theor Popul Biol 60:135–139
Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genetics 4 7(8):e1002221
Garcia M, Eckermann P, Haefele S, Satija S, Sznajder B, Timmins A, Baumann U, Wolters P, Mather D, Fleury D (2019) Phenotypic and genotypic data from a wheat diversity panel. https://adelaide.figshare.com/articles/dataset/Phenotypic_and_genotypic_data_from_a_wheat_diversity_panel/7344407
Geshnizjani N, Snoek BL, Willem LA, Rienstra JA, Nijveen H, Hilhorst HW, Ligterink W (2020) Detection of QTLs for genotype × environment interactions in tomato seeds and seedlings. Plant Cell Environ 43:1973–1988
Haley SD, Quick JS, Johnson JJ, Peairs FB, Stromberger JA, Clayshulte SR, Clifford BL, Rudolph JB, Seabourn BW, Chung OK (2005) Registration of “Hatcher” wheat. Crop Sci 45:2654–2656
Hochman Z, Horan H (2018) Causes of wheat yield gaps and opportunities to advance the water-limited yield frontier in Australia. Field Crop Res 228:20–30
Hoffman V, Kucera H (1978) Grain harvest losses. Circular AE-627, Cooperative Extension Service, North Dakota State University
Holland JB (2007) Genetic architecture of complex traits in plants. Curr Opin Plant Biol 10:156–161
Honsdorf N, Becker HC, Ecke W (2010) Association mapping for phenological, morphological, and quality traits in canola quality winter rapeseed (Brassica napus L.). Genome 53:899–907
Hunt JR, Lilley JM, Trevaskis B, Flohr BM, Peake A, Fletcher A, Zwart AB, Gobbett D, Kirkegaard JA (2019) Early sowing systems can boost Australian wheat yields despite recent climate change. Nat Clim Change 9:244–247
Jantasuriyarat C, Vales MI, Watson CJW, Riera-Lizarazu O (2004) Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat (Triticum aestivum L.). Theor Appl Genet 108:261–273
Jatayev S, Kurishbayev A, Zotova L, Absattarova A, Serikbay D, Sukhikh I, Vavilova V, Smolenskaya SE, Goncharov NP, Hu YG, Borisjuk N (2020) Green revolution ‘stumbles’ in a dry environment: Dwarf wheat with Rht genes fails to produce higher grain yield than taller plants under drought. Plant Cell Environ 43:2355–2364
Jermstad KD, Bassoni DL, Jech KS, Ritchie GA, Wheeler NC, Neale DB (2003) Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas fir III Quantitative trait loci-by-environment interactions. Genetics 165:1489–1506
Johnson EB, Nalam VJ, Zemetra RS, Riera-Lizarazu O (2008) Mapping the compactum locus in wheat (Triticum aestivum L.) and its relationship to other spike morphology genes of the Triticeae. Euphytica 163:193–201
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:1–15
Kandemir N, Kudrna DA, Ullrich SE, Kleinhofs A (2000) Molecular marker assisted genetic analysis of head shattering in six-rowed barley. Theor Appl Genet 101:203–210
Kerber RE, Rowland GG (1974) Origin of the threshing character in hexaploid wheat can. J Genet Cytol 16:145–154
Kiszonas AM, Morris CF (2018) Wheat breeding for quality: a historical review. Cereal Chem 95:17–34
Kozak M, Piepho HP (2018) What’s normal anyway? Residual plots are more telling than significance tests when checking ANOVA assumptions. J Agron Crop Sci 204:86–98
Li CB, Zhou AL, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Lou Q, Chen L, Mei H, Wei H, Feng F, Wang P, Xia H, Li T, Luo L (2015) Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J Exp Bot 66:4749–4757
Lukens LN, Doebley J (1999) Epistatic and environmental interactions for quantitative trait loci involved in maize evolution. Genet Res 74:291–302
Martinez AF, Lister C, Freeman S, Ma J, Berry S, Wingen L, Griffiths S (2021) Resolving a QTL complex for height, heading, and grain yield on chromosome 3A in bread wheat. J Exp Bot 72:2965–2978
Marza FE, Bai GH, Carver BF, Zhou WC (2006) Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor Appl Genet 112:688–698
Nakamura A, Komatsu S, Xra BS, Hirano H (1995) Linkage analysis of the gene loci for seed glutelin (Glu-1), semi-dwarfism (sd-1) and shattering habit (sh-2) in rice (Oryza sativa L.). Jpn J Breed 45:185–188
Nalam VJ, Vales MI, Watson CJ, Kianian SF, Riera-Lizarazu O (2006) Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat (Triticum turgidum L.). Theor Appl Genet 112:373–381
Oba S, Kikuchi F, Maruyama K (1990) Genetic analysis of semi-dwarfness and grain shattering of Chinese rice variety “Ai-Jio-Nan-Te.” Jpn J Breed 40:13–20
Peleg Z, Fahima T, Korol AB, Abbo S, Saranga Y (2011) Genetic analysis of wheat domestication and evolution under domestication. J Exp Bot 62:5051–5061
Platt AW, Wells SA (1949) Shattering, breaking and threshability in barley varieties. Sci Agric 29:453–464
Porter KB (1959) The inheritance of shattering in wheat. Agron J 51:173–177
Pourkheirandish M, Hensel G, Kilian B, Senthil N, Chen G, Sameri M, Azhaguvel P, Sakuma S, Dhanagond S, Sharma R, Mascher M (2015) Evolution of the grain dispersal system in barley. Cell 162:527–539
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Richards RA (2006) Physiological traits used in the breeding of new cultivars for water-scarce environments. Agric Water Manag 80:197–211
Schefler WC (1979) Statistics for the biological sciences, 2nd edn. Addison-Wesley, New York
Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M (2012) An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet 44:825–830
Shahbandeh M (2022) Wheat - statistics and facts. https://wwwstatistacom/topics/1668/wheat/. Assessed 25 Apr 22
Shirdelmoghanloo H, Taylor JD, Lohraseb I, Rabie H, Brien C, Timmins A, Martin P, Mather DE, Emebiri L, Collins NC (2016) A QTL on the short arm of wheat (Triticum aestivum L.) chromosome 3B affects the stability of grain weight in plants exposed to a brief heat shock early in grain filling. BMC Plant Biol 16:1–5
Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai Y-S, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172:547–555
Sissons M, Pleming D, Taylor JD, Emebiri L, Collins NC (2018) Effects of heat exposure from late sowing on the agronomic and technological quality of tetraploid wheat. Cereal Chem 95:274–287
Sood S, Kuraparthy V, Bai G, Gill BS (2009) The major threshability genes soft glume (sog) and tenacious glume (Tg), of diploid and polyploid wheat, trace their origin to independent mutations at non-orthologous loci. Theor Appl Genet 119:341–351
Stekhoven DJ, Bühlmann P (2011) MissForest—nonparametric missing value imputation for mixed-type data. Bioinformatics 28:112–118
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Verbyla AP, Taylor JD, Verbyla KL (2012) RWGAIM: an efficient high-dimensional random whole genome average (QTL) interval mapping approach. Genet Res 94:291–306
Vogel OA (1941) Relation of glume strength and other characters to shattering in wheat. Agron J 33:583–589
Vogel OA (1938) The relation of lignification of the outer glume to resistance to shattering in wheat. Agron J 30:599–603
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Watanabe N, Fujii Y, Kato N, Ban T, Martinek P (2006) Microsatellite mapping of the genes for brittle rachis on homoeologous group 3 chromosomes in tetraploid and hexaploid wheats. J Appl Genet 47:93–98
Weinig C, Stinchcombe JR, Schmitt J (2003) QTL architecture of resistance and tolerance traits in Arabidopsis thaliana in natural environments. Mol Ecol 12:1153–1163
Wheeler T, Von Braun J (2013) Climate change impacts on global food security. Science 341(6145):508–513
Würschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL (2017) A modern green revolution gene for reduced height in wheat. Plant J 92:892–903
Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhang G, Mergoum M (2007a) Developing evaluation methods for kernel shattering in spring wheat. Crop Sci 47:1841–1850
Zhang G, Mergoum M (2007b) Molecular mapping of kernel shattering and its association with Fusarium head blight resistance in a Sumai3 derived population. Theor Appl Genet 115:757–766
Zhang G, Mergoum M, Kianian S, Meyer DW, Simsek S, Singh PK (2009) Genetic relationship and QTL association between kernel shattering and agronomic traits in wheat. Crop Sci 49:451–458
Zhang L, Liu D, Wang D, Zhang R, Geng S, Wu L, Li A, Mao L (2013) Over expression of the wheat BEL1-like gene TaqSH1 affects floral organ abscission in Arabidopsis thaliana. J Plant Biol 56:98–105
Zhu J (1995) Analysis of conditional genetic effects and variance components in developmental genetics. Genetics 141:1633–1639
Acknowledgements
We thank the anonymous reviewers of this paper for their invaluable suggestions to improve the original manuscript. We are grateful to Dr. Nicholas Collins for access to the Crusader × RT812 genotype data.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. The data used for this paper were derived from research projects funded by the Grains Research and Development Corporation (GRDC), and the NSW Department of Primary Industries, Australia, which is gratefully acknowledged. Development of the Drysdale × Waagan doubled haploid population and 9 K SNP genotype data was funded by the NSW BioFirst initiative (Project E2985); Development of the Crusader × RT812 doubled haploid population and DArTseq genotype data was funded by GRDC project UA00147.
Author information
Authors and Affiliations
Contributions
LE: Conceptualisation, Methodology, Field experimentations, Data collection, Data analyses, Writing. SH: Field experimentations, Trial field operations, Data collection.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval & consent to participate
Not applicable.
Consent for publication
The authors give their consent for publication in Molecular Breeding.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Emebiri, L., Hildebrand, S. Natural variation and genetic loci underlying resistance to grain shattering in standing crop of modern wheat. Mol Genet Genomics 298, 1211–1224 (2023). https://doi.org/10.1007/s00438-023-02051-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-023-02051-z