Abstract
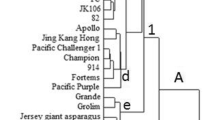
Vigna is a large, pan-tropic and highly variable group of the legumes family which is known for its > 10 cultivated species having significant commercial value for their nutritious grains and multifarious uses. The wild vignas are considered a reservoir of numerous useful traits which can be deployed for introgression of resistance to biotic and abiotic stresses, seed quality and enhanced survival capability in extreme environments. Nonetheless, for their effective utilization through introgression breeding information on their genetic diversity, population structure and crossability is imperative. Keeping this in view, the present experiment was undertaken with 119 accessions including 99 wild Vigna accessions belonging to 19 species and 18 cultivated genotypes of Vigna and 2 of Phaseolus. Total 102 polymorphic SSRs were deployed to characterize the material at molecular level which produced 1758 alleles. The genotypes were grouped into four major clusters which were further sub-divided in nine sub-clusters. Interestingly, all cultivated species shared a single cluster while no such similarities were observed for the wild accessions as these were distributed in different groups of sub-clusters. The co-dominant allelic data of 114 accessions were then utilized for obtaining status of the accessions and their hybrid forms. The model-based population structure analysis categorized 114 accessions of Vigna into 6 genetically distinct sub-populations (K = 6) following admixture-model based simulation with varying levels of admixture. 91 (79.82%) accessions resembled their hierarchy and 23 (20.18%) accessions were observed as the admixture forms. Maximum number of accessions (25) were grouped in sub-population (SP) 6 and the least accessions were grouped in SP3 and SP5 (11 each). The population genetic structure, therefore, supported genetic diversity analysis and provided an insight into the genetic lineage of these species which will help in effective use of germplasm for development of cultivars following selective prebreeding activities.



Similar content being viewed by others
References
Abbott RJ, Ritchie MG, Hollingsworth PM (2008) Introduction. Speciation in plants and animals: pattern and process. Phil Trans R Soc B 363:2965–2969
Aidbhavi R, Pratap A, Verma P, Lamichaney A, Bandi SM, Nitesh SD, Akram M, Rathore M, Singh B, Singh NP (2021) Screening of endemic wild Vigna accessions for resistance to three bruchid species. J Stored Prod Res 93:101864
Anderson E (1948) Hybridization of the habitat. Evolution 2:1–9
Arora RK, Nayar ER (1984) Wild relatives of crop plants in India NBPGR Sci. Monograph No. 7. National Bureau of Plant Genetic Resources, New Delhi, India
Arora RK, Chandel KPS, Joshi BS (1973) Morphological diversity in Phaseolus sublobatus Roxb. Curr Sci 42:359–361
Badiane FA, Gowda BS, Cissé N, Diouf D, Sadio O, Timko MP (2012) Genetic relationship of cowpea (Vigna unguiculata) varieties from Senegal based on SSR markers. Genet Mol Res 11:292–304. https://doi.org/10.4238/2012
Basu PS, Pratap A, Gupta S, Sharma K, Tomar R, Singh NP (2019) Physiological traits for shortening crop duration and improving productivity of greengram (V. radiata (L.) Wilczek) under high temperature. Front Plant Sci. https://doi.org/10.3389/fpls.2019.01508
Baudoin JP, Marechal R (1988) Taxonomy and evolution of the genus Vigna. In: Shanmugasundaram S, McLean BT (eds) Mungbean: Proceedings of the Second International Symposium. Taiwan: Asian Vegetable Research and Development Center, pp 2–12
Bisht IS, Bhat KV, Lakhanpaul S, Latha M, Jayan PK, Biswas BK, Singh AK (2005) Diversity and genetic resources of wild Vigna species in India. Genet Res Crop Evol 52:53–68
Blair MW, Pedraza F, Buendia HF, Gaitan-Solis E, Beebe SE, Gepts P, Tohme J (2003) Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L). Theor Appl Genet 107:1362–1374
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Chandel KPS, Laster RN (1991) Origin and evolution of Asiatic Vigna species. In: Sharma B, Mehra RB (eds) Golden jubilee celebration symposium on grain legumes. IARI, New Delhi, India, pp 25–45
Chandel KPS, Lester RN, Starling RJ (1984) The wild ancestors of urd and mungbeans [Vigna mungo (L.) Hepper and V. radiata (L.) Wilczek]. Bot J Linn Soc 89:85–96
Chankaew S, Somta P, Isemura T, Tomooka N, Kaga A, Vaughan DA, Srinives P (2013) Quantitative trait locus mapping reveals conservation of major and minor loci for powdery mildew resistance in four sources of resistance in mungbean [Vigna radiata (L.) Wilczek]. Mol Breed 32(1):121–130
Chankaew S, Isemura T, Isobe S, Kaga A, Tomooka N, Somta P, Hirakawa H, Shirasawa K, Vaughan DA, Srinives P (2014) Detection of genome donor species of neglected tetraploid crop Vigna reflexo-pilosa (creole bean) and genetic structure of diploid species based on newly developed EST-SSR markers from Azuki bean (Vigna angularis). PLoS ONE 9(8):e104990
Chapman MA, Burke JM (2007) Genetic divergence and hybrid speciation. Evolution 61:1773–1780. https://doi.org/10.1111/j.1558-5646.2007.00134.x
Chavan VM, Patil GD, Bhapkar DG (1966) Improvement of cultivated Phaseolus species-need for interspecific hybridization. Indian J Genet Plant Breed 26:152–154
Chen HK, Mok MC, Shanmugasundaram S, Mok DWS (1989) Interspecific hybridization between Vigna radiata (L.) Wilczek and V. glabrescens. Theor Appl Genet 78:641–647
Dachapak S, Somta P, Poonchaivilaisak S, Yimram T, Srinives P (2017) Genetic diversity and structure of the zombi pea (Vigna vexillata (L.) A. Rich) gene pool based on SSR marker analysis. Genetica 145:189–200. https://doi.org/10.1007/s10709-017-9957-y
Dana S (1964) Interspecific cross between tetraploid Phaseolus species and P. ricciardianus. Nucleus 7:1–10
Dana S (1968) Hybrid between Phaseolus mungo and tetraploid Phaseolus species. Japan J Genet 43:153–155
Dana S, Karmakar PG (1990) Species relation in Vigna subgenus Ceratotropis and its implications in breeding. Plant Breed Rev 8:19–42
de Candolle A (1884) Origin of cultivated plants. Hafner, New York
Douglas C, Pratap A, HanumanthaRao B, Manu B, Dubey S, Singh P, Tomar R (2020) Breeding progress and future challenges: abiotic stresses. In. Nair RM et al (eds) The mungbean genome, compendium of plant genomes. https://doi.org/10.1007/978-3-030-20008-4_6
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Gaitan-Solis E, Duque MC, Edwards KJ, Tohme J (2002) Microsatellite repeats in ommon bean (Phaseolus vulgaris): isolation, characterization and cross-species amplification in Phaseolus ssp. Crop Sci 42:2128–2136
Goel S, Raina SN, Ogihara Y (2001) Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of nuclear ribosomal DNA in the Phaseolus-Vigna complex. Mol Phylogenet Evol 1037:1–19
Gomathinayagam P, Ram SG, Rathnaswamy R, Ramaswamy NM (1998) Interspecific hybridization between Vigna unguiculata and V. vexillata through in vitro embryo culture. Euphytica 102:203–209
Gore PG, Tripathi K, Pratap A, Bhat KV, Umdale SD, Gupta V, Pandey A (2019) Delineating taxonomic identity of two closely related Vigna species of section Aconitifoliae: V. trilobata (L.) Verdc. and V. stipulacea (Lam.) Kuntz in India. Genet Resour Crop Evol. https://doi.org/10.1007/s10722-019-00767-9
Gwag JG, Dixit A, Park YJ, Ma KH, Kwon SJ, Cho GT, Lee GA, Lee SY, Kang HK, Lee SH (2010) Assessment of genetic diversity and population structure in mungbean. Genes Genomics 32:299–308
HanumanthaRao B, Nair RM, Nayyar H (2016) Salinity and high temperature tolerance in Mungbean [Vigna radiata (L.) Wilczek] from a physiological perspective. Front Plant Sci. https://doi.org/10.3389/fpls.2016.00957
Harlan JR (1971) Agricultural origins: centers and noncenters. Science 174:468–474
Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T, Jo U, Vaughan DA, Tomooka N (2012) Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean. PLoS ONE 7:41304. https://doi.org/10.1371/journal.pone.0041304
John KJ, Latha M, Senthil KR, Asokan NR, Abraham Z, Mishra SK (2009) Vigna dalzelliana (O. Kuntz) Verdc: a new distributional record from Andaman Islands, India. Indian J Plant Genet Resour 22:138–140
Kaewwongwal A, Chen J, Somta P, Kongjaimun A, Yimram T, Chen X, Srinives P (2017) Novel alleles of two tightly linked genes encoding polygalacturonase-inhibiting proteins (VrPGIP1 and VrPGIP2) associated with the Br locus that Confer Bruchid (Callosobruchus spp) resistance to Mungbean (Vigna radiata) accession V2709. Front Plant Sci 8:1692. https://doi.org/10.3389/fpls.2017.01692
Kajonphol T, Sangsiri C, Somta P, Toojinda T, Srinives P (2012) SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mung bean (Vigna radiata (L.) Wilczek). SABRAO J Breed Genet 44:71–86. https://doi.org/10.1556/019.70.2019.09
Kempf K, Mora-Ortiz M, Smith LM, Kölliker R, Skøt L (2016) Characterization of novel SSR markers in diverse sainfoin (Onobrychis viciifolia) germplasm. BMC Genet 17(1):1–4
Kitsanachandee R, Somta P, Chatchawankanphanich O, Akhtar KP, Shah TM, Nair RM, Bains TS, Sirari A, Kaur L, Srinives P (2013) Detection of quantitative trait loci for mungbean yellow mosaic India virus resistance in mungbean in India and Pakistan. Breed Sci 63:367–373
Krishnan R, De DN (1968) Cytogenetical studies in Phaseolus II. Phaseolus mungo × tetraploid phaseolus species and the amphidiploid. Indian J Genet Plant Breed 28:23–30
Kumar SV, Tan SG, Quah SC, Yusoff K (2002a) Isolation of microsatellite markers in mungbean, Vigna radiata. Mol Ecol Notes 2:96–98
Kumar SV, Tan SG, Quah SC, Yusoff K (2002b) Isolation and characterization of seven tetranucleotide microsatellite loci in mungbean, Vigna radiata. Mol Ecol Notes 2:293–295
Kumar S, Gupta S, Chandra S, Singh BB (2004) How wide is the genetic base of pulse crops? In: Ali M, Singh BB, Kumar S, Dhar V (eds) Pulses in new perspective. Indian Society of Pulses Research and Development, Kanpur, pp 211–221
Li CD, Fatokun CA, Ubi B, Singh BB, Scoles GJ (2001) Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci 41:189–197
Nair RM, Pandey AK, War AR, Bindumadhava H, Shwe T, Alam AKMM, Pratap A, Malik SR, Karimi R, Mbeyagala EK, Douglas CA, Rane J, Schafleitener R (2019) Biotic and abiotic constraints in mungbean production—progress in genetic improvement. Front Plant Sci 10:1340. https://doi.org/10.3389/fpls.2019.01340
Palmgren MG, Edenbrandt AK, Vedel SE, Andersen MM, Landes X, Østerberg JT, Falhof J, Olsen LI, Christensen SB, Sandøe P, Gamborg C (2015) Are we ready for back-to-nature crop breeding? Trends Plant Sci 20(3):155–164
Pandiyan M, Senthil N, Ramamoorthi N, Muthiah AR, Tomooka N, Duncan V et al (2010) (2010) Interspecific hybridization of Vigna radiata × 13 wild Vigna species for developing MYMV donor. Electron J Plant Breed 1:600–610
Parker PG, Snow AA, Schug MD, Booton GC, Fuerst PA (1998) What molecules can tell us about populations: choosing and using a molecular marker. Ecology 79:361–382
Pratap A, Kumar J (2011) History origin and evolution. In: Pratap A, Kumar J (eds) Biology and breeding of food legumes. CABI, Oxfordshire, p 432
Pratap A, Malviya N, Tomar R, Gupta DS, Kumar J (2014a) Vigna. In: Pratap A, Kumar J (eds) Alien gene transfer in crop plants, vol 2. Achievements and impacts. Springer Business+ Science Media, New York, pp 163–190
Pratap A, Basu PS, Gupta S, Malviya N, Rajan N, Tomar R, Latha M, Nadarajan N, Singh NP (2014b) Identification and characterization of sources for photo- and thermo-insensitivity in Vigna species. Plant Breeding 133:756–764
Pratap A, Gupta S, Malviya N, Rajan N, Tomar R, Latha M, John JK, Singh NP (2015) Genome scanning of asiatic Vigna species for discerning population genetic structure based on microsatellite variation. Mol Breed 35:178
Pratap A, Chaturvedi SK, Tomar R, Rajan N, Malviya N, Thudi M, Saabale PR, Prajapati U, Varshney RK, Singh NP (2017) Marker-assisted introgression of resistance to fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol Genet Genomics 292:137–1245. https://doi.org/10.1007/s00438-017-1343-z
Pratap A, Douglas C, Prajapati U, Kumari G, Was AR, Tomar R, Pandey AK, Dubey S (2020) Breeding progress and future challenges: Biotic stresses. In. Nair RM et al (eds) The Mungbean genome, compendium of plant genomes. https://doi.org/10.1007/978-3-030-20008-4_5
Pratap A, Gupta S, Rathore M, Basavaraja T, Singh CM, Prajapati U, Singh P, Singh Y, Kumari G (2021a) Mungbean. In: Pratap A, Gupta S (eds) The beans and the peas: from orphan to mainstream crops. Elsvier, Duxdorf, pp 1–32
Pratap A, Das A, Kumar S, Gupta S (2021b) Current perspectives on introgression breeding in food legumes. Front Plant Sci. https://doi.org/10.3389/fpls.2020.589189
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rawal KM (1975) Natural hybridization among wild, weedy and cultivated Vigna unguiculata L. Walp. Euphytica 24:699–707
Sangiri C, Kaga A, Tomooka N (2007) Genetic diversity of the mungbean (Vigna radiata) gene pool on the basis of microsatellite analysis. Aust J Bot 55:837–847
Sarr A, Bodian A, Gbedevi KM et al (2020) Genetic Diversity and population structure analyses of wild relatives and cultivated cowpea (Vigna unguiculata (L.) Walp.) from senegal using simple sequence repeat markers. Plant Mol Biol Rep. https://doi.org/10.1007/s11105-020-01232-z
Singh DP, Singh BB, Pratap A (2017) Genetic improvement of mungbean and urdbean and their role in enhancing pulse production in India. Indian J Genet Plant Breed 76:550–567
Singh B, Das A, Parihar AK, Bhagawati B, Singh D, Pathak KN, Dwivedi K, Das N, Keshari N, Midha RL, Kumar R, Pratap A, Kumar V, Gupta S (2020) Delineation of Genotype-by-Environment interactions for identification and validation of resistant genotypes in root-knot nematode (Meloidogyne incognita) using GGE biplot. Sci Rep 10(1):4108. https://doi.org/10.1038/s41598-020-60820-x
Smartt J (1985) Evolution of grain legumes. III. Pulses in the genus Vigna. Exp Agric 21:87–100
Soares BM, Ferreira PAA, de Oliveira-Longatti SM, Marra LM, Rufini M, de Andrade MJB, de Souza-Moreira FM (2014) Cowpea symbiotic efficiency, pH and aluminium tolerance in nitrogen-fixing bacteria. Sci Agric 71:171–180
Somta P, Seehalak W, Srinives P (2009) Development, characterization and cross-species amplification of mungbean (Vigna radiata) genic microsatellite markers. Conserv Genet 10:1939–1943
Sonnante G, Piergiovanni AR, Ng QN, Perrino P (1996) Relationships of Vigna unguiculata (L.) Walp., V. vexillata (L.) A. Rich. and species of section Vigna based on isozyme variation. Genet Resour Crop Evol 43:157–165
Takahashi Y, Somta P, Muto C, Iseki K, Naito K, Pandiyan M et al (2016) Novel genetic resources in the genus Vigna unveiled from gene bank accessions. PLoS ONE 11(1):e0147568. https://doi.org/10.1371/journal.pone.0147568
Tomooka N (2009) The origin of rice bean (Vigna umbellata) and azuki bean (V. angularis): the evolution of two lesser-known Asian beans. In: Akimiti T (ed) An illustrated eco-history of the Mekong River Basin. White Lotus Co, Bangkok
Tomooka N, Vaughan DA, Moss H, Maxted N (2002) The Asian Vigna: Genus Vigna subgenus Ceratotropis genetic resources. Kluwer Academic Publishers
Tomooka N, Naito K, Kaga A, Sakai H, Isemura T, Ogiso-Tanaka E, Iseki K, Takahashi Y (2014) Evolution, domestication and neo-domestication of the genus Vigna. Plant Genet Resour 2(S1):S168–S171
Varshney RK, Mohan SM, Gaur PM, Chamarthi SK, Singh VK, Srinivasan S, Swapna N, Sharma M, Pande S, Singh S, Kaur L (2014) Marker-assisted backcrossing to introgress resistance to fusarium wilt race 1 and Ascochyta blight in C 214, an elite cultivar of chickpea. Plant Genome 7:1–11
Vasconcelos EV, de Andrade Fonsêca AF, Pedrosa-Harand A, de Andrade Bortoleti KC, Benko-Iseppon AM, da Costa AF, Brasileiro-Vidal AC (2015) Intra- and interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosome Res 23(2):253–266. https://doi.org/10.1007/s10577-014-9464-2
Vavilov NI (1926) Studies on the origin of cultivated plants. Leningrad. 1951
Vaz Patto MC, Amarowicz R, Aryee AN, Boye JI, Chung HJ, Martín-Cabrejas MA, Domoney C (2015) Achievements and challenges in improving the nutritional quality of food legumes. Crit Rev Plant Sci 134:105–143
Wang XW, Kaga A, Tomooka N, Vaughan DA (2004) The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi]. Theor Appl Genet 109:352–360
Wang ML, Barkley NA, Gillaspie GA, Pederson GA (2008) Phylogenetic relationships and genetic diversity of the USDA Vigna germplasm collection revealed by gene-derived markers and sequencing. Genet Res 90:467–480. https://doi.org/10.1017/S0016672308009889
Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388
Yoshida Y, Marubodee R, Ogiso-Tanaka E et al (2016) Salt tolerance in wild relatives of adzuki bean, Vigna angularis (Willd.) Ohwi et Ohashi. Genet Resour Crop Evol 63:627–637. https://doi.org/10.1007/s10722-015-0272-0
Zukovskij PM (1962) Cultivated plants and their wild relatives. Commonwealth Agriculture Bureau, London
Funding
The authors acknowledge the funding support received from Department of Biotechnology, Government of India (Grant no. BT/Ag/Network/Pulses-I/2017-18) and the Uttar Pradesh Council of Agricultural Research, Lucknow (745/AP&AS/CROPS/RF/2014) in the form of funded research projects.
Author information
Authors and Affiliations
Contributions
GK and AP conceptualized, planned and executed the experiment. GK, SPS and AP analyzed the data and interpreted the results. GK, YS, BP and PS undertook field and laboratory experiments, LM provided seeds of some of the accessions. SG, GRP, NPS provided suggestions for improvement of the experiments and the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumari, G., Roopa Lavanya, G., Shanmugavadivel, P.S. et al. Genetic diversity and population genetic structure analysis of an extensive collection of wild and cultivated Vigna accessions. Mol Genet Genomics 296, 1337–1353 (2021). https://doi.org/10.1007/s00438-021-01825-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01825-7