Abstract
Winter wheats require a long exposure to cold temperatures (vernalization) to accelerate flowering. However, varieties differ in the length of the period of cold required to saturate the vernalization response. Here we show that single nucleotide polymorphisms (SNP) at the binding site of the GRP2 protein in the VRN-A1 first intron (henceforth, RIP3) are associated with significant differences in heading time after a partial vernalization treatment. The ancestral winter VRN-A1 allele in ‘Triple Dirk C’ has one SNP in the RIP3 region (1_SNP) relative to the canonical RIP3 sequence, whereas the derived ‘Jagger’ allele has three SNPs (3_SNPs). Both varieties have a single VRN-A1 copy encoding identical proteins. In an F2 population generated from a cross between these two varieties, plants with the 3_SNPs haplotype headed significantly earlier (P < 0.001) than those with the 1_SNP haplotype, both in the absence of vernalization (17 days difference) and after 3-weeks of vernalization (11 days difference). Plants with the 3_SNPs haplotype showed higher VRN-A1 transcript levels than those with the 1_SNP haplotype. The 3_SNPs haplotype was also associated with early heading in a panel of 127 winter wheat varieties grown in three separate controlled-environment experiments under partial vernalization (36 to 54 days, P < 0.001) and one experiment under field conditions (21 d, P < 0.0001). The RIP3 polymorphisms can be used by wheat breeders to develop winter wheat varieties adapted to regions with different duration or intensity of the cold season.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is one of the most widely cultivated cereals and an important staple food worldwide. Almost 70% of 740M tonnes of annual production are used for direct human consumption making wheat yields crucial for human nutrition and global food security (FAOSTAT 2015). Recent estimates of the impact of climate change on crops yield have raised the interest in understanding how plant development is modulated by environmental cues (Cang et al. 2016; Cook et al. 2012; Franks et al. 2007; Liu et al. 2016).
Wheat varieties are divided into two major categories (winter and spring) based on their growth habit. Winter wheats require a long exposure to low temperatures (vernalization) to accelerate the transition from the vegetative to the reproductive phase. This requirement prevents exposure of delicate floral meristems to damaging freezing temperatures (Distelfeld et al. 2009a; Woods et al. 2016). By contrast, spring wheats have reduced or no vernalization requirement. Winter wheats are sown in fall in regions where wheat can tolerate winter freezing temperatures. However, where winter temperatures are too low, spring wheats are sown in the spring to avoid freezing. In Mediterranean regions, with mild and rainy winters, spring wheats are planted in the fall to take advantage of water availability during the winter.
Wheat varieties sown in these contrasting regions show different allele profiles at the main vernalization gene VERNALIZATION1 (VRN1), a MADS-box transcription factor homologous to the meristem identity gene APETALA 1 (AP1) in Arabidopsis (Yan et al. 2003; Trevaskis et al. 2003). Winter wheats carry the ancestral VRN1 alleles, whereas spring wheats show deletions or mutations in regulatory regions located in the promoter or first intron. Changes in these regions have been observed in all three VRN1 homologs (VRN-A1, VRN-B1 and VRN-D1), as well as in the relatively recent duplicated paralog VRN-D4 (Fu et al. 2005; Konopatskaia et al. 2016; Muterko et al. 2015, 2016; Yan et al. 2004a, 2003; Kippes et al. 2015; Chu et al. 2011; Pidal et al. 2009).
Additional differences in wheat vernalization requirement have been detected at the VERNALIZATION2 (VRN2) and VERNALIZATION3 (VRN3) loci. VRN2 encodes a protein with a putative zinc finger and a CCT domain (ZCCT) that acts as a dominant long-day repressor of VRN3 and flowering. VRN2 deletions or loss-of-function mutations result in spring growth habit in diploid wheat and barley accessions (Yan et al. 2004b), but have not been observed so far in natural polyploid wheat species. Combination of deletions or non-functional vrn2 alleles at all homologs in tetraploid and hexaploid wheat (e.g., by marker assisted selection) can generate spring wheats (Distelfeld et al. 2009b; Kippes et al. 2016).
During the fall, VRN2 represses the expression of VRN3 (= FT1), a homolog of the Arabidopsis flowering promoter FLOWERING LOCUS T (FT) (Yan et al. 2006). FT encodes a mobile protein that travels from the leaves to the apex (Corbesier et al. 2007; Tamaki et al. 2007) where it forms a complex with FD-like and 14-3-3 proteins. This complex binds to the VRN1 promoter and induces flowering (Taoka et al. 2011; Li et al. 2015). During the winter, a slight induction of VRN1 is sufficient to repress VRN2, which favors FT1 up-regulation when days get longer during spring (Chen and Dubcovsky 2012). These interactions result in a positive feedback-loop that irreversibly promotes wheat flowering in the spring (Loukoianov et al. 2005).
The natural allelic variation in VRN-A1 responsible for differences between spring and winter wheats is well characterized. However, less is known about the role of VRN-A1 on the differences among winter wheats in the duration of the cold period required for saturating the vernalization response. Three independent studies found that a large proportion of the variation in vernalization requirement among winter wheat varieties is linked to the VRN-A1 locus. However, the three studies propose alternative explanations for these differences. Diaz et al. (2012) suggested that the differences were caused by the presence of a single copy of VRN-A1 in ‘Claire’ and three in ‘Hereward’. Li et al. (2013) argued that the differences in heading time were caused by an amino acid polymorphism at position 180 between ‘Jagger’ (alanine) and ‘2174’ (valine). More recently, Kippes et al. (2015) found that both ‘Claire’ and ‘Jagger’ also differed from ‘Hereward’ and ‘2174’ by single nucleotide polymorphisms (SNPs) at the binding site of a GLYCINE RICH RNA-BINDING PROTEIN 2 (GRP2) in a region of the VRN-A1 first intron designated as RNA Immune Precipitation fragment 3 (RIP3, Xiao et al. 2014 and; Kippes et al. 2015). RIP3 natural polymorphisms were shown to disrupt the binding of GRP2 to the RIP3 site (Kippes et al. 2015), but their effects on VRN1 expression and heading time were not characterized. No variation in the RIP3 site was detected in the VRN-B1 or VRN-D1 genes (Kippes et al. 2015).
In this study, we show that polymorphisms in the VRN-A1 RIP3 region segregating in an F2 population are associated with differences in VRN-A1 transcript levels and heading time independently of differences in VRN-A1 copy number and protein sequence. We also show that polymorphisms in the RIP3 region in a panel of winter wheat varieties from different geographic regions are associated with differences in heading time. The distribution of RIP3 alleles and VRN-A1 copy number among winter wheat varieties and the utilization of these variants in winter wheat breeding are discussed.
Materials and methods
Plant material
The parental lines of the F2 population analyzed in this study were the winter wheat lines Triple Dirk C (TDC) and CS5402 (Kippes et al. 2014, 2015). TDC is part of a set of near isogenic lines for vernalization alleles described in Pugsley 1971 and 1972. CS5402 is a substitution line of chromosome 5D of Chinese Spring (dominant Vrn-D1 allele) by the Ae. tauschii chromosome 5D (recessive vrn-D1 allele). Both lines have a single copy of the VRN-A1 gene homozygous for the recessive vrn-A1 allele, which encodes for identical VRN-A1 proteins (alanine residue at position 180, Li et al. 2013). The two accessions differ in the sequence of the RIP3 region in intron 1. Relative to the RIP3 canonical sequence, the VRN-A1 haplotype from TDC has one SNP (henceforth 1_SNP, GenBank AY747600) and the one from CS5402 three SNPs (henceforth 3_SNPs, GenBank KR422423, Kippes et al. 2015).
For the association study, we used a panel of 127 winter lines from the USDA National Germplasm Collection from diverse geographical origin. Sixty-five percent of the lines were collected from 31 different countries in Europe, 15% from 9 countries in Asia, 18% from 4 countries in the Americas, and 1% from Australia. These 127 accessions were selected from a larger set after filtering out accessions carrying alleles associated with a spring growth habit using available molecular markers (Yan et al. 2004b, 2006; Fu et al. 2005).
Molecular markers and VRN1 polymorphisms
Genomic DNA was isolated from individual plants for the F2 study and from bulks of leaves from four seedlings of each accession for the winter wheat panel (MAG-Bind Plant DNA Plus 96 kit, Omega Bio-Tek, Norcross, GA). Kompetitive Allele Specific PCR (KASP) fluorescent assays were used to detect polymorphism among the 127 accessions at wheat vernalization loci. PCR was run according to manufacturer’s instructions, using a reaction volume of 4 µL, which consisted of 2 µL 2× KASPar reaction mix, 0.05 µL 72× assay mix, and 2 µL of template DNA (10 ng µL−1). Endpoint genotyping was conducted using the software KlusterCaller (LGC Genomics, Hoddeson, UK).
Assays developed from published sequences of causal genes were used to distinguish lines in the core collection possessing spring alleles at Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 loci (Supplemental Table 1). The exception was the KASP assay wMAS000033 used for detection of the Vrn-A1a spring allele developed from the contextual sequences of iSelect SNP marker IWA0001 determined to be associated with Vrn-A1a. Accessions carrying alleles for spring growth habit at any VRN locus were excluded from further analysis. The VRN-A1 RIP3 region was sequenced in the 127 winter accessions by Sanger sequencing using primers listed in Table 1. Genotyping of the RIP3 alleles in the F2 population was conducted with a KASP assay (Table 1) run on a 7500 Fast Real-Time PCR system (Applied Biosystems). VRN-A1 copy number was determined using a TaqMan assay from four biological replications per genotype as described in Kippes et al. (2015). Differences in flowering time between VRN-A1 copy number classes were analyzed using Tukey’s test (P < 0.05). The VRN-A1 promoter from Triple Dirk C (TDC) was sequenced by Sanger sequencing (GenBank MH347747) using four overlapping PCR products amplified with primers listed in Table S3.
Principal component analysis
Accessions from the winter panel were previously genotyped with the Illumina Infinium 9K iSelect platform as part of the characterization of the NSGC core collection of winter and facultative common wheat (Bonman et al. 2015). Genotypes of 4,483 markers for all accessions were obtained from The Triticeae Toolbox (https://triticeaetoolbox.org/wheat). Principal component analysis was conducted in Tassel v5.0 (http://www.maizegenetics.net/tassel) using the covariance matrix for markers having minor allele frequency greater than or equal to 0.05. Lines missing more than 10% of data were removed.
RT-qPCR
The last expanded leaf of four biological replications per genotype was collected in liquid nitrogen, and RNA was extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). RNA samples were treated with DNase I (RQ1 RNase-Free DNase, Promega) and first strand cDNAs were synthesized from 1 µg of total RNA using the High Capacity Reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR was performed using SYBR Green and a 7500 Fast Real-Time PCR system (Applied Biosystems). ACTIN was used as an endogenous control. Primers are detailed in Table 1; primers for ACTIN were previously described (Dubcovsky et al. 2006). Transcript levels were expressed as linearized fold-ACTIN levels calculated by the formula 2(ACTIN CT − TARGET CT) ± standard error (SE) of the mean. The resulting number indicates the ratio between the initial number of molecules of the target gene and the number of molecules of ACTIN.
Phenotyping
Vernalization
Seeds were germinated in 2 × 2 plastic inch pots filled with vermiculite in a greenhouse with 16-h light and 8-h dark photoperiod. One week after planting, seedlings were moved to a growth chamber for 3 weeks of vernalization at 4 °C under 16-h light/8-h dark photoperiod. Similar conditions were used in the facilities at the University of California, Davis (UCD) and North Carolina State University (NCSU).
After vernalization, up to 10 seedlings were transplanted into 0.7 L black plastic containers (Stuewe and Sons, Tangent, OR) with 1:1 Metro Mix 2 and soil. Two grams of slow release fertilizer (Multicote 14–14–16, N-P-K) was incorporated with the soil in each cone. Cones were placed in greenhouses or growth chambers under the conditions described below.
Growth chamber
After vernalization, four replications of each entry were grown in a controlled-environment chamber at NCSU in a completely randomized design. Plants were grown under 20/18 °C day/night temperature and 16-h light/8-h dark photoperiod. Heading date was noted as the number of days after transplanting when the spike fully emerged from the boot (Zadoks 60, Zadoks et al. 1974). The experiment was ended 175 days after planting.
Greenhouse
After the vernalization treatment described above, plants were grown at greenhouse facilities at UCD (four replications) and NCSU (up to six replications) in a completely randomized design. Supplemental lighting was used to provide 16-h light/8-h dark photoperiod. During the growth period, the minimum temperatures during the night varied from 10 to 18 °C and the maximum temperatures during the day ranged from 21 to 32 °C. The experiment was ended 210 days after planting. Heading date is reported as the number of days after transplanting when the spike fully emerged from the boot (Zadoks 60, Zadoks et al. 1974).
Field
Three grams of seed of each entry were sown in a single 1.5 m row at the NCSU Research Farm at Raleigh, NC (35.73°N, 78.68°W; elevation: 116.5 m) on October 17, 2015. Heading date was recorded as the number days after January 1st, when 50% of the plants in a plot were at Zadoks 60 growth stage.
Results
The RIP3 region at the first intron of VRN-A1 is associated with differences in flowering time
To determine the effect of the two VRN-A1 RIP3 natural alleles on heading time, we intercrossed TDC (1_SNP allele) with CS5402 (3_SNPs allele). These two lines have a single VRN-A1 copy encoding identical proteins. We generated an F2 population of 142 plants segregating for these haplotypes, genotyped them for the RIP3 alleles and recorded heading time under three different vernalization treatments (no vernalization, 3 weeks vernalization and 7 weeks vernalization).
In the absence of vernalization, the parental line CS5402 headed 30.2 days earlier than TDC, whereas F2 plants homozygous for the 3_SNPs haplotype flowered 16.9 days earlier than plants homozygous for the 1_SNP haplotype (P < 0.0001, Fig. 1a). When plants were vernalized for 3 weeks, the difference in heading time between parental lines was reduced to 12 days (TDC later than CS5402) and those between F2 homozygous plants to 11.3 days (1_SNP haplotype later than 3_SNPs haplotype, P < 0.0001, Fig. 1b). After 7 weeks of vernalization, the difference in heading time between the parental lines was reduced to only 2 days (TDC later than CS5402, P = 0.0128) and those between homozygous F2 plants for the two RIP3 homozygous plants were no longer significant (0.9 d, P = 0.0578). These results show that there are significant differences in heading time linked to polymorphisms at the RIP3 intronic region.
Difference in heading time between TDC (1_SNP) and CS5402 (3_SNPs) and between F2 plants homozygous for the RIP3 haplotypes. Data represents heading time means of at least eight plants per genotype under three different treatments (no vernalization and 3 or 7 weeks of vernalization). Different letters indicate significant differences (Tukey’s test P < 0.05)
Flowering time differences are linked to differences in VRN-A1 expression levels
To study the effects of RIP3 polymorphisms on VRN-A1 transcript levels, we sampled plants from the F2 population homozygous for the 1_SNP and 3_SNPs haplotypes at different time points of the partial vernalization treatment. Leaf samples were collected from 3-week-old plants immediately before vernalization (3w), 48 h after the plants were transfer to 4 °C (48 h), after 1 and 3 weeks of vernalization (1wV and 3wV) and 3 weeks after the plants were returned to room temperature (3wR).
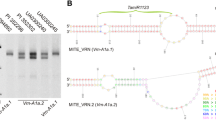
Since two alternative splice variants of VRN-A1 were detected in the study of Xiao et al. (2014), we designed specific primers to amplify each variant separately. The long VRN-A1 transcript variant (henceforth, long variant) encodes the complete gene, whereas the short VRN-A1 transcript variant (600 bp, henceforth, short variant) includes the complete first exon (185 bp) and a small portion of the first intron. The short transcript ends a few base pairs downstream of the RIP3 region, located 2,767 bp downstream of the VRN-A1 start codon in TDC (Fig. 2a). Figure 2b presents the transcript levels of the long variant, Fig. 2c the short variant, and Fig. 2d the ratio between the two splice variants.
Transcript levels of VRN-A1 alternative splice variants during vernalization. Transcript levels of VRN-A1 alternative splice variants were studied in F2 plants homozygous for the two RIP3 haplotypes. Leaf samples were collected from 3-week-old plants immediately before vernalization (3w), 48 h after the plants were transfer to 4 °C (48 h), after 1 and 3 weeks of vernalization (1 and 3wV) and 3 weeks after the plants were returned to room temperature (3wR). (A) Schematic representation of the different VRNA1 transcripts studied. Arrowheads indicate regions complementary to the qRT-PCR primers utilized. The VRN-A1 alternative splice variant includes the RIP3 region (red). VRNA1-long correspond to the complete gene (B) and VRNA1-short to the alternative splice variant (C). Average ratios of short/long transcript versions are presented in D. Bars represent means of four biological replications and error bars correspond to SEM (* P < 0.05)
For the VRN-A1 long variant, the transcript levels of the F2 plants carrying the 3_SNPs haplotype were 2- to 13-fold larger than those of plants carrying the 1_SNP haplotype. However, the differences were not significant at any of the time points or in the combined repeated measurements ANOVA (P = 0.18, Fig. 2b).
For the VRN-A1 short variant, transcript levels of the F2 plants carrying the 3_SNPs haplotype were significantly higher than those of plants carrying the 1_SNP haplotype in the combined repeated measures ANOVA (P = 0.0003). However, for the individual time points the difference between haplotypes was significant only at 3 weeks after vernalization (P = 0.0177, Fig. 2c 3wV).
The ratio between the short and long VRN-A1 variants showed a decrease during and after vernalization (Fig. 2d). This was the result of faster increases of the long variant relative to the short variant (Fig. 2d). After vernalization, when the plants were returned to room temperature, the short/long variant ratio was from 40- to 60-fold smaller than the same ratio before vernalization (Fig. 2d), suggesting a decreasing importance of the short variant at this time point. Before vernalization, the ratio between the short and long VRN-A1 variants was three times higher in the plants carrying the 1_SNP RIP3 haplotype than in those carrying the 3_SNPs haplotype (Fig. 2d, P = 0.0462). No significant differences between haplotypes were detected for the other time points (Fig. 2d).
Frequency and effect of the RIP3 haplotypes on heading time in a winter wheat panel
Based on the observed effect of the RIP3 haplotypes on heading time in the F2 population, we decided to study their effect in a panel of 127 winter lines from diverse geographical origins. The 1_SNP haplotype was found in 90.5% of the accessions whereas the 3_SNPs haplotype was found in only 9.5% of the accessions. A principal component analysis showed that lines with the 3_SNPs haplotype tend to cluster together in two separate groups of Asian or European origin (Fig. 3, Supplemental Table S2).
Principal component analysis (PCA) of the winter panel used in this study. The PCA was based on 4483 SNP markers. Colors indicate geographical origin and RIP3 genotypes are indicated by circles (1_SNP) or open diamonds (3_SNPs). a First (PC1) and second (PC2) principal components. b First (PC1) and third (PC3) principal components
To study the effect of the RIP3 haplotypes on heading time the winter wheat panel was grown in three independent experiments with partial vernalization conditions and one field experiment. The first experiment was conducted in a grown chamber, where plants were first exposed to 3 weeks of vernalization and then moved to room temperature conditions. In two additional experiments performed at the University of California Davis (UCD) and North Carolina State University (NCSU), plants were transferred to greenhouses after 3 weeks of vernalization. The final experiment was conducted under field conditions in Raleigh, North Carolina. In the three experiments grown under controlled-environments, plants carrying the 3_SNPs haplotype flowered significantly earlier (36–54 days, P < 0.0001) than plants with the 1_SNP haplotype (Fig. 4a–c; Table 2). In the field experiment, plants with the 3_SNPs haplotype flowered 21 days earlier than the plants with the 1_SNP haplotype (Fig. 4d; Table 2).
Heading time differences between RIP3 haplotypes in a winter wheat panel. Independent experiments were conducted under a–c control environmental conditions (3-week vernalization, or d) under natural vernalization in the field (Raleigh, NC). a, b Plants transferred to greenhouses at UCD and NCSU after 3-weeks vernalization. c Plants were vernalized in a growth chamber at 4 °C for 3 weeks and then temperature settings were switched to warm conditions (20/18 °C day/night) until heading
Variation in VRN-A1 copy number
To study the effect of VRN-A1 copy number variation on heading time we characterized all lines in the winter wheat panel with a TaqMan assay developed by Diaz et al. (2012). VRN-A1 copy number varied from one (10.2% of the lines) to four copies (3.9% of the lines), with most lines carrying two (35.4%) or three copies (50.4%). Most of the winter lines with a single VRN-A1 copy have the RIP3 3_SNPs haplotype. The only exception was PI 627798, which has a single VRN-A1 copy and the 1_SNP haplotype (same as TDC). PI 627798 heading time was more similar to plants with the 1_SNP haplotype and multiple VRN-A1 copies, than to plants with the 3_SNPs haplotype and a single VRN-A1 copy. Compared with the plants in the last class, PI 627798 was the latest flowering in two experiments (Fig. 4a, d) and the second latest in the remaining one (Fig. 4c, not tested in b). These results are consistent with the late flowering of the plants carrying the 1_SNP haplotype in the F2 population.
We detected no significant differences in heading time among varieties carrying two, three or four copies of VRN-A1 in any of the three pairwise comparisons. Taken together, these results suggest that VRN-A1 copy number variation has limited effect on heading time under the conditions used in our four experiments.
Discussion
VRN-A1 is linked to differences in vernalization requirement in winter wheat
The length of the vernalization period required to saturate the acceleration of flowering varies widely among winter wheat varieties. Some varieties reach the saturation point after only 3 weeks of vernalization (sometimes called ‘facultative’ types), but most varieties require approximately 6 weeks of vernalization to reach this point. In some exceptional cases, vernalization treatments of up to 8 weeks are necessary to saturate the acceleration of flowering (Brooking 1996; Kosner and Pankova 2002).
The genetic factors controlling differences in vernalization requirement among winter varieties are not as well understood as those controlling differences between winter and spring varieties (Distelfeld et al. 2009a). However, studies that genetically mapped genes controlling heading time in winter wheat using partial vernalization treatments (3–4 weeks of cold treatment) found that at least part of these differences were linked to the VRN-A1 locus (Diaz et al. 2012; Li et al. 2013). A similar conclusion is supported by this study.
One limitation of linkage studies using small segregating populations is that they cannot rule out the possibility of other linked SNP or genes affecting the trait. This is particularly critical for the VRN-A1 locus that is tightly linked with the PHYC gene (0.02 cM, Yan et al. 2003). PHYC mutants affect heading time in wheat (Chen et al. 2014) and natural variation in this gene is associated with variation in flowering time in Arabidopsis (Balasubramanian et al. 2006) and pearl millet (Saïdou et al. 2014). Fortunately, Li et al. (2013) were able to find recombination events between VRN-A1 and PHYC that demonstrated that the differences in vernalization requirement between their parental lines ‘Jagger’ and ‘2174’ were linked to VRN-A1 and not to PHYC.
Different polymorphisms in VRN-A1 may contribute to flowering time differences
Although there is agreement on the contribution of VRN-A1 to the differences in vernalization requirement among winter wheats, there is no agreement on the current interpretation of the causal polymorphisms. Diaz et al. (2012) suggested that the differences in heading time linked to VRN-A1 were caused by differences in VRN-A1 copy number among Claire (1 copy), Malacca (2 copies) and Hereward (3 copies). However, the F2 population they generated from the cross between Claire and Hereward segregated also for the Ala/Val polymorphism at position 180 of the VRN-A1 protein and for the 3_SNPs/1_SNP polymorphism in the RIP3 site of the first intron, complicating the interpretation of these results.
Diaz et al. (2012) also analyzed a double haploid population from the cross between Malacca and Hereward, and found that the plants carrying three VRN-A1 copies tended to head later than the plants carrying two VRN-A1 copies after 4 weeks of vernalization. In this population, both parental lines have the valine residue at position 180 and the 1_SNP haplotype at the RIP3 site, increasing the chances that the observed differences in heading time were caused by the differences in VRN-A1 copy number. Similarly, Guedira et al. (2016) observed in a RIL population from a cross between cultivars 26R61 and AGS 2000, both having the Val 180, that after 2 or 4 weeks of vernalization lines having three VRN-A1 copies flowered later than those having two VRN-A1 copies. However, it is not possible to rule out completely the effect of a linked gene given the small size of these segregating populations.
Li et al. (2013) proposed that the early heading time observed after partial vernalization of the plants carrying the Jagger VRN-A1 allele (Ala 180) relative to 2174 (Val 180) were caused by different amino acid residues at position 180. However, these two winter wheat varieties differed also in VRN-A1 copy number (Jagger one VRNA1 copy vs. 2174 two VRNA1 copies) and the RIP3 haplotype in the first intron (Jagger 3_SNPs vs. 2174 1_SNP), complicating the interpretation of the results.
The VRN-A1 allele in CS5402 is almost identical to the alleles present in Jagger and Claire, all carrying a single VRN-A1 copy, the 3_SNPs RIP3 haplotype and Ala 180. Therefore, it is likely that the contrasting RIP3 haplotypes segregating in the Jagger (3_SNPs) × 2174 (1_SNP) (Li et al. 2013) and Claire (3_SNPs) × Hereward (1_SNP) (Diaz et al. 2012) populations could have contributed to the differences in heading time observed after partial vernalization in these studies. This conclusion, does not rule out the possibility that the polymorphisms at position 180 or the differences in copy number could have also contributed to the observed differences in heading time in the previous studies.
The plants from the F2 population segregating for the RIP3 haplotypes have a single VRN-A1 copy encoding identical proteins, but differ in VRN-A1 transcript levels. Therefore, polymorphisms at the VRN-A1 regulatory regions are good candidates to explain the differences in heading time linked to this gene. A comparison of the VRN-A1 promoter regions (2254 bp upstream from the start codon) from the 1_SNP and 3_SNPs haplotypes revealed no-polymorphisms in the first 436 bp (Supplemental Figure S1). The rest of the promoter region (437–2254 bp) showed seven SNPs and four indels (1–2 bp), but none of them were located within known regulatory elements (Pidal et al. 2009; Kane et al. 2007; Li and Dubcovsky 2008; Li et al. 2015), predicted binding sites of transcription factors, or evolutionary conserved regions (Supplemental Figure S1). By contrast, the RIP3 polymorphisms have been shown to affect the binding of GRP2 proteins to the pre-mVRN-A1 transcripts (Kippes et al. 2015), a result consistent with the differences in relative abundance of alternative splice variants described in the following section.
Differences in VRN-A1 expression are consistent with the proposed RIP3/GRP2 molecular mechanisms
Diaz et al. (2012) observed faster and higher VRN-A1 transcript levels in Claire (3_SNPs haplotype) than in the two varieties carrying the 1_SNP haplotype. Re-analysis of the expression data from Li et al. (2013) using a two-way ANOVA with time (3 weeks and 6 weeks) and genotypes (Jagger and 2174) as factors, revealed higher VRN-A1 transcript levels in Jagger (3_SNPs) than in 2174 (1_SNP, P < 0.0001, Supplemental Table S2). These results are similar to the ones presented here, and suggest that at least part of the differences in heading time between these two VRN-A1 alleles are regulated at the transcriptional level.
We have recently shown that the natural polymorphisms found in the 3_SNPs haplotype result in reduced binding of the GRP2 protein to the RIP3 site in the VRN-A1 first intron (Kippes et al. 2015). GRP2 has been previously shown to be a repressor of flowering that binds the pre-mVRN-A1 transcript (Xiao et al. 2014). During vernalization, GRP2 is O-GlcNAc modified and its levels in the nucleus decrease, allowing higher VRN1 mRNA accumulation (Xiao et al. 2014).
Differences in the speed and processivness of transcription affects the selection of alternative splicing sites (de la Mata et al. 2003). Therefore, the stronger binding of the GRP2 protein to the translated RIP3 sites with the 1_SNP haplotype may favor the VRN-A1 short alternative splice variant, first described by Xiao et al. (2014). We detected in silico a short alternative splice variant for VRN-B1, which showed the same structure as the one described for VRN-A1 (http://plants.ensembl.org, TGACv1, Traes_5BL_89636D032.1). However, we did not find this short variant in the D genome of Chinese Spring, a result that is consistent with a deletion encompassing the RIP3 site in the first intron of VRN-D1.
The binding of the GRP2 protein to the pre-mRNA RIP3 site may explain the significantly higher short/long variant ratio observed before vernalization in the F2 plants carrying the 1_SNP haplotype relative to those carrying the 3_SNPs haplotype in this study (Fig. 2d). The reduced GRP2 levels during and after vernalization would favor the long transcript variant and explain the decrease in the short/long variant ratio in these later time points. The higher relative abundance of the short splice variant before vernalization may contribute to maintain low levels of functional VRN-A1 until the vernalization requirement is satisfied. Alternatively, the 155 amino acids encoded by the short variant (including the MADS-box domain MADS_MEF2_like, cd00265) may interact with other MADS-Box proteins altering their function. Transgenic experiments overexpressing this short variant will be required to test this hypothesis. Once the role of these alternative splice variants is better understood, it may provide an additional entry point to modulate the vernalization requirement in wheat.
Based on the previous results and discussion, we favor the hypothesis that the polymorphisms in the RIP3 region are responsible for the differences in VRN-A1 transcript levels and heading time between the lines carrying the 1_SNP and 3_SNPs haplotypes. However, we cannot rule the possibility of effects caused by linked polymorphisms in the promoter region or outside the sequenced region.
Winter wheat varieties carrying the 3_SNPs haplotype were detected at low frequencies
A recent study of 1,100 winter wheat lines from different regions of the world (with emphasis in European varieties) found a single VRN-A1 copy in only 7% of the varieties (Würschum et al. 2015). This percentage is similar to the 10.2% of varieties with a single VRN-A1 copy found in this study. Unfortunately, Würschum et al. (2015) did not have information about the RIP3 haplotypes.
Since 92.3% of the accessions with a single VRN-A1 copy in our winter panel have the 3_SNPs haplotype, we will assume for the following discussion that in the winter panel from Würschum et al. (2015) most of the accessions carrying a single VRN-A1 copy also carry the 3_SNPs allele. Würschum et al. (2015) found that the frequency of the varieties with a single VRN-A1 copy was larger in Southern Europe and the UK where the winters are milder (Würschum et al. 2015). This distribution is consistent with the milder vernalization requirement of the varieties carrying the 3_SNPs haplotype. We present the geographical origins of 12 accessions with 3_SNPs haplotypes detected in this study in Supplemental Table S4, but due to the small sample size, it is difficult to draw any solid conclusion.
The only accession in this survey with one VRN-A1 copy and the 1_SNP haplotype (PI 627798) was the latest or second latest flowering line when compared with the accessions with a single VRN-A1 copy and the 3_SNPs haplotype, and within the range of the accession with multiple VRN-A1 copies and the 1_SNP haplotype. The late flowering of PI 627798 is also consistent with the late flowering of the plants carrying the 1_SNP haplotype in our F2 population, which suggests that the RIP3 haplotypes correlate better with heading time in winter wheat than the differences in VRN-A1 copy number.
VRN-A1 copy number variants within the 1_SNP haplotype showed limited association with differences in heading time
Once we removed the effect of the 3_SNPs haplotype, we did not detect differences in heading time among the winter wheat varieties with two, three or four VRN-A1 copies in any of the field or controlled environment experiments with partial vernalization (Fig. 5). Even the single variety found with one VRN-A1 copy with the 1_SNP haplotype (PI 627798) flowered within the range of the varieties with multiple VRN-A1 copies.
Heading time differences among winter wheats with different VRN-A1 copy number. The relative copy number and VRN-A1 haplotypes are described in supplemental Table S4. Lines were grouped based on the estimated VRN-A1 copy number and average heading times among groups were compared using a Tukey’s test. Different letters above the box-plots indicate significant differences among groups (P < 0.05)
The study by Würschum et al. (2015) also failed to detect differences in heading time among varieties with two, three or four VRN-A1 copies. However, their heading time studies were performed under field conditions in Germany (> 48°N), where winter conditions were likely sufficient to satisfy completely the vernalization requirement. We cannot rule out a role of VRN-A1 copy number under different conditions. In fact, in a double haploid population from the cross between Malacca (two VRN-A1 copies) and Hereward (three VRN-A1 copies) subjected to partial vernalization, Diaz et al. (2012) showed that the lines with three VRN-A1 copies flowered later than those with two copies. Guedira et al. (2016) observed that RILs from the AGS 2000 × 26R61 RIL population with two VRN-A1 copies flowered earlier than the lines with three copies when the population was grown in the field at locations with mild winters in the southeastern United States. It would be interesting to test if the same effect can be detected in other biparental populations segregating for VRN-A1 copy number variants.
Although the frequencies of varieties with different VRN-A1 copy number alleles in different geographical regions suggest a possible adaptive role, additional studies using isogenic lines or biparental populations will be necessary to quantify better the effect of the number of VRN-A1 copies on the adaptation to different environments. Adaptation to these environments may depend not only on the effect of the different VRN-A1 copy number variants on heading time, but also in their interactions with FR-A2 alleles for frost tolerance. Zhu et al. (2014) showed that winter wheat varieties carrying three VRN-A1 copies were more frost tolerant than varieties with two VRN-A1 copies when the FROST TOLERANCE 2 allele T (FR-A2-T) was present.
Conclusions and practical applications
It is still not clear if the relatively low frequency of the 3_SNPs allele in the Western breeding programs is a result of its recent introduction or the effect of a narrow adaptive value, limited to a small range of environments. If the 3_SNPs allele introgression happened recently, it is possible that this VRN-A1 allele has not reached yet its optimal frequency. It would be interesting to test the value of this allele in modern winter wheat breeding programs, particularly in regions with mild winters. It is also possible, that as global temperatures increase and winters become milder, the 3_SNPs allele will became more valuable in certain regions.
The presence of the 3_SNPs haplotype in the successful winter wheat variety Jagger may represent an example of the potential of this allele. Jagger was grown in more than 25% of the Oklahoma acreage for 11 years and was the dominant wheat variety in Kansas between 1998 and 2010 (USDA/NASS Oklahoma Field Office wheat.okstate.edu and kswheat.com). Although we do not know how much the 3_SNPs VRN-A1 haplotype contributed to Jagger success, it would be interesting to characterize the presence of this allele in the multiple varieties derived from Jagger. Additionally, it will be informative to monitor the changes in the 3_SNPs allele frequency as new varieties are released in this region.
In summary, we have shown a significant effect of the RIP3 haplotypes on wheat heading times, both under controlled environments with partial vernalization and in field experiments. Our results and those from Würschum et al. (2015) suggest that one VRN-A1 copy with the 3_SNPs haplotype may have an adaptive value in regions with mild winters. The confirmation of the role of the 3_SNPs allele on heading time and in the modulation of the vernalization requirement can provide winter wheat breeders new genetic tools to improve wheat adaptation to new or changing environments.
Abbreviations
- FT1 :
-
FLOWERING LOCUS T1
- GRP2:
-
GLYCINE RICH RNA-BINDING PROTEIN 2
- RIP3:
-
RNA immune precipitation fragment 3
- SNP:
-
Single nucleotide polymorphism
- VRN1 :
-
VERNALIZATION1 gene
- VRN2 :
-
VERNALIZATION2 gene
- VRN3 :
-
VERNALIZATION3 gene
- VRN-D4 :
-
VERNALIZATION4 gene D genome
References
Balasubramanian S, Sureshkumar S, Agrawal M et al (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38:711–715
Bonman JM, Babiker EM, Cuesta-Marcos A, Esvelt-Klos K, Brown-Guedira G, Chao S, See D, Chen J, Akhunov E, Zhang J, Bockelman HE (2015) Genetic diversity among wheat accessions from the USDA National Small Grains Collection. Crop Sci 55:1243–1253
Brooking IR (1996) Temperature response of vernalization in wheat: A developmental analysis. Ann Bot London 78:507–512
Cang AF, Wilson AA, Wiens JJ (2016) Climate change is projected to outpace rates of niche change in grasses. Biol Lett 12:20160368
Chen A, Dubcovsky J (2012) Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet 8:e1003134
Chen A, Li C, Hu W, Lau M, Lin H, Rockwell NC, Martin SS, Jernstedt JA, Lagarias JC, Dubcovsky J (2014) PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long days. Proc Natl Acad Sci USA 111:10037–10044
Chu CG, Tan CT, Yu GT, Zhong S, Xu SS, Yan L (2011) A novel retrotransposon inserted in the dominant Vrn-B1 allele confers spring growth habit in tetraploid wheat (Triticum turgidum L.). G3-Genes Genom Genet 1:637–645
Cook BI, Wolkovich EM, Davies JT, Ault TR, Betancourt JL, Allen JM, Bolmgren K, Cleland EE, Crimmins TM, Kraft NJB, Lancaster LT, Mazer SJ, McCabe GJ, McGill BJ, Parmesan C, Pau S, Regetz J, Salamin N, Schwartz MD, Travers SE (2012) Sensitivity of spring phenology to warming across temporal and spatial climate gradients in two independent databases. Ecosystems 15:1283
Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distan ce signaling in floral induction of Arabidopsis. Science 316:1030–1033
de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR (2003) A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12:525–532
Diaz A, Zikhali M, Turner AS, Isaac P, Laurie DA (2012) Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). Plos One 7:e33234
Distelfeld A, Li C, Dubcovsky J (2009a) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184
Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J (2009b) Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol 149:245–257
Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L (2006) Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol 60:469–480
FAOSTAT (2015) Food and Agriculture Organization (FAO Statistical Pocketbook 2015, http://www.fao.org/3/a-i4691e.pdf)
Franks SJ, Sim S, Weis AE (2007) Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci USA 104:1278–1282
Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65
Guedira M, Xiong M, Hao YF, Johnson J, Harrison S, Marshall D, Brown-Guedira G, Zhang A (2016) Heading Date QTL in Winter Wheat (Triticum aestivum L.) Coincide with Major Developmental Genes VERNALIZATION1 and PHOTOPERIOD1. PLoS One 11(5):e0154242
Kane NA, Agharbaoui Z, Diallo AO, Adam H, Tominaga Y, Ouellet F, Sarhan F (2007) TaVRT2 represses transcription of the wheat vernalization gene TaVRN1. Plant J 51:670–680
Kippes N, Zhu J, Chen A, Vanzetti L, Lukaszewski A, Nishida H, Kato K, Dvorak J, Dubcovsky J (2014) Fine mapping and epistatic interactions of the vernalization gene VRN-D4 in hexaploid wheat. Mol Genet Genomics 289:47–62
Kippes N, Debernardi J, Vasquez-Gross HA, Akpinar BA, Budak H, Kato K, Chao S, Akhunov E, Dubcovsky J (2015) Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc Natl Acad Sci USA 112:E5401-E5410
Kippes N, Chen A, Zhang X, Lukaszewski AJ, Dubcovsky J (2016) Development and characterization of a spring hexaploid wheat line with no functional VRN2 genes. Theor Appl Genet 129:1417–1428
Konopatskaia I, Vavilova V, Kondratenko EY, Blinov A, Goncharov NP (2016) VRN1 genes variability in tetraploid wheat species with a spring growth habit. BMC Plant Biol S3:2
Kosner J, Pankova K (2002) Vernalisation response of some winter wheat cultivars (Triticum aestivum L.). Czech J Genet Plant Breed 38:97–103
Li C, Dubcovsky J (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55:543–554. https://doi.org/10.1111/j.1365-313X.2008.03526.x
Li G, Yu M, Fang T, Cao S, Carver BF, Yan L (2013) Vernalization requirement duration in winter wheat is controlled by TaVRN-A1 at the protein level. Plant J 76:742–753
Li C, Lin H, Dubcovsky J (2015) Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J 84:70–82
Liu B, Asseng S, Müller C, Ewert F, Elliott J, Lobell D, Martre P, Ruane A, Wallach D, Jones J, Rosenzweig C, Aggarwal P, Alderman P, Anothai J, Basso B, Biernath C, Cammarano D, Challinor A, Deryng D, Sanctis G, Doltra J, Fereres E, Folberth C, Garcia-Vila M, Gayler S, Hoogenboom G, Hunt L, Izaurralde R, Jabloun M, Jones C, Kersebaum K, Kimball B, Koehler A, Kumar S, Nendel C, O’Leary G, Olesen J, Ottman M, Palosuo T, Prasad P, Priesack E, Pugh T, Reynolds M, Rezaei E, Rötter R, Schmid E, Semenov M, Shcherbak I, Stehfest E, Stöckle C, Stratonovitch P, Streck T, Supit I, Tao F, Thorburn P, Waha K, Wall G, Wang E, White J, Wolf J, Zhao Z, Zhu Y (2016) Similar estimates of temperature impacts on global wheat yield by three independent methods. Nat Clim Change 6:1130–1136
Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol 138:2364–2373
Muterko A, Balashova I, Cockram J, Kalendar R, Sivolap Y (2015) The new wheat vernalization response allele Vrn-D1s is caused by DNA transposon insertion in the first intron. Plant Mol Biol Rep 33:294–303
Muterko A, Kalendar R, Salina E (2016) Novel alleles of the VERNALIZATION1 genes in wheat are associated with modulation of DNA curvature and flexibility in the promoter region. BMC Plant Biol 16:9
Pidal B, Yan L, Fu D, Zhang F, Tranquilli G, Dubcovsky J (2009) The CArG-box in the promoter region of wheat vernalization gene VRN1 is not necessary to mediate the vernalization response. J Hered 100:355–364
Pugsley AT (1971) A genetic analysis of the spring-winter habit of growth in wheat. Aust J Agric Res 22:21–31
Pugsley AT (1972) Additional genes inhibiting winter habit in wheat. Euphytica 21:547–552
Saïdou AA, Clotault J, Couderc M, Mariac C, Devos KM, Thuillet AC, Amoukou IA, Vigouroux Y (2014) Association mapping, patterns of linkage disequilibrium and selection in the vicinity of the PHYTOCHROME C gene in pearl millet. Theor Appl Genet 127:19–32
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036
Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476:332–335
Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100:13099–13104
Woods D, Mckeown M, Dong Y, Preston JC, Amasino RM (2016) Evolution of VRN2/GhD7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol 170:2124–2135
Würschum T, Boeven PHG, Langer SM, Longin CFH, Leiser WL (2015) Multiply to conquer: Copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genet 16:96
Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, Gao C, Chong K (2014) O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun 5:4572
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004a) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109:1677–1686
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004b) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586
Zadoks J, Chang T, Konzak C (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Zhu J, Pearce S, Burke A, See DR, Skinner DZ, Dubcovsky J, Campbell KG (2014) Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor Appl Genet 127:1183–1197
Funding
This project was supported by the Agriculture and Food Research Initiative Competitive Grants 2016-67013-24617 and 2017-67007-25939 (WheatCAP) from the USDA National Institute of Food and Agriculture and by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kippes, N., Guedira, M., Lin, L. et al. Single nucleotide polymorphisms in a regulatory site of VRN-A1 first intron are associated with differences in vernalization requirement in winter wheat. Mol Genet Genomics 293, 1231–1243 (2018). https://doi.org/10.1007/s00438-018-1455-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1455-0