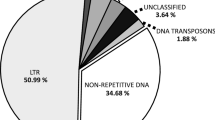
Abstract
Satellite DNA (satDNA) is a major component of the heterochromatic regions of eukaryote genomes and usually shows a high evolutionary dynamic, even among closely related species. Section Arachis (genus Arachis) is composed of species belonging to six different genomes (A, B, D, F, G and K). The most distinguishing features among these genomes are the amount and distribution of the heterochromatin in the karyotypes. With the objective of gaining insight into the sequence composition and evolutionary dynamics of the heterochromatin fraction in Arachis, we investigated here the sequence diversity, genomic abundance, and chromosomal distribution of a satDNA family (ATR-2) among seven diploid species of section Arachis. All of the isolated sequences were AT-rich and highly conserved at both intraspecific and interspecific levels, without any species-specific polymorphism. Pairwise comparisons of isolated ATR-2 monomers revealed that most of the nucleotide sites were in the first two transitional stages of Strachan’s model. However, the abundance of ATR-2 was significantly different among genomes according to the ‘library hypothesis’. Fluorescent in situ hybridization revealed that ATR-2 is a main component of the DAPI+ centromeric heterochromatin of the A, F, and K genomes. Thus, the evolution of the different heterochromatin patterns observed in Arachis genomes can be explained, at least in part, by the differential representation of ATR-2 among the different species or even among the chromosomes of the same complement. These findings are the first to demonstrate the participation of satDNA sequences in the karyotype diversification of wild diploid Arachis species.




Similar content being viewed by others
References
Ansari HA, Ellison NW, Griffiths AG, Williams WM (2004) A lineage-specific centromeric satellite sequence in the genus Trifolium. Chromosome Res 12:357–367
Bachmann L, Sperlich D (1993) Gradual evolution of a specific satellite DNA family in Drosophila ambigua, D. tristis and D. obscura. Mol Biol Evol 10:647–659
Bechara MD, Moretzsohn MC, Palmieri DA, Monteiro JP, Bacci M Jr, Martins J Jr, Valls JFM, Lopes CR, Gimenes MA (2010) Phylogenetic relationships in genus Arachis based on ITS and 5.8S rDNA sequences. BMC Plant Biol 10:255
Bertioli DJ, Vidigal B, Nielen S, Ratnaparkhe MB, Lee T-H, Leal-Bertioli SCM, Kim C, Guimarães PM, Seijo G, Schwarzacher T, Paterson AH, Heslop-Harrison P, Araujo ACG (2013) The repetitive component of the A genome of peanut (Arachis hypogaea) and its role in remodelling intergenic sequence space since its evolutionary divergence from the B genome. Ann Bot 112:545–559
Biscotti MA, Olmo E, Heslop-Harrison JP (2015) Repetitive DNA in eukaryotic genomes. Chromosome Res 23:415–420
Blattes R, Monod C, Susbielle G, Cuvier O, Wu JH, Hsieh TS et al (2006) Displacement of D1, HP1 and topoisomerase II from satellite heterochromatin by a specific polyamide. EMBO J 25:2397–2408
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Čížková J, Hřibová E, Humplíková L, Christelová P, Suchánková P, Doležel J (2013) Molecular analysis and genomic organization of major DNA satellites in banana (Musa spp.). PLoS One 8:e54808
De Jong JH, Fransz P, Zabel P (1999) High resolution FISH in plants—techniques and applications. Trends Plant Sci 4:258–263
Dover G (1982) Molecular drive a cohesive mode of species evolution. Nature 299:111–117
Dover G (2002) Molecular drive. Trends Genet 18:587–589
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fernandez A, Krapovickas A (1994) Cromosomas y evolución en Arachis (Leguminosae). Bonplandia 8:187–220
Ferree PM, Prasad S (2012) How can satellite DNA divergence cause reproductive isolation? Let us count the chromosomal ways. Genet Res Int 2012:430136
Flavell RB (1986) Repetitive DNA and chromosome evolution in plants. Phil Trans R Soc Lond B 312:227–242
Friend SA, Quandt D, Tallury SP, Stalker HT, HiluK W (2010) Species, genomes, and section relationships in the genus Arachis (Fabaceae): a molecular phylogeny. Plant Syst Evol 290:185–199
Fry K, Salser W (1977) Nucleotide sequence of HS—a satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 12:1069–1084
Garrido-Ramos MA, de la Herran R, Ruiz-Rejon M, Ruiz- Rejon C (1999) A subtelomeric satellite DNA family isolated from the genome of the dioecious plant Silene latifolia. Genome 42:442–446
Gindullis F, Desel C, Galasso I, Schmidt T (2001) The large-scale organization of the centromeric region in Beta species. Genome Res 11:253–265
Glinka S, De Lorenzo D, Stephan W (2006) Evidence of gene conversion associated with a selective sweep in Drosophila melanogaster. Mol Biol Evol 23:1869–1878
Grabiele M, Chalup L, Robledo G, Seijo G (2012) Genetic and geographic origin of domesticated peanut as evidenced by 5S rDNA and chloroplast DNA sequences. Plant Syst Evol 298:1151–1165
Grattapaglia D, Sederoff R (1994) Genetic linkage Maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudotestcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Gregory MP, Gregory WC (1979) Exotic germoplasm of Arachis L. interspecific hybrids. J Hered 70:185–193
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Heslop-Harrison JS, Brandes A, Schwarzacher T (2003) Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species. Chromosome Res 11:241–253
Hizume M, Shibata F, Matsusaki Y, Garajova Z (2002) Chromosome identification and comparative karyotypic analyses of four Pinus species. Theor Appl Genet 105:491–497
Horvath JE, Willard HF (2007) Primate comparative genomics: lemur biology and evolution. Trends Genet 23:173–182
Hudakova S, Michalek W, Presting GG et al (2001) Sequence organization of barley centromeres. Nucleic Acids Res 29:5029–5035
Husted L (1936) Cytological studies of peanut, Arachis. II Chromosome number, morphology and behavior and their application to the origin of cultivated forms. Cytologia 7:396–423
Koukalova B, Moraes AP, Renny-Byfield S, Matyasek R, Leitch AR, Kovarik A (2010) Fall and rise of satellite repeats in allopolyploids of Nicotiana over c. 5 million years. New Phytol 186:148–160
Krapovickas A, Gregory WC (1994) Taxonomía del género Arachis (Leguminosae). Bonplandia 8:1–186
Kuhn GCS, Sene F, Moreira-Filho O, Schwarzacher T, Heslop-Harrison JS (2008) Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Res 16:307–324
Kurbis S, Schimdt T, Heslop Harrison JS (1998) Repetitive DNA elements as major componenet of plant genomes. Ann Bot 82:45–55
Langdon T, Seago C, Jones RN, Ougham H, Thomas H, Forster JW, Jenkins G (2000) De Novo evolution of satellite DNA on the rye B chromosome. Genetics 154:869–884
Leal-Bertioli SC, Santos SP, Dantas KM et al (2015) Arachis batizocoi: a study of its relationship to cultivated peanut (A. hypogaea) and its potential for introgression of wild genes into the peanut crop using induced allotetraploids. Ann Bot 115:237–249
Lee HR, Zhang WL, Langdon T et al (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA 102:11793–11798
Lim KY, Matyasek R, Lichtenstein CP, Leitch AR (2000) Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma 109:245–258
Luchetti A, Cesari M, Carrara G, Cavicchi S, Passamonti M, Scali V, Mantovani B (2003) Unisexuality and molecular drive: bag320 sequence diversity in Bacillus taxa (Insecta Phasmatodea). J Mol Evol 56:587–596
Luchetti A, Marini M, Mantovani B (2006) Non-concerted evolution of the RET76 satellite DNA family in Reticulitermes taxa (Insecta, Isoptera). Genetica 128:123–132
Macas J, Pozrkovaa D, Navratilova A, Nouzova M, Neumann P (2000) Two new families of tandem repeats isolated from genus Vicia using genomic self-priming PCR. Mol Gen Genet 263:741–751
Macas J, Meszaros T, Nouzova M (2002) PlantSat: a specialized database for plant satellite repeats. Bioinformatics 18:28–35
Mantovani B (1998) Satellite sequence turnover in parthenogenetic systems: the apomictic triploid hybrid Bacillus lynceorum (Insecta, Phasmatodea). Mol Biol Evol 15:1288–1297
Melters DP, Bradnam KR, Young HA et al (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14:R10
Menzel G, Dechyeva D, Wenke T, Holtgräwe D, Weisshaar B, Schmidt T (2008) Diversity of a complex centromeric satellite and molecular characterization of dispersed sequence families in sugar beet (Beta vulgaris). Ann Bot 102:521–530
Meštrovic N, Castagnone-Sereno P, Plohl M (2006) High conservation of the differentially amplified MPA2 satellite DNA family in parthenogenetic root-knot nematodes. Gene 376:260–267
Mlinarec J, Chester M, Siljak-Yakovlev S, Papeš D, Leitch AR, Besendorfer V (2009) Molecular structure and chromosome distribution of three repetitive DNA families in Anemone hortensis L. (Ranunculaceae). Chromosome Res 17:331–346
Moretzsohn MC, Gouvea EG, Inglis PW, Leal-Bertioli SCM, Valls JFM, Bertioli DJ (2013) A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Ann Bot 111:113–126
Mravinac B, Plohl M, Mestrovic N, Ugarkovic D (2002) Sequence of PRAT satellite DNA ‘frozen’ in some Coleopteran species. J Mol Evol 54:774–783
Mravinac B, Plohl M, Ugarkovic D (2005) Preservation and high sequence conservation of satellite DNA suggest functional constraints. J Mol Evol 61:542–550
Mukai Y, Friebe B, Gill BS (1992) Comparison of C-banding patterns and in situ hybridization sites using highly repetitive and total genomic rye DNA probes of ‘Imperial’ rye chromosomes added to ‘Chinese Spring’ wheat. Jpn J Genet 67:71–83
Navajas-Pérez R, Rubio-Escudero C, Aznarte JL, Rejón MR, Garrido-Ramos MA (2007) SatDNA Analyzer: a computing tool for satellite-DNA evolutionary analysis. Bioinformatics 23:767–768
Navajas-Pérez R, Del Bosque MQ, Garrido-Ramos MA (2009) Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Mol Genet Genom 282:395–406
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nielen S, Campos-Fonseca F, Leal-Bertioli S, Guimaraes P, Seijo G, Town C, Arrial R, Bertioli D (2010) FIDEL—a retrovirus-like retrotransposon and its distinct evolutionary histories in the A and B-genome components of cultivated peanut. Chromosome Res 18:227–246
Nielen S, Vidigal B, Leal-Bertioli S, Ratnaparkhe M, Paterson A, Garsmeur O, D’Hont A, Guimaraes P, Bertioli D (2012) Matita, new retroelement from peanut: characterization and evolutionary context in the light of the Arachis A-B genome divergence. Mol Genet Genomics 287:21–38
Pérez-Gutiérrez MA, Suárez-Santiago VN, López-Flores I, Romero AT, Garrido-Ramos MA (2012) Concerted evolution of satellite DNA in Sarcocapnos: a matter of time. Plant Mol Biol 78:19–29
Pich U, Fritsch R, Schubert I (1996) Closely related Allium species (Alliaceae) share a very similar satellite sequence. Plant Syst Evol 202:255–264
Plohl M, Luchetti A, Mestrovic N, Mantovani B (2008) Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero) chromatin. Gene 409:72–82
Plohl M, Petrovic V, Luchetti A, Ricci A, Satovic E, Passamonti M, Mantovani B (2010) Long-term conservation vs high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity 104:543–551
Plohl M, Meštrović N, Mravinac B (2012) Satellite DNA evolution. Genome Dyn 7:126–152
Radic MZ, Lundgren K, Hamkalo BA (1987) Curvature of mouse satellite DNA and condensation of heterochromatin. Cell 50:1101–1108
Robledo G, Seijo JG (2008) Characterization of Arachis D genome using physical mapping of heterochromatic regions and rDNA loci by FISH. Genet Mol Biol 31:717–724
Robledo G, Seijo G (2010) Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theor Appl Genet 121:1033–1046
Robledo G, Lavia GI, Seijo G (2009) Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theor Appl Genet 118:1295–1307
Robles F, de la Herran R, Ludwig A, Ruiz-Rejon C, Ruiz-Rejon M, Garrido-Ramos MA (2004) Evolution of ancient satellite DNAs in sturgeon genomes. Gene 338:133–142
Rosato M, Galian JA, Rossello JA (2012) Amplification, contraction and genomic spread of a satellite DNA family (E180) in Medicago (Fabaceae) and allied genera. Ann Bot 109:773–782
Rudd MK, Wray GA, Willard HF (2006) The evolutionary dynamics of alpha-satellite. Genome Res 16:88–96
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Samoluk SS, Chalup L, Robledo G, Seijo JG (2015a) Genome sizes in diploid and allopolyploid Arachis L. species (section Arachis). Genet Res Crop Evol 62:747–763
Samoluk SS, Robledo G, Podio M, Chalup L, Ortiz JPA, Pessino SC, Seijo JG (2015b) First insight into divergence, representation and chromosome distribution of reverse transcriptase fragments from L1 retrotransposons in peanut and wild relative species. Genetica 143:113–125
San Miguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot 82:37–44
Schmidt T, Heslop-Harrison JS (1998) Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends Plant Sci 3:195–199
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN: a software for population genetics data analysis, version 2.000. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Seijo G, Lavia GI, Fernández A, Krapovickas A, Ducasse D, Moscone EA (2004) Physical mapping of 5S and 18S-25S rRNA genes evidences that Arachis duranensis and A. ipaensis are the wild diploid species involved in the origin of A. hypogaea (Leguminosae). Am J Bot 91:2293–2303
Seijo G, Lavia GI, Fernández A, Krapovickas A, Ducasse D, Bertioli DJ, Moscone EA (2007) Genomic relationships between the cultivated peanut (Arachis hypogaea- Leguminosae) and its close relatives revealed by double GISH. Am J Bot 94:1963–1971
Sharma S, Raina SN (2005) Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet Genome Res 109:15–26
Silvestri MC, Ortiz AM, Lavia GI (2014) rDNA loci and heterochromatin positions support a distinct genome type for ‘x = 9 species’ of section Arachis (Arachis, Leguminosae). Plant Syst Evol 301:555–562
Smartt J, Gregory WC, Gregory MP (1978) The genomes of Arachis hypogaea. 1. Cytogenetic studies of putative genome donors. Euphytica 27:665–675
Stalker HT (1991) A new species-section Arachis of peanuts with D genome. Am J Bot 78:630–637
Stalker HT, Dhesi JS, Parry DC, Hahn JH (1991) Cytological and interfertility relationships of Arachis section Arachis. Amer J Bot 78:238–246
Stephan W (1986) Recombination and the evolution of satellite DNA. Genet Res 47:167–174
Strachan T, Webb D, Dover G (1985) Transition stages of molecular drive in multiple-copy DNA families in Drosophila. EMBO J 4:1701–1708
Suárez-Santiago VN, Blanca G, Ruiz-Rejón M, Garrido-Ramos MA (2007) Satellite-DNA evolutionary patterns under a complex evolutionary scenario: the case of Acrolophus subgroup (Centaurea L., Compositae) from the western Mediterranean. Gene 404:80–92
Tallury SP, Hilu KW, Milla SR, Friend SA, Alsaghir M, Stalker HT, Quandt D (2005) Genomic affinities in Arachis section Arachis (Fabaceae): molecular and cytogenetic evidence. Theor Appl Genet 111:1229–1237
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Ugarkovic D (2005) Functional elements residing within satellite DNAs. EMBO Rep 6:1035–1039
Ugarkovic D, Plohl M (2002) Variation in satellite DNA profiles-causes and effects. EMBO J 2:5955–5959
Urdampilleta JD, de Souza AP, Schneider DR, Vanzela AL, Ferrucci MS, Martins ER (2009) Molecular and cytogenetic characterization of an AT-rich satellite DNA family in Urvillea chacoensis Hunz. (Paullinieae, Sapindaceae). Genetica 136:171–177
Valls JFM, Simpson CE (2005) New species of Arachis (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia 14:35–64
Vittorazzi SE, Lourenço LB, Recco-Pimentel SM (2014) Long-time evolution and highly dynamic satellite DNA in leptodactylid and hylodid frogs. BMC Genet 15:1–11
Zhang L, Xu C, Yu W (2012) Cloning and characterization of chromosomal markers from a Cot-1 library of peanut (Arachis hypogaea L.). Cytogenet Genome Res 137:31–41
Zhang T, Talbert PB, Zhang W, Wu Y, Yang Z, Henikoff JG et al (2013) The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc Natl Acad Sci 110:4875–4883
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2012-1875 and PICTO 2011-0260) and the Consejo Nacional de Investigaciones Científicas y Técnicas (PIP 11220120100192CO), Argentina.
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Samoluk, S.S., Robledo, G., Bertioli, D. et al. Evolutionary dynamics of an at-rich satellite DNA and its contribution to karyotype differentiation in wild diploid Arachis species. Mol Genet Genomics 292, 283–296 (2017). https://doi.org/10.1007/s00438-016-1271-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1271-3