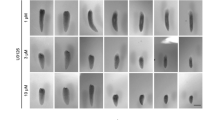
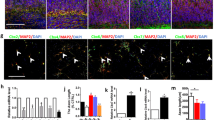
Abstract
The myosin essential light chain (ELC) is a structure component of the actomyosin cross-bridge, however, the functions in the central nervous system (CNS) development and regeneration remain poorly understood. Planarian Dugesia japonica has revealed fundamental mechanisms and unique aspects of neuroscience and neuroregeneration. In this study, the cDNA DjElc, encoding a planarian essential light chain of myosin, was identified from the planarian Dugesia japonica cDNA library. It encodes a deduced protein with highly conserved functionally domains EF-Hand and Ca2+ binding sites that shares significant similarity with other members of ELC. Whole mount in situ hybridization studies show that DjElc expressed in CNS during embryonic development and regeneration of adult planarians. Loss of function of DjElc by RNA interference during planarian regeneration inhibits brain lateral branches regeneration completely. In conclusion, these results demonstrated that DjElc is required for maintenance of neurons and neurite outgrowth, particularly for involving the brain later branch regeneration.







Similar content being viewed by others
References
Adell T, Cebria F, Salo E (2010) Gradients in planarian regeneration and homeostasis. Cold Spring Harb Perspect Biol 2:1–13
Agata K, Umesono Y (2008) Brain regeneration from pluripotent stem cells in planarian. Philos T R Soc B 363:2071–2078
Agata K, Watanabe K (1999) Molecular and cellular aspects of planarian regeneration. Semin Cell Dev Biol 10:377–383
Agata K, Tanaka T, Kobayashi C, Kato K, Saitoh Y (2003) Intercalary regeneration in planarians. Dev Dynam 226:308–316
Brown ME, Bridgman PC (2004) Myosin function in nervous and sensory systems. J Neurobiol 58:118–130
Burland TG (2000) DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol 132:71–91
Burnette DT, Lin J, Schaefer AW, Medeiros NA, Danuser G, Forscher P (2008) Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev Cell 15:163–169
Cardona A, Fernandez J, Solana J, Romero R (2005a) An in situ hybridization protocol for planarian embryos: monitoring myosin heavy chain gene expression. Dev Genes Evol 215:482–488
Cardona A, Hartenstein V, Romero R (2005b) The embryonic development of the triclad Schmidtea polychroa. Dev Genes Evol 215:109–131
Cebria F (2007) Regenerating the central nervous system: how easy for planarians! Dev Genes Evol 217:733–748
Cebria F (2008) Organization of the nervous system in the model planarian Schmidtea mediterranea: an immunocytochemical study. Neurosci Res 61:375–384
Diefenbach TJ, Latham VA, Yimlamai D, Liu CA, Herman IM, Jay DG (2002) Myosin 1c and myosin IIB serve opposing roles in lamellipodial dynamics of the neuronal growth cone. J Cell Biol 158:1207–1217
Falkenthal S, Parker VP, Mattox WW, Davidson N (1984) Drosophila melanogaster has only one myosin alkali light-chain gene which encodes a protein with considerable amino acid sequence homology to chicken myosin alkali light chains. Mol Cell Biol 4:956–965
Goodwin EB, Szent-Gyorgyi AG, Leinwand LA (1987) Cloning and characterization of the scallop essential and regulatory myosin light chain cDNAs. J Biol Chem 262:11052–11056
Handberg-Thorsager M, Fernádez E, Saló E (2008) Stem cells and regeneration in planarians. Front Biosci 13:6374–6394
Hernandez OM, Jones M, Guzman G, Szczesna-Cordary D (2007) Myosin essential light chain in health and disease. Am J Physiol-Heart C 292:H1643–H1654
Hirokawa N, Niwa S, Tanaka Y (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68:610–638
Holland LZ, Pace DA, Blink ML, Kene M, Holland ND (1995) Sequence and expression of amphioxus Alkali Myosin Light-Chain (Amphimlc-Alk) throughout development—implications for vertebrate myogenesis. Dev Biol 171:665–676
Iglesias M, Almuedo-Castillo M, Aboobaker AA, Saló E (2011) Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/β-catenin pathway. Dev Biol 358:68–78
Ikura M (1996) Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci 21:14–17
Inoue T, Kumamoto H, Okamoto K, Umesono Y, Sakai M, Alvarado AS, Agata K (2004) Morphological and functional recovery of the planarian photosensing system during head regeneration. Zool Sci 21:275–283
Kazmierczak K, Xu YY, Jones M, Guzman G, Hernandez OM, Kerrick WGL, Szczesna-Cordary D (2009) The role of the N-Terminus of the Myosin essential light chain in cardiac muscle contraction. J Mol Biol 387:706–725
Lenz S, Lohse P, Seidel U, Arnold HH (1989) The alkali light chains of human smooth and nonmuscle myosins are encoded by a single gene. Tissue-specific expression by alternative splicing pathways. J Biol Chem 264:9009–9015
Ma XF, Kawamoto S, Uribe J, Adelstein RS (2006) Function of neuron-specific alternatively spliced isoforms of nonmuscle mysoin II-B during mouse brain development. Mol Biol Cell 17:2138–2149
Martin-Duran JM, Amaya E, Romero R (2010) Germ layer specification and axial patterning in the embryonic development of the freshwater planarian Schmidtea polychroa. Dev Biol 340:145–158
Newmark PA, Alvarado AS (2002) Not your father’s planarian: a classic model enters the era of functional genomics. Nat Rev Genet 3:210–219
Newmark PA, Reddien PW, Cebria F, Alvarado AS (2003) Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci USA 100:11861–11865
Nishimura K, Kitamura Y, Inoue T, Umesono Y, Sano S, Yoshimoto K, Inden M, Takata K, Taniguchi T, Shimohama S, Agata K (2007) Reconstruction of dopaminergic neural netwrok and locomotion function in planarian regenerates. Dev Neurobiol 67:1059–1078
Nishimura K, Kitamura Y, Umesono Y, Takeuchi K, Takata K, Taniguchi T, Agata K (2008) Identification of glutamic acid decarboxylase gene and distribution of GABAergic nervous system in the planarian, Dugesia japonica. Neuroscience 153:1103–1114
Park IHC, Jin S, Lee B, Choi H, Kwon JT, Kim D, Kim J, Lifirsu E, Park WJ, Park ZY, Kim DH, Cho C (2011) Myosin regulatory light chains are required to maintain the stability of myosin II and cellular integrity. Biochem J 434:171–180
Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Alvarado AS (2009) Formaldehyde-Based whole-mount in situ hybridization method for planarians. Dev Dynam 238:443–450
Reddien PW, Alvarado AS (2004) Fundamentals of planarian regeneration. Annual Rev Cell Dev Biol 20:725–757
Sakai T, Kato K, Watanabe K, Orii H (2002) Planarian pharynx regeneration revealed by the expression of myosin heavy chain-A. Int J Dev Biol 46:329–332
Salo E, Baguna J (2002) Regeneration in Planarians and other worms: new findings, new tools, and new perspectives. J Exp Zool 292:528–539
Sánchez Alvarado A, Kang H (2005) Multicellularity, stem cells, and the neoblasts of the planarian Schmidtea mediterranea. Exp Cell Res 305:299–308
Umesono Y, Agata K (2009) Evolution and regeneration of the planarian central nervous system. Dev Growth Differ 51:185–195
Ushakov DS (2008) Structure and function of the essential light chain of myosin. Biophys 53:505–509
Woolner S, Bement WM (2009) Unconventional myosins acting unconventionally. Trends Cell Biol 19:245–252
Yuan ZQ, Zhao BS, Zhang JY, Zhang SC (2010a) Characterization and expression of DjPreb gene in the planarian Dugesia japonica. Mol Biol 44:8–13
Yuan ZQ, Zhao BS, Zhang JY, Zhang SC (2010b) Expression pattern of DjPreb gene during the planarian Dugesia japonica embryonic development. Mol Biol 44:571–576
Acknowledgments
This work was supported by grants from Shandong Province Natural Science Foundation of China (ZR2013CM011) and State Key Laboratory of Genetic Resource and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences (GREKF09-04) and a Project of Shandong Province Higher Educational Science and Technology Program, China (J11LC09).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
S. Yu and X. Chen contributed equally to this work.
GenBank Accession Number: HQ585080.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2015_990_MOESM2_ESM.tif
Alignment of ELCs, including DjELC using the MegAlign program (DNASTAR) by the CLUSTAL W method. Shaded (with solid black) residues are the amino acids that match the consensus. Gaps introduced into sequences to optimize alignment are represented by (−). “#” means Ca2+ binding sites. ELC amino acid sequences were obtained from NCBI: Apis mellifera (XP_393544); Ascaris suum (ERG85616); Branchiostoma floridae (XP_002603233.1); Bos taurus (NP_001071124); Caenorhabditis brenneri (ACD86909); Caenorhabditis elegans (P53014); Dictyostelium discoideum (P09402); Danio rerio (NP_001005955); Dugesia japonica (HQ585080); Drosophila melanogaster (AAF56733); Echinococcus granulosus (CDJ16649); Gallus gallus (NP_001038097); Hymenolepis microstoma (CDJ14659); Homo sapiens (NP_524144); Marsupenaeus japonicas (ADD70028); Mus musculus (NP_067260); Pan troglodytes (XP_516064); Rattus norvegicus (NP_001071124); Setaria digitata (ACT15365); Schistosoma japonicum (CAX70946); Schistosoma mansoni (AAD41591); Xenopus tropicalis (NP_001121402) (TIFF 10759 kb)
438_2015_990_MOESM3_ESM.tif
Expression level of DjElc mRNA in regenerating trunk fragments of DjElc-RNAi-treated planarians by quantitative real-time PCR (TIFF 699 kb)
438_2015_990_MOESM4_ESM.tif
Ventral view of defects of DjElc-RNAi-treated trunk fragments to regenerate head by immunostaining with anti-synaptotagmin. (A) Normal regenerating head at 7 days as a control. (B) Loss of DjElc disrupted the brain lateral branches at 7 days. Scale bars: 50 μm (TIFF 1329 kb)
Rights and permissions
About this article
Cite this article
Yu, S., Chen, X., Yuan, Z. et al. Planarian myosin essential light chain is involved in the formation of brain lateral branches during regeneration. Mol Genet Genomics 290, 1277–1285 (2015). https://doi.org/10.1007/s00438-015-0990-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-0990-1