Abstract
The genus Dasypyrum contains two species: the annual and widespread D. villosum (2x = 2n = 14) and the perennial and generally rare D. breviaristatum (2x = 2n = 14 and 4x = 2n = 28). The origin of the latter and its genome constitution have been subject of several studies. There is agreement that the genome of the diploid D. villosum (VV) is different from the diploid cytotype of D. breviaristatum (VbVb), but there is no agreement of the constitution of the tetraploid cytotype, specifically whether is it an autotetraploid or an allotetraploid. This is a long-standing disagreement that this study aims to resolve using the 5S nrDNA as a genomic marker. Our studies suggest that the 4x D. breviaristatum is an allotetraploid (VVVbVb).


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altschul SF, Madden TL, Schoffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Appels R, Baum BR, Clarke BC (1992) The 5S DNA units of bread wheat (Triticum aestivum L.). Plant Syst Evol 183:183–194
Baum BR, Johnson DA (2004) Differences between South American H haplome diploids and I haplome diploids from the perspective of the 5S rDNA gene in the genus Hordeum. Czech. J Plant Breed 40:45–50
Baum BR, Johnson DA (2008) Molecular confirmation of the genomic constitution of Douglasdeweya (Triticeae: Poaceae). Demonstration of the utility of the 5S DNA sequence as a genomic tool. MGG 279:621–628. doi:10.1007/s00438-008-0338-1
Baum BR, Feldman M (2010) Elimination of 5S DNA unit classes in newly formed allopolyploids of the genera Aegilops and Triticum. Genome 53:430–438. doi:10.1139/G10-017
Baum BR, Johnson DA (1994) The molecular diversity of the 5S rRNA gene in barley (Hordeum vulgare). Genome 37:992–998
Baum BR, Johnson DA (1996) The 5 s rRNA gene units in ancestral two rowed barley (Hordeum spontaneum C. Koch) and bulbous barley (H. bulbosum L.): sequence analysis and phylogenetic relationships with the 5 s rDNA units of cultivated barley (H. vulgare L.). Genome 39:140–149
Baum BR, Johnson DA (1998) The 5S rRNA gene in sea barley (Hordeum marinum Hudson sensu lato): sequence variation among repeat units and relationship to the X haplome in barley (Hordeum). Genome 41:652–661
Baum BR, Johnson DA (2002) A comparison of the 5S rDNA diversity in the Hordeum brachyantherum–californicum complex with those of the eastern Asiatic Hordeum roshevitzii and the South American Hordeum cordobense (Triticeae: Poaceae). Can J Bot 80:752–762. doi:10.1139/B02-057
Baum BR, Johnson DA (2007) The 5S DNA sequences in Hordeum bogdanii and in the H. brevisubulatum complex, and the evolution and the geographic dispersal of the diploid Hordeum species (Triticeae: Poaceae). Genome 60:1–14
Baum BR, Johnson DA, Bailey LG (2001) Defining orthologous groups among multicopy genes prior to inferring phylogeny, with special emphasis on the Triticeae (Poaceae). Hereditas 135:123–138
Baum BR, Edwards T, Johnson DA (2008) Loss of 5S rDNA units in the evolution of Agropyron, Pseudoroegneria, and Douglasdeweya. Genome 51:589–598. doi:10.1139/G08-045
Baum BR, Edwards T, Mamuti M, Johnson DA (2012) Phylogenetic relationships among the polyploid and diploid Aegilops species inferred from 5S rDNA units (Triticeae: Poaceae). Genome 55:1–17
Baum BR, Edwards T, Johnson DA (2013) What does the 5S rRNA multigene family tell us about the origin of the annual Triticeae (Poaceae)? Genome 56:1–23. doi:10.1139/gen-2012-0195
Birchler JA, Veitia RA (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19:395–402
Blanco A, Resta P, Simeone R, Parmar S, Shewry PR, Sabelli P, Lafiandra D (1991) Chromosomal location of seeds storage protein genes in the genome of Dasypvrum villosum (L.) Candargy. Theor Appl Genet 82:358–362
Blanco A, Simeone R, Resta P, Pace CD, Delre V, Caccia R, Mugnozza GT, Frediani M, Cremonini R, Cionini PG (1996) Genomic relationships between Dasypyrum villosum (L.) Candargy and D. hordeaceum (Cosson et Durieu) Candargy. Genome 39:83–92
Cremonini R (1996) Cytological studies on Dasypyrum villosum. Haynaldia BIO TAR Electronic 5.2, pp 3–14
De Pace C, Vaccino P, Giorgio P, Pasquini M, Qualset CO (2011) Dasypyrum. Chpt 4. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, cereals. Springer, Berlin, pp 185–292. doi: 10.1007/978.3-642-14228-4-4
De Smet R, Adams KL, Vanderpoele K, Montagu MCE, Maere S, Van de Peer Y (2013) Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. PNAS 110:2898–2903. doi:10.1073/pnas.1300127110
Edgard RG (2004) MUSCLE: a multiple sequence alignment method with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Felsenstein J (1984) The statistical approach to inferring evolutionary trees and what it tells us about parsimony and compatibility Chpt.10. In: Duncan T, Stuessy TF (eds) Cladistics: perspectives on the reconstruction of evolutionary history. Columbia Univ. Press, New York, pp 169–191
Frederiksen S (1991) Taxonomic studies in Dasypyrum (Poaceae). Nord J Bot 11:135–142
Galasso I, Blanco A, Katsiotis A, Pignone D, Heslop-Harrison JS (1997) Genomic organization and phylogenetic relationships in the genus Dasypyrum analysed by Southern and in situ hybridization of total genomic and cloned DNA probes. Chromosoma 106:53–61
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224
Gradzielewska A (2006) The genus Dasypyrum—part 1. The taxonomy and relationships within Dasypyrum and with Triticeae species. Euphytica 152:429–440. doi:10.1007/s10681-006-9232-2
Linde-Laursen I, Frederiksen S (1991) Comparison of the Giemsa C-banded karyotypes of Dasypyrum villosum (2x) and D. breviaristatum (4x) from Greece. Hereditas 114:237–244
Liu C, Li GR, Sehgal SK, Jia JQ, Yang ZJ, Friebe B, Gill BS (2010) Genome relationships in the genus Dasypyrum: evidence from molecular phylogenetic analysis and in situ hybridization. Plant Syst Evol 288:149–156
Löve Á (1984) Conspectus of the Triticeae. Feddes Repert 95:425–521
Morris JJ, Lenski RE, Zinser ER (2012) The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3(2):1–8. doi:10.1128/mBio.00036-12
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152. doi:10.1146/annurev.genet.39.073003.112240
Nicholas KB, Nicholas HB Jr (1997) GeneDoc©: a tool for editing and annotating multiple sequence alignments Distributed by the authors Available from http://www.psc.edu/biomed/genedoc/ or http://www.nrbsc.org/downloads/
Ohta S, Morishita M (2001) Genome relationships in the genus Dasypyrum (Gramineae). Hereditas 135:101–110
Ohta S, Koto M, Osada T, Matsuyama A, Furuta Y (2002) Rediscovery of a diploid cytotype of Dasypyrum breviaristatum in Morocco. Genet Res Crop Evol 49:305–312
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Posada D (2009) Selection of models of DNA evolution with jMODELTEST. In: Posasda D (ed) Bioinformatics for DNA sequence analysis. Humana Press, Totowa, pp 93–112
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53: pp 793–798, 8
Rambaut A, (2009) FigTree Figure Drawing Tool Version 1.3.1. Available online from http://tree.bio.ed.ac.uk/. Accessed 5 Dec 2012
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna A, Larget B, Liu L, Suchard MA, Hulsenbeck JP (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542. doi:10.1093/sysbio/sys029 (Version 3.2.1 x64 accessed 2012)
Sabbagh A, Marin J, Veyssiere C, Lecompte E, Boukouvala S, Poloni E, Darlu P, Crouau-Roy B (2013) Rapid birth-and-death evolution of the xenobiotic metabolizing NAT gene family in vertebrates with evidence of adaptive selection. BMC Evol Biol 13:62. doi:10.1.186/1471.2148.13.62
Sakamoto S (1986) Genome analysis of a polyploid form of Haynaldia hordeacea in the tribe Triticeae, Gramineae. In: Li Z, Swaminathan MS (eds) Proceedings of the 1st international symposium on chromosome engineering in plants. Xian, China, pp 52–53
Sarkar P (1957) A new diploid from Haynaldia hordeacea Hack. Wheat Inform Serv 6:22
Sears ER (1953) Additions of the genome of Haynaldia villosa to Triticum aestivum. Ann J Bot 40:168–174
Swofford DL (2001) PAUP* Phylogenetic analysis using parsimony (*and other methods) Version 4 Sinauer Associates, Sunderland. Massachusetts
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Uslu E, Reader SM, Miller TE (1999) Characterization of Dasypyrum villosum (L.) Candargy chromosomes by fluorescence in situ hybridization. Hereditas 131:129–134
von Bothmer R, Claesson L (1998) The hybrid Triticum turgidum ssp. dicoccum Dasypyrum villosum and the cross and backcrosses to bread wheat, T. aestivum. Hereditas 128:47–52
Yang ZJ, Li GR, Feng J, Jiang HR, Ren ZL (2005) Molecular cytogenetic characterization and disease resistance observation or wheat-Dasypyrum breviaristatum partial amphiploid and its derivatives. Hereditas 142:80–85
Yang ZJ, Liu C, Feng J, Li GR, Zhou JP, Deng KJ, Ren ZL (2006) Studies on genome relationship and species-specific PCR marker for Dasypyrum breviaristatum in Triticeae. Hereditas 143:47–54
Zuccon A, Zuccon D 2013 MrEnt v.2.4. Program distributed by the authors http://www.mrent.org. Accessed 01 Mar 2013
Acknowledgments
We thank Dr. Shoji Ohta; Plant Germplasm Institute, Faculty of Agriculture, Kyoto University, Japan, for sending us material of Dasypyrum breviaristatum that also included the identified diploid and tetraploid cytotypes. We also thank the two referees for helpful suggestions and D. Laczko for technical assistance. This research was funded in part by grants from the Natural Sciences and Engineering Research Council of Canada and the University of Ottawa, Research Development Program to D. A. J.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Tian.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Baum BR, Edwards T, Johnson DA XXXX What does the nr5S DNA multigene family tell us about the genomic relationship between Dasypyrum breviaristatum and D. villosum (Triticeae: Poaceae)?
438_2014_825_MOESM1_ESM.pdf
Supplementary material Fig. S1.Sequence alignment of the Long V1 unit class in the Dasypyrum study. DV = Dasypyrum villosum; DB = Dasypyrum breviaristatum. Sequence label prefix: 2 = diploid, 4 = tetraploid. (PDF 8 kb)
438_2014_825_MOESM2_ESM.pdf
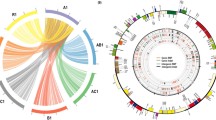
Supplementary material Fig.S2. Circular phylogram to complement Fig. 1. In this rendition the labels of the sequences are made clearer and along with the unit classes that were visually defined in the alignments. Disposition of the four unit classes follows. Long V1: starting from 2XDB1454_11 to DB1455_6; Long Vb1: from DB1456_10 to DB1427_11; Short Vb1: from DB1460_21 to DB1460_5; and Short V1: DB746_6 to DV748_16. (PDF 125 kb)
Rights and permissions
About this article
Cite this article
Baum, B.R., Edwards, T. & Johnson, D.A. What does the nr5S DNA multigene family tell us about the genomic relationship between Dasypyrum breviaristatum and D. villosum (Triticeae: Poaceae)?. Mol Genet Genomics 289, 553–565 (2014). https://doi.org/10.1007/s00438-014-0825-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0825-5