Abstract
Enterocytozoon bieneusi is an obligate intracellular pathogen that infects livestock, companion animals, and wildlife and has the potential to cause severe diarrhea especially in immunocompromised humans. In the underlying study, fecal samples from 177 calves with diarrhea and 174 adult cows originating from 70 and 18 farms, respectively, in Austria were examined for the presence of E. bieneusi by polymerase chain reaction targeting the Internal Transcribed Spacer 1 (ITS1) region. All positive samples were further sequenced for genotype determination. Overall, sixteen of the 351 (4.6%) samples were positive for E. bieneusi, two of the 174 samples from cows (1.2%) and 14 of the 177 samples from calves (7.9%). In total, four genotypes, J (n = 2), I (n = 12), BEB4 (n = 3), and BEB8 (n = 1), were identified. The uncorrected p-distance between the four ITS1 lineages (344 bp) ranges from 0.3% to 2.9%. The lineages differ by 1 bp (I and J), 2 bp (J and BEB4), and 3 bp (I and BEB4), respectively, and BEB8 differs by 7 to 10 bp from the latter three lineages. Two of the E. bieneusi-positive calves showed an infection with two different genotypes. E. bieneusi occurred significantly more often in calves > 3 weeks (8/59) than in calves ≤ 3 weeks (6/118), respectively (p = 0.049). Calves with a known history of antimicrobial treatment (50 of 177 calves) shed E. bieneusi significantly more often than untreated calves (p = 0.012). There was no statistically significant difference in E. bieneusi shedding in calves with or without a medical history of antiparasitic treatment (p = 0.881). Calves showing a co-infection with Eimeria spp. shed E. bieneusi significantly more often than uninfected calves (p = 0.003). To our knowledge, this is the first report of E. bieneusi in cattle in Austria. Cattle should be considered as a reservoir for human infection since potentially zoonotic E. bieneusi genotypes were detected.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microsporidia are obligate intracellular pathogens that infect a wide range of invertebrates and vertebrates and have the potential to infect humans. This taxon contains at least 1700 species in approximately 220 genera, of which ten genera (17 species) have been described in humans (Han et al. 2021). The two taxa that frequently infect humans are Enterocytozoon bieneusi and Encephalitozoon spp., which have also been described in wild animals, domestic pets, and food-producing animals (Didier 2005, Hinney et al. 2016). In particular, immunocompromised humans, e.g., AIDS patients or organ transplant recipients, are at higher risk for infection (Han et al. 2021). Enterocytozoon bieneusi is the microsporidian species that most frequently causes human infection and is responsible for 90% of cases of chronic diarrhea in AIDS patients (Weber et al. 1994; Sak et al. 2011).
However, E. bieneusi can also lead to asymptomatic and symptomatic infections in immunocompetent humans (Didier and Weiss 2011; Sak et al. 2011). The main transmission route is oral ingestion of food and/or water contaminated with spores (Mathis et al. 2005). Since microsporidian spores are very small (0.7 to 1.0 μm by 1.1 to 1.6. μm), detection in samples by light microscopy is difficult and requires staining. For the identification of. E. bieneusi to the genus or species level, molecular methods have the highest sensitivity and specificity (Franzen and Müller 1999; Zhang et al. 2021). Among the molecular detection methods, amplification and sequencing based on the Internal Transcribed Spacer 1 (ITS1) region was established as a reliable tool to identify species and genotypes of Microsporidia (Dong et al. 2010). Molecular genetics also allow differentiation between host-specific and zoonotic E. bieneusi genotypes. Genotypes can be divided into separate groups according to host specificity. Groups 1 and 2 have been detected in multiple species including humans and are considered to be zoonotic; groups 3 to 11 seem to be more specific to animals (Li et al. 2019). Nevertheless, suggested ruminant-specific genotypes (J, I, BEB4, and BEB6) have also been reported in humans (Jiang et al. 2015; Yu et al. 2019). Enterocytozoon bieneusi was first reported in cattle by Rinder et al. (2000), who detected genotypes J and I in cattle in Germany. The ITS1 sequences of genotypes J and I from samples from cattle from Germany were identical to BEB1 and BEB2 originating from cattle in the USA, respectively (Rinder et al. 2000; Sulaiman et al. 2004). A recent meta-analysis showed that genotypes BEB4, J (BEB1), and I (BEB2) were the most frequently reported genotypes in cattle globally (Taghipour et al. 2022). Genotype BEB8 belongs to group 2 and has been described in bovines, non-human primates, and bats (Li et al. 2019).
To date, there is no information on the occurrence of E. bieneusi in cattle in Austria. The objective of the study was to describe the occurrence of E. bieneusi in fecal samples from diarrheic calves and clinically healthy cows in Austria, and we hypothesized that E. bieneusi frequently occurs in these samples.
Materials and methods
Animal selection and sample collection
Calves
During the years 2017 and 2018, veterinarians and farmers from all over Austria were asked to participate in the study. If the farmer was willing to participate, one of the first authors (KL) or the local veterinarian visited the farms and collected the fecal samples. Additionally, specific information on the included calves (medical history, clinical examination) was collected. Clinical examination was performed according to Baumgartner and Wittek (2018). The inclusion criteria for the calves were signs of diarrhea at the time of the herd visit (soft, liquid, or watery feces) and less than 180 days of age. At least 10 g or 10 ml of feces was collected during the farm visit using a urine collection cup. In total, 177 calves with diarrhea originating from 70 farms all over Austria were included. The fecal samples were immediately transported to the University Clinic for Ruminants Vienna by one of the first authors (KL). All samples were analyzed for the following calf diarrhea-causing pathogens: protozoa (Cryptosporidium spp., Giardia intestinalis, and Eimeria spp.), viruses (bovine Rotavirus A and bovine Coronavirus), and bacteria (Clostridium perfringens). The methods implemented for the detection of other enteropathogens (viruses, bacteria, and protozoa) have been described elsewhere (Lichtmannsperger et al. 2019, Lichtmannsperger et al. 2022). The remaining fecal and DNA samples were stored at minus 80 °C for further investigations.
Cows
In 2020, one hundred seventy-four cows from 18 conveniently selected dairy farms from the province of Salzburg, Austria, were included. All of the dairy farmers were customers of one local veterinary practice, and samples were taken on occasion of a farm visit for herd health checkup. During the farm visit, general information on farm characteristics and detailed information on the cows were gathered. All cows were clinically healthy. Fecal samples were collected using disposable gloves and transported to the University Clinic for Ruminants Vienna under cooling conditions within one day. Upon arrival, the samples were immediately frozen at minus 80 °C until further investigations.
DNA extraction
All fecal samples were thawed at room temperature, transferred into a 20 ml collection cup, and homogenized using a wooden mouth spatula before approximately 200 mg aliquots were used for DNA extraction. Extraction was performed using the NucleoSpin® Soil kit (Macherey–Nagel GmbH & Co. KG, USA) according to the manufacturer’s specifications. In brief, lysis buffer (SL1) was used for sample preparation without lysis condition adjustment, and DNA elution was carried out using 100 μl of elution buffer. DNA was stored at minus 20 °C until used in PCR analysis.
PCR for genotyping
Enterocytozoon bieneusi was screened based on the ITS1 region by nested PCR under the following conditions: the cycling protocol for both reactions included an initial cycle of 94 °C for 2 min, followed by 20 cycles (nest 1) or 35 cycles (nest 2) of 94 °C for 30 s, 57 °C for 30 s, 72 °C for 60 s, and a final extension of 72 °C for 10 min. The reaction volume was 25 μl containing 2 μl of genomic DNA template, standard PCR buffer (5xGreen GoTaq® Reaction Buffer, Promega USA), 10 mM of each dNTP, 1.25 U of Taq polymerase (GoTaq® G2 DNA Polymerase, Promega, USA), and 10 pmol of each oligonucleotide primer (Table 1).
Sequencing of PCR products
The PCR products were sequenced in both directions at LGC Genomics GmbH (Berlin, Germany). The raw forward and reverse sequences (and electropherograms) were carefully checked and aligned with Bioedit v.7.0.8.0 (Hall 1999). Sequences were compared with reference sequences in GenBank® using the Basis Local Alignment Searching Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast). The obtained sequences are deposited in GenBank® under the accession numbers OP455917-OP455934.
Statistical analysis
Statistical analysis was performed using IBM® SPSS® Statistics Version 27 (IBM®, New York, USA) and Microsoft Excel 2010. To describe the differences in parasite occurrence between age groups, antimicrobial treatment, antiparasitic treatment, and the additional diseases, chi-square tests were applied. All tests were calculated with a significance level of p < 0.05.
Results
Calves
General information
In total, 177 calves with diarrhea originating from 70 farms were included. The animals were between one and 164 days old (median = 12; mean = 27). On average, 2.5 calves were sampled per farm (range = 1 to 10 calves). Female (108) and male (69) calves were included. The calves were Simmental (141), Holstein (8), Brown Swiss (1), or crossbreeds (27).
PCR results
In total, fourteen of the 177 included samples were positive for E. bieneusi by PCR. Eight of the 14 E. bieneusi-positive calves originated from dairy farms, four from fattening units, and two from cow-calf operations. The positive calves originated from 12 different farms; two farms had two positive calves each. The 14 positive calves were between 7 and 164 days of age (mean = 41.8 days, median = 29.5). Enterocytozoon bieneusi occurred significantly more often in calves > 3 weeks (8/59) than in calves ≤ 3 weeks (6/118) days, respectively (p = 0.049). Six of the positive animals were female (6/108) and eight were male (8/69) (p = 0.244). Nine of the calves were born on the farm and five were purchased at an auction or from calf dealers. The duration of diarrhea before the farm visit was for 1 to 3 days and > 3 days in ten and four positive calves, respectively. Eight animals had a very good and six animals had a reduced feed intake at the time of sample collection. The body condition was good in 13 calves; one calf was in poor body condition. The hydration status was normal in 12 of the investigated calves; two animals showed moderate dehydration.
Medical history of diarrheic calves
Based on the medical history, fifty diarrheic calves received antibiotics prior to sampling. Of the 50 treated animals, 31 (31/50) were less or equal than three weeks of age and 19 (19/50) were older than 3 weeks of age. There was no significant difference between calves receiving antibiotics ≤ 3 and > 3 weeks of age (p = 0.409). Table 2 gives an overview on the implemented antimicrobial classes. Enterocytozoon bieneusi infection status (PCR results) and the differences in medical history regarding the treatment with antimicrobial, antiprotozoal, and antiphlogistic drugs are shown in Table 3. Information on the E. bieneusi status (positive or negative) and concurrent infections with other calf diarrhea-causing pathogens is summarized in Table 4.
Cows
All fecal samples of cows (n = 174) originated from dairy farms (n = 18) from the province of Salzburg, Austria. On average, thirty-six dairy cows were housed on the dairy farms (median = 33.5, range = 13–70). The majority of animals were Simmental breed (83.3%) followed by Holstein (3.9%) and the remaining 12.8% of cows were Brown Swiss, Pinzgauer, and crossbreeds. Fecal samples from two cows showed a positive PCR result. The two positive Simmental cows originated from the same dairy farm. At the time of sample collection, all the dairy cows were lactating.
Distribution and genetic characterization of E. bieneusi genotypes
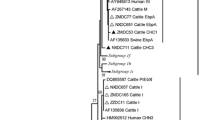
The analysis of the ITS1 sequences revealed the presence of four genotypes (J, I, BEB4, and BEB8). The most prevalent genotype in the present study was I (66.7%, 12/18), followed by BEB4 (16.7%, 3/18), J (11.1%, 2/18), and BEB8 (5.6%, 1/18). Two genotype I-positive calves showed a concurrent infection with genotype J and BEB4. The two positive cows harbored genotype I. We downloaded all ITS1 sequences from NCBI GenBank and found 677 unique E. bieneusi lineages, of which 107 were isolated from humans and mostly also from various animal species. Most of these lineages are from unpublished studies, and the data is available on GenBank only. We therefore decided to only show the information on host species and country for the four ITS1 lineages found in the present study (Fig. 1). BEP4 is the only one of the four lineages which was isolated from five humans, all sampled in China.
Pie charts showing the number of records per country and host species for the documented four Enterocytozoon bieneusi genotypes (I, J, BEB4, and BEB8). The country codes (ISO 3166–1) stand for Austria (AT), Australia (AU), Bangladesh (BD), Brazil (BR), China (CN), Egypt (ET), Germany (DE), Ireland (I) Portugal (PT), South Korea (KR), Slovakia (SK), United States of America (US), and Zaire (ZA). The numbers in brackets following the country codes refer to the number of records per animal species and country
Discussion
This survey was part of a larger study on the occurrence of enteropathogens (viruses, bacteria, and protozoa) in calves with diarrhea in Austria (Lichtmannsperger et al. 2019, 2020, 2022). To our knowledge, this is the first report of E. bieneusi in calves and cows in Austria.
Recently, a meta-analysis has been published on the global prevalence of E. bieneusi in cattle. A pooled prevalence of cattle microsporidiosis of 13.9% has been described (Taghipour et al. 2022). The average E. bieneusi infection rates ranged from 2.0% to 21.4% (Zheng et al. 2020). The cumulative prevalence in the present investigation was 4.6%, which is comparatively low compared to these studies.
A higher prevalence has been described in pre-weaned calves (less than 6 months of age), female cattle, and cattle with diarrhea, but without statistical significance (Taghipour et al. 2022). In contrast to other studies, where it has been described that weaned calves had a higher prevalence than pre-weaned calves with 15.9% positive and 9.1% positive, respectively (p < 0.01) (Wang et al. 2019), in the present study, the occurrence of E. bieneusi in calves (14 of 177 calves positive) was 7.9% and in cows (2 of 174 cows positive) 1.1%, respectively (p = 0.005). E. bieneusi occurred significantly more often in calves > 3 weeks than in calves ≤ 3 weeks days, respectively (p = 0.049).
All calves were suffering from diarrhea and cows were healthy. The discrepant values among the studies might result from various factors such as differences in the animal management systems (e.g., loose housing, tethered housing, and outdoor and indoor housing), sample sizes (random selection or convenient sampling), climatic and environmental conditions, and the health status of the animals. For instance, the majority of available literature on the occurrence of E. bieneusi originate from Asia, especially China, and North America, the USA (Taghipour et al. 2022). Therefore, the comparison between these studies and the underlying investigation is limited.
The occurrence of E. bieneusi has been investigated in 314 pre-weaned diarrheic and healthy calves in Korea (Hwang et al. 2020). They found an overall E. bieneusi prevalence of 16.9% and a significantly higher prevalence in diarrheic calves aged from one to 20 days old. A longitudinal study carried out in China investigated 250 fecal samples from 25 calves with differing fecal consistency (formed, loose, and liquid feces) throughout a sampling period of ten weeks (Zhao et al. 2020). The highest infection rate was recorded at six weeks of age which is also in accordance to the present investigation where the mean age of the infected animals was 41.8 days (median = 29.5 days). Nevertheless, in both studies, E. bieneusi has also been detected in calves with normal fecal consistency (Hwang et al. 2020; Zhao et al. 2020). In previous investigations, no difference in E. bieneusi infection status (positive or negative) and Cryptosporidium spp. or Giardia intestinalis co-infections was detected (Hwang et al. 2020). In the present study, a significant difference has been found only between calves infected with Eimeria spp. The 27 Eimeria spp.-positive calves were on average 65.4 days old (range = 19 to 164, median = 56.0). Since only diarrheic animals were included, it cannot be concluded if there is a real difference in calves positive for E. bieneusi with/without Eimeria infection.
Calves with a medical history of antimicrobial treatment shed E. bieneusi significantly more often than the animals with no history of antimicrobial treatment. In total, fifty calves were treated with antibiotics and nine of these received the highest priority critically important antimicrobials (WHO 2019). There is an urgent need for more scientific data allowing to define the best practices for the treatment of neonatal calf diarrhea (Eibl et al. 2021). In healthy calves, it has been well known that the gut microbiota shows a higher richness, evenness, and diversity in comparison to diarrheic calves (Gomez et al. 2022). Additionally, the administration of antimicrobials (e.g., fluoroquinolones or penicillins) has been described to alter the fecal microbiota in calves (Beyi et al. 2021; Grønvold et al. 2011). The role of parasites and fungi in the gut microbiota of diarrheic calves and the combined use of antimicrobial treatment have to be further investigated.
It has been described that the E. bieneusi genotypes shed by young calves changed in the first ten weeks of life (Zhao et al. 2020); therefore, it might be possible that some genotypes might be underreported. The initially suggested cattle-specific genotype BEB4, detected in three samples from calves in Austria, is also found in humans (Zhang et al. 2011). In dairy cows, the suggested host-specific genotype I was detected. Genotype I has also been detected in multiple counties and species in China (n = 72) (Wang et al. 2016; Tao et al. 2020), Australia (18) (Zhang et al. 2019a), the USA (8) (Sulaiman et al. 2004), Germany (2) (Rinder et al. 2000), Slovakia (2) (Valenčáková and Danišová 2019), Ethiopia (1) (Wegayehu et al. 2020), and South Africa (1) (Samra et al. 2012). In China, genotype I was found in the yak Bos grunniens (7) (Wu et al. 2019), the takin Budorcas taxicolor (6) (Zhao et al. 2015), the sika deer Cervus nippon (1) (Tao et al. 2020), the cat Felis catus (1) (Karim et al. 2014a), the rhesus macaque Macaca mulatta (Karim et al. 2014b), and the meerkat Suricata suricatta (2) (Li et al. 2015).
Genotype J was isolated from cattle in Austria (2), China (144) (Wang et al. 2016), Australia (14) (Zhang et al. 2019b), Germany (1) (Rinder et al. 2000), Portugal (1) (Lobo et al. 2006), and the USA (9) (Sulaiman et al. 2004). In China, genotype J was also found in the horse Equus ferus caballus (7) (Li et al. 2020), the goat Capra aegagrus (1) (Shi et al. 2016), the donkey Equus asinus (1) (Yue et al. 2017), the palm civet Paguma larvata (1) (Yu et al. 2020), the zebra Equus quagga (1), the lama Vicugna pacos (2), the sheep Ovis aries (1), and the meerkat Suricata suricatta (1) (Li et al. 2015). The genotype was also isolated from the Malayan pangolin Manis javanica in Bangladesh (1) (Karim et al. 2020), the white-tailed deer Odocoileus virginianus in the USA (1) (Santin and Fayer 2015), and the sambar Rusa unicolor (1) in Australia (Zhang et al. 2018). Genotype BEB4 was isolated from cattle in Austria (3), China (30) (Tao et al. 2020), the USA (8) (Sulaiman et al. 2004), and South Africa (3) (Samra et al. 2012). Moreover, it was found in the yak Bos grunniens (11) (Wu et al. 2019), the pig Sus scrofa (4), and humans in China (5) (Zhang et al. 2011). Genotype BEB8 was isolated from cattle in Austria (1), Brazil (23) (da Silva Fiuza et al. 2016), South Africa (4) (Samra et al. 2012), and Ethiopia (1) (Wegayehu et al., unpublished; accession no. MT231510). The genotype was also found in the greater tube-nosed bat Murina leucogaster (1) in South Korea (Lee et al. 2018) and the golden snub-nosed monkey Rhinopithecus roxellana (2) in China (Yu et al. 2017).
Since zoonotic genotypes of E. bieneusi have been detected in cattle, they need to be recognized as reservoirs of zoonotic E. bieneusi in Austria. Since the detection of microsporidia is not part of the routine water analysis nor the routine diarrhea-diagnostic panel in humans, it is unknown to what extent the pathogen is present in the environment and the population in Austria.
One of the limitations of our study was that we performed convenient and not randomized sampling and included only diarrheic calves and not healthy ones and also no other age groups, so the true prevalence of E. bieneusi in Austrian cattle is still unknown. The intermittent shedding of E. bieneusi needs to be taken into account since the cross-sectional study design with single individual samples might still underreport the prevalence and the range of genotypes found.
In conclusion, the present study is the first report of E. bieneusi in diarrheic calves and healthy cows from Austria. Fourteen (7.9%) of the 177 calves and two (1.1%) of the 174 included cows were positive for E. bieneusi, respectively. The observed occurrence of potential zoonotic genotypes emphasizes the possible role of cattle in the transmission of E. bieneusi to humans in Austria.
Data availability
The obtained sequences are deposited in GenBank® under the accession numbers OP455917-OP455934.
References
Baumgartner W, Wittek T (2018) Klinische Propädeutik der Haus- und Heimtiere, 9th edn. Enke, Stuttgart
Beyi AF, Brito-Goulart D, Hawbecker T, Slagel C, Ruddell B, Hassall A, Dewell R, Dewell G, Sahin O, Zhang Q, Plummer PJ (2021) Danofloxacin treatment alters the diversity and resistome profile of gut microbiota in calves. Microorganisms 9:2023
da Silva Fiuza VR, Lopes CWG, de Oliveira FCR, Fayer R, Santin M (2016) New findings of Enterocytozoon bieneusi in beef and dairy cattle in Brazil. Vet Parasitol 216:46–51
Didier ES (2005) Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop 94:61–76
Didier ES, Weiss LM (2011) Microsporidiosis: not just in AIDS patients. Curr Opin Infect Dis 24(5):490–495
Dong SN, Shen ZY, Xu L (2010) Sequence and phylogenetic analysis of SSU rRNA gene of five microsporidia. Curr Microbiol 60:30–37
Eibl C, Bexiga R, Viora L, Guyot H, Félix J, Wilms J, Tichy A, Hund A (2021) The antibiotic treatment of calf diarrhea in four European countries: a survey. Antibiotics 10:910
Franzen C, Müller A (1999) Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin Microbiol Rev 12(2):243–285
Gomez DE, Li L, Goetz H, MacNicol J, Gamsjaeger L, Renaud DL (2022) Calf diarrhea is associated with a shift from obligated to facultative anaerobes and expansion of lactate-producing bacteria. Front Vet Sci 9:846383
Grønvold AMR, Mao Y, Abee-Lund TML, Sørum H, Sivertsen T, Yannarell AC, Mackie RI (2011) Fecal microbiota of calves in the clinical setting: effect of penicillin treatment. Vet Microbiol 153:354–360
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acidy Symposium Series 41:95–98
Han B, Pan G, Weiss LM (2021) Microsporidiosis in humans. Clin Microbiol Rev 34(4):e0001020
Hinney B, Sak B, Joachim A, Kváč M (2016) More than a rabbit’s tale - Encephalitozoon spp. in wild mammals and birds. Int J Parasitol Parasites Wildl 5(1):76–87
Hwang S, Shin SU, Kim S, Ryu JH, Choi KS (2020) Zoonotic potential of Enterocytozoon bieneusi in pre-weaned Korean native calves. Parasit Vector 13:300
Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, Zhang S, Li W (2015) Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in Northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl Environ Microbiol 81:3326–3335
Karim MR, Dong H, Yu F, Jian F, Zhang L, Wang R, Zhang S, Rume FI, Ning C, Xiao L (2014a) Genetic diversity in Enterocytozoon bieneusi isolates from dogs and cats in China: host specificity and public health implications. J Clin Microbiol 52(9):3297–3302
Karim MR, Wang R, Dong H, Zhang L, Li J, Zhang S, Rume FI, Qi M, Jian F, Sun M, Yang G, Zou F, Ning C, Xiao L (2014b) Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl Environ Microbiol 80(6):1893–1898
Karim MR, Rume FI, Rahman ANMA, Zhang Z, Li J, Zhang L (2020) Evidence for zoonotic potential of Enterocytozoon bieneusi in its first molecular characterization in captive mammals at Bangladesh national zoo. J Eukaryot Microbiol 67(4):427–435
Lee SH, Oem JK, Lee SM, Son K, Jo SD, Kwak D (2018) Molecular detection of Enterocytozoon bieneusi from bats in South Korea. Med Mycol 56(8):1033–1037
Li J, Qi M, Chang Y, Wang R, Li T, Dong H, Zhang L (2015) Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in captive wildlife at Zhengzhou Zoo China. J Eukaryotic Microbiol 62(6):833–839
Li W, Feng Y, Santin M (2019) Host specificity of and public health implications. Trends Parasitol 35(6):436–451
Li F, Wang R, Guo Y, Li N, Feng Y, Xiao L (2020) Zoonotic potential of Enterocytozoon bieneusi and Giardia duodenalis in horses and donkeys in northern China. Parasitol Res 119(3):1101–1108
Lichtmannsperger K, Hinney B, Wittek T, Joachim A (2019) Molecular characterization of Giardia intestinalis and Cryptosporidium parvum from calves with diarrhoea in Austria and evaluation of point-of-care tests. Comp Immunol Microbiol Infect Dis 66:101333
Lichtmannsperger K, Harl J, Freudenthaler K, Hinney B, Wittek T, Joachim A (2020) Cryptosporidium parvum, Cryptosporidium ryanae, and Cryptosporidium bovis in samples from calves in Austria. Parasitol Res 119(12):4291–4295
Lichtmannsperger K, Freudenthaler K, Hinney B, Joachim A, Auer A, Rümenapf T, Spergser J, Tichy A, Wittek T (2022) Evaluation of immunochromatographic point-of-care tests for the detection of calf diarrhoea pathogens in faecal samples. Wien Tierärztl Monat – Vet Med Austria 109:Doc11
Lobo ML, Xiao L, Cama V, Stevens T, Antunes F, Matos O (2006) Genotypes of Enterocytozoon bieneusi in mammals in Portugal. J Eukaryotic Microbiol 53:61–64
Mathis A, Weber R, Deplazes P (2005) Zoonotic potential of the microsporidia. Clin Microbiol Rev 18(3):423–445
Rinder H, Thomschke A, Dengjel B, Gothe R, Löscher T, Zahler M (2000) Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J Parasitol 86(1):185–188
Sak B, Kvac M, Kucerova Z, Kvetonova D, Sakova K (2011) Latent microsporidial infection in immunocompetent individuals—a longitudinal study. PLoS Negl Trop Dis 5:e1162
Samra NA, Thompson PN, Jori F, Zhang H, Xiao L (2012) Enterocytozoon bieneusi at the wildlife/livestock interface of the Kruger National Park, South Africa. Vet Parasitol 190(3–4):587–590
Santin M, Fayer R (2015) Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. J Eukaryotic Microbiol 62(1):34–43
Shi K, Li M, Wang X, Li J, Karim MR, Wang R, Zhang L, Jian F, Ning C (2016) Molecular survey of Enterocytozoon bieneusi in sheep and goats in China. Parasite Vectors 9(1):1–8
Sulaiman IM, Fayer R, Yang C, Santin M, Matos O, Xiao L (2004) Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol Res 92(4):328–334
Taghipour A, Bahadory S, Abdoli A (2022) A systematic review and meta-analysis on the global prevalence of cattle microsporidiosis with focus on Enterocytozoon bieneusi: an emerging zoonotic pathogen. Prev Vet Med 200:105581
Tao WF, Ni HB, Du HF, Jiang J, Li J, Qiu HY, Zhang XX (2020) Molecular detection of Cryptosporidium and Enterocytozoon bieneusi in dairy calves and sika deer in four provinces in Northern China. Parasitol Res 119(1):105–114
Valenčáková A, Danišová O (2019) Molecular characterization of new genotypes Enterocytozoon bieneusi in Slovakia. Acta Trop 191:217–220
Wang XT, Wang RJ, Ren GJ, Yu ZQ, Zhang LX, Zhang SY, Lu H, Peng XQ, Zhao GH (2016) Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi province, northwestern China. Parasitol Res 115(3):1355–1361
Wang HY, Qi M, Sun MF, Li DF, Wang RJ, Zhang SM, Zhao JF, Li JQ, Cui ZH, Chen YC, Jian FC, Xiang RP, Ning CS, Zhang LX (2019) Prevalence and population genetics analysis of Enterocytozoon bieneusi in dairy cattle in China. Front Microbiol 10:1399
Weber R, Bryan RT, Schwartz DA, Owen RL (1994) Human microsporidial infections. Clin Microbiol Rev 7(4):426–461
Wegayehu T, Li J, Karim M, Zhang L (2020) Molecular characterization and phylogenetic analysis of Enterocytozoon bieneusi in lambs in Oromia Special Zone, Central Ethiopia. Front Vet Sci 7:6
World Health Organization (2019) Critically important antimicrobials for human medicine, 6th revision
Wu Y, Chang Y, Zhang X, Chen Y, Li D, Wang L, Zheng S, Wang R, Zhang S, Jian F, Ning C, Li J, Zhang L (2019) Molecular characterization and distribution of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi from yaks in Tibet. China. BMC Vet Res 15(1):1–9
Yu F, Wu Y, Li T, Cao J, Wang J, Hu S, Zhu H, Zhang S, Wang R, Ning C, Zhang L (2017) High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Vet Res 13(1):1–7
Yu F, Li D, Chang Y, Wu Y, Guo Z, Jia L, Xu J, Li J, Qi M, Wang R, Zhang L (2019) Molecular characterization of three intestinal protozoans in hospitalized children with different disease backgrounds in Zhengzhou, central China. Parasit Vectors 12:543
Yu Z, Wen X, Huang X, Yang R, Guo Y, Feng Y, Xiao L, Li N (2020) Molecular characterization and zoonotic potential of Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium sp. in farmed masked palm civets (Paguma larvata) in southern China. Parasit Vectors 13(1):1–10
Yue DM, Ma JG, Li FC, Hou JL, Zheng WB, Zhao Q, Zhang XX, Zhu XQ (2017) Occurrence of Enterocytozoon bieneusi in donkeys (Equus asinus) in China: a public health concern. Front Microbiol 8:565
Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, Lu H, Jiang N, Yin J, Xiang M, Chen Q (2011) Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol 49(5):2006–2008
Zhang Y, Koehler AV, Wang T, Haydon SR, Gasser RB (2018) First detection and genetic characterisation of Enterocytozoon bieneusi in wild deer in Melbourne’s water catchments in Australia. Parasit Vectors 11(1):1–9
Zhang Y, Koehler AV, Wang T, Haydon SR, Gasser RB (2019a) Enterocytozoon bieneusi genotypes in cattle on farms located within a water catchment area. J Eukaryotic Microbiol 66(4):553–559
Zhang Q, Zhang Z, Ai S, Wang X, Zhang R, Duan Z (2019b) Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis from animal sources in the Qinghai-Tibetan Plateau Area (QTPA) in China. Comp Immunol Microbiol Infect Dis 67:101346
Zhang Y, Koehler AV, Wang T, Gasser RB (2021) Chapter One - Enterocytozoon bieneusi of animals—With an ‘Australian twist.’ Adv Parasitol 111:1–73
Zhao GH, Du SZ, Wang HB, Hu XF, Deng MJ, Yu SK, Zhang LX, Zhu XQ (2015) First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect Genet Evol 34:394–401
Zhao A, Zhang K, Xu C, Wang T, Qi M, Li J (2020) Longitudinal identification of Enterocytozoon bieneusi in dairy calves on a farm in Southern Xinjiang, China. Comp Immunol Microbiol Infect Dis 73:101550
Zheng XL, Zhou HH, Ren G, Ma TM, Cao ZX, Wie LM, Liu XW, Wang F, Zhang Y, Liu HL (2020) Genotyping and zoonotic potential of Enterocytozoon bieneusi in cattle farmed in Hainan Province, the southernmost region of China. Parasite 27:65
Acknowledgements
The authors would like to thank the local veterinarians for their cooperation and the laboratory technicians at the Institute of Parasitology for their constructive work during this study.
Funding
Open access funding provided by University of Veterinary Medicine Vienna. The work of the veterinary undergraduate student (Sarah Rosa Roehl) was financially supported by the Austrian Association for Buiatrics (ÖBG).
Author information
Authors and Affiliations
Contributions
All authors jointly planned the study and analysis. Katharina Lichtmannsperger, Julia Schoiswohl, and Cassandra Eibl took part in the sample collection. Josef Harl, Sarah Rosa Roehl, Barbara Hinney, and Sandra Wiedermann analyzed the samples. Katharina Lichtmannsperger and Josef Harl drafted the first version of the manuscript. Thomas Wittek, Anja Joachim, Barbara Hinney, Sandra Wiedermann, Cassandra Eibl, Sarah Rosa Roehl, and Julia Schoiswohl revised it. All authors approved of the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This trial was evaluated and approved by the institutional Ethics and Animal Welfare Committee of the University of Veterinary Medicine Vienna.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Disclaimer
The sponsor did not participate in the study planning, data analysis, and manuscript compilation.
Additional information
Section Editor: Lihua Xiao.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lichtmannsperger, K., Harl, J., Roehl, S.R. et al. Enterocytozoon bieneusi in fecal samples from calves and cows in Austria. Parasitol Res 122, 333–340 (2023). https://doi.org/10.1007/s00436-022-07733-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-022-07733-y