Abstract
Purpose
Brain metastasis formation is a rare and late event in colorectal cancer (CRC) patients and associated with poor survival. In contrast to other metastatic sites, the knowledge on chromosomal aberrations in brain metastases is very limited.
Methods
Therefore, we carried out single nucleotide polymorphism (SNP) array analyses on matched primary CRC and brain metastases of four patients as well as on liver metastases of three patients.
Results
Brain metastases showed more chromosomal aberrations than primary tumors or liver metastases. Commonly occurring aberrations were gain of 8q11.1-q24.3 (primary CRC), gain of 13q12.13-q12.3 (liver metastases), and gain of 20q11.1-q13.33 (brain metastases). Furthermore, we found one copy-neutral loss of heterozygosity (cn-LOH) region on chromosome 3 in primary CRC, three cn-LOH regions in liver metastases and 23 cn-LOH regions in brain metastases, comprising 26 previously undescribed sites.
Conclusion
The more frequent occurrence of cn-LOHs and subsequently affected genes in brain metastases shed light on the pathophysiology of brain metastasis formation. Further pairwise genetic analyses between primary tumors and their metastases will help to define the role of affected genes in cn-LOH regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most common cancers and the third leading cause of death in the western hemisphere (Ferlay et al. 2015). Each year, approximately 1.4 million cases of colon cancer are diagnosed worldwide (Ferlay et al. 2015; Sefrioui et al. 2017). The standard curative therapy of CRC comprises surgical resection with or without neoadjuvant or adjuvant chemo- or radio chemotherapy. The most frequent localization of primary metastasis is the liver, followed by the lung (Vatandoust et al. 2015; Weiss et al. 1986). In rare cases (0.6–3.2%), brain metastases develop as late events of CRC. Despite aggressive neurosurgical therapy and radiation, the prognosis of brain metastases remains poor. The overall survival of patients with metachronous brain metastases is 2–8 months after diagnosis (Damiens et al. 2012).
In CRC, three processes have been described to influence pathogenesis: chromosomal instability (CIN), microsatellite instability (MSI), and the CpG island methylator phenotype (CIMP) (Jasmine et al. 2012; Mauri et al. 2019; Pino and Chung 2010). Most CRC cases arise through the CIN pathway. Characteristic features are chromosomal rearrangements and copy number variations. Consequences of CIN could be loss of tumor suppressor genes and amplification of oncogenes in the affected chromosomal regions (Jasmine et al. 2012). Recent investigations suggest the accumulation of genetic alterations as an essential step for the tumor evolution of CRC. The timing of specific genetic events could influence the metastatic potential, whereby copy number aberrations already acquire early in tumor development (Golas et al. 2022; Nguyen et al. 2022). In comparison to CIN, MSI is less common and seems to be associated with a better prognosis (Christensen et al. 2016; Guinney et al. 2015; Knösel et al. 2002; Watanabe et al. 2001).
However, colorectal carcinogenesis involves both genetic and epigenetic events, which are still not completely understood (Fearon and Vogelstein 1990; Grady and Carethers 2008; Sefrioui et al. 2017). The genetic profile of CRC comprises several hundred genetic aberrations, such as chromosomal gains on 6p, 7p, 7q, 8q, 13q, 17q and 20q, as well as losses of 4p, 4q, 5q, 8p, 14q, 17p, 18p, 18q, and 20p (Bacolod and Barany 2011; Gutenberg et al. 2010; Sefrioui et al. 2017). In liver metastases of CRC, copy number variations (CNV) have been identified at 1q, 11, 12qter, 17q12-q21, 19, and 22q and deletions have been described at 2q, 5q, 8p, 9p, 10q, and 21q21 (Knosel et al. 2005; Knösel et al. 2002, 2004). Using comparative genomic hybridization (CGH), Gutenberg et al. found significantly more chromosomal aberrations in brain metastases than in the corresponding primary CRC. In detail, gains on 8q, 12p, 12q, and 20p and loss of 5q were identified only in brain metastasis but not in the primary CRC, suggesting a further selection of different genetic alterations during brain metastasis formation (Gutenberg et al. 2010). Recently, accumulation of Her2 amplification has been described in brain metastases of CRC (Mitra et al. 2019). Furthermore, KRAS and BRAF mutations were significantly correlated with brain and lung metastasis formation (Liu et al. 2018).
Previous studies, reviewed by Diep et al. and Cardoso et al., showed limitations due to intertumoral heterogeneity when analyzing unmatched sample groups by gene expression or CGH (Cardoso et al. 2007; Diep et al. 2006). Thus, especially intraindividual comparisons of genetic aberrations between the primary CRC tissue and brain metastases are scarce.
In the present study we analyzed the genetic profile of matched pairs of primary CRC, liver metastases and brain metastases. In comparison to CGH, SNP array enables additional determination of copy-neutral loss of heterozygosity (cn-LOH). To the best of our knowledge, this is the first analysis of the genetic profile of CRC and the corresponding liver metastases and brain metastases using SNP array.
Material and methods
Patient material
Ethics approval was obtained from the ethics committee of the University of Leipzig (Az.: 005/17-ek). The study confirms the provisions of the Declaration of Helsinki (as revised in Fortaleza, Brazil, in 2013). Informed consent was given by patients and appropriate anonymity considerations were taken into account.
Patients with CRC, liver metastasis and brain metastasis who were diagnosed at the University Hospital of Leipzig (in a time slot of 7 years) were included into our study. Data were collected with regard to patient characteristics including overall survival, localization of the primary colorectal cancer and metastases, their extent and treatment modalities.
In total, tissue samples of 4 patients were collected from primary colorectal cancer, hepatic and brain metastases (see Fig. 1).
The average age of the patients was 55 years at the diagnosis of the primary CRC with tumor localization in the rectum (n = 2) and descending colon (n = 2). Surgical resection or biopsy of liver metastasis was performed at a mean of 22.25 months after first diagnosis of CRC. Brain metastases occurred at an average of ~ 64.5 months after first diagnosis. The mean overall survival after first diagnosis was ~ 75.25 months and after resection of brain metastasis ~ 9,5 months. The mean age at the time of death was 60.75 years (Table 1).
Isolation and conservation of tumor tissue
For tumor cell isolation, fresh non-necrotic surgical specimens were collected, fixed in formalin, embedded in paraffin and tissue sections were mounted onto glass slides.
DNA isolation and molecular karyotyping using SNP array
After pathological investigations, the samples from primary colorectal cancer tissue, liver metastasis and brain metastasis were used for the analyses. The samples were subjected to genome-wide copy number variation (CNV) analysis and assessment of copy number neutral loss of heterozygosity (cn-LOH) chromosomal regions using SNP array (OncoScan® FFPE Assay Kit, ThermoFisher Scientific, Dreieich, Hesse, Germany). Genomic DNA was extracted from tumor samples of colorectal cancer, liver metastasis (except from patient 4 due to technical reasons) and brain metastasis according to the manufacturers’ protocols (QIAamp® DNA FFPE Tissue Handbook, QIAGEN, Hilden, North Rhine-Westphalia, Germany). Paraffin was dissolved in Xylene and the samples were lysed under denaturing conditions using Proteinase K. After binding of DNA to the column, residual contaminants were removed and pure DNA was eluted. Subsequently, we performed OncoScan® array according to the manufacturers’ protocol (UserGuide OncoScan® FFPE Assay Kit, P/N 703175 Rev. 2, Thermo Fisher Scientific, Affymetrix, USA; Waltham). The evaluation was carried out using the software Chromosome Analysis Suite, Version. 3.3.0.139 (Thermo Fisher Scientific Inc., USA, Waltham) and based on the User Guide for Chromosome Analysis Suite 3.3 (ChAS 3.3), Publication Number 702943, Revision 12, available by Thermo Fisher Scientific (Thermo Fisher Scientific Inc., USA, Waltham). The following reference models were used: OncoScan.FFPE.na33.r2.REF_MODEL (as copy number reference) and OncoScan.FFPE.na33.r2.SOM_REF_MODEL (as somatic mutation reference). Pre-processing steps includes the Dual Quantile Normalization followed by Array Data QC Metrics and TuScan Algorithm (see User Guide for Chromosome Analysis Suite 3.3 – Appendix G). According to the literature, we considered chromosomal aberrations ≥ 3 Mb as reliable gain or loss and cn-LOH regions ≥ 5 Mb as representing (segmental) uniparental disomy (UPD) (Beroukhim et al. 2006; Žilina et al. 2015). Subsequent statistical analyses were performed using Prism 6 (GraphPad Software, La Jolla, USA). A detailed workflow is given in S1 Appendix.
Results
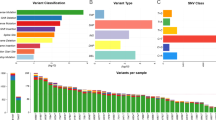
In total (primary tumors, liver metastases and brain metastases of all patients), we identified 283 genetic aberrations. The number of aberrations, including 152 gains, 104 losses, and 27 cn-LOH regions for all sample materials are listed in Table 2.
Primary colorectal carcinoma
For primary tumor, we identified an average of 17.3 (± 6.3) chromosomal aberrations. Frequent gains were identified on 8q11.1-q24.3, 13q14.3-q34, and 20q11.21-q13.33 (4/4 patients). Common losses comprised 8p23.3-p12 (3/4 patients) and 18q21.1-q22.2 (4/4 patients). The detected gains 10p11.23-q11.21, 10q24.31-q26.3 and 15q11.2-q15.1 are not previously described.
Liver metastases
The mean number of chromosomal aberrations for liver metastases are 18.7 (± 5.0). Consistent chromosomal aberrations were gains of 8q11.23-q24.3, 13q12.13-q12.3, and 20q11.21-q13.33 as well as loss of 8p23.3-p12 (3/3 patients, liver metastasis tissue available only from 3 patients). Not previously described aberrations were identified for gains on 2q33.1-q33.3, 5q33.3-q35.3, and 17q24.1-q24.3.
Brain metastases
Mean chromosomal aberrations were 39.5 (± 22.3). Common gains were identified on chromosomal regions 8q22.3-q24., 13q11.1-q13.2, 13q14.3–32.3 and 20q11.21-q13.33 (4/4 patients). Furthermore, losses were frequently detected of 21q11.2-q21.1 (4/4 patients) as well as of chromosomal regions 4p16.3-p15.33, 4q23-q25, 4q32.3-q35.2, 8p23.3-p12, and 18q21.1-q21.33 for three out of four patients. Totally, 30 out of 135 detected CNV in brain metastases are not previously described: gains on chromosomal regions 3p26.3-p22.1, 4p15.33-p11, 5q14.3-q21.1, 10p14-p11.1, 10p11.23-p11.21, 10p11.23-p11.1, 10q24.31-q26.3, 14q11.2-q24.1, 14q32.12-q32.33, 15q11.2-q21.1, 15q11.2-q26.3, 15q21.2-q21.3, 15q24.2-q26.3, 16p12.2-q24.3, 18p11.32-p11.21, 18q11.1-q21.1, 18q21.33-q23, and 19p13.3-p12 as well as losses on chromosomal regions 2p25.3-p23.1, 2q13-q22.3, 9q21.11-q31.2, 9q22.33-q34.3, 10p15.3-p11.23, 10p15.2-p11.23, 11p15.5-p12, 11q14.1-q25, 12p13.33-p11.1, 19p13.3-q13.43, 20p13-p11.1, and 20p12.3-p11.21. (detailed chromosomal sequences are shown in Table 5, appendix). An overview of all detected chromosomal aberrations for CRC, liver metastasis and brain metastasis of each patient is given in Fig. 4 and Table 5 (including not previously described CNV, marked in bold) in the appendix.
Comparison of primary colorectal carcinoma, liver metastasis and brain metastasis
In accordance to the description of Sylvester et al. 2015, we detected genetic aberrations displaying a heterogeneity between the different sample materials (intertumoral heterogeneity, intra-individual) and between different patients (intratumoral, inter-individual) (Sylvester and Vakiani 2015). Intertumoral heterogeneity was detected for chromosomal region 9q31.2-q34.3 (Primary tumor and liver metastasis: loss; brain metastasis: gain and loss), chromosomal region 10q24.31-q26.3 (primary tumor: gain; liver metastasis: loss; brain metastasis: gain and loss) as well as chromosomal region 16q23.1–24.1 (primary tumor and liver metastasis: gain; brain metastasis: gain and loss). Intratumoral heterogeneity was identified for chromosomal region 4p15.33-p11 in brain metastasis (patient 1 and 4: gain; patient 2 and 3: loss) and chromosomal region 12p13.33-p12.1 in brain metastasis (patient 1 and 4: gain; patient 2 and 3: loss). Gains on chromosome 5p15.33-q11.2, 5q34-q35.3, 7p22.3-q36.3, 8q11.23-q24.3, 12q11-q24.33, 13q11-q34, 16p13.3-q24.1, and 20q11.21-q13.33 as well as losses on chromosome 5q11.2-q35.3 and 15q11.2-q26.3 were identified in both primary tumor tissue and liver metastases. Occasionally, genetic differences of primary tumor vs. both metastases were identified too. Gain in 11q14.3-q24.2 as well as losses in 1p36.33-p11.1, 3p26.3-p14.1, 4p13-q13.3, 4q21.23-q22.2, 10q23.33-q26.3, 19p13.3-q13.43, and 22q11.1-q13.33 were exclusively identified in liver and brain metastases. Some aberrations were only identified in brain metastases. Our results of SNP array detected gains on chromosomal regions 3p26.3-p22.1, 4p16.3-q35.2 (except the chromosomal region 4q28.2-q31.21), 5q14.3-q21.1, 5q33.2-q33.3, 9p21.3-p21.1, 10q21.2–22.1, 11q13.2-q14.1, 14q11.2-q24.1, and 17q24.3-q25.3 as well as losses on the chromosomal regions 2q13-q22.3, 3p12.2-q29, 4p16.3-q35.2 (except chromosomal regions 4p13-q13.3 and 4q21.3-q22.2), 6q27, 8q21.11-q21.13, 10p15.3-p11.23, 11p15.5-p12, 11q14.1-q25, 12p11.1-q24.33, 13q32.3-q34, 14q24.2-q32.33, and 16q23.1-q24.3.
Interestingly, cn-LOH aberrations were mainly detected in brain metastases compared with primary tumor and liver metastases (primary tumor: one cn-LOH aberration, liver metastases: three cn-LOH aberration, brain metastases: 23 cn-LOH aberrations) (Table 3). Most chromosomal cn-LOH aberrations for liver metastasis (LM) and brain metastasis (BM) have not been previously described.
Based on the detected cn-LOH regions of the CRC-based brain metastases, we identified a total of 902 different cancer genes within the 23 chromosomal cn-LOH regions using NCG7.0 Network of Cancer Genes & Healthy Drivers (Dressler et al. 2022). Subsequently performed pathway analyses of these cancer genes revealed possible impacts on transcriptional processes as well as receptor-mediated pathways. The most relevant pathways are listed in Table 4.
Less frequent, the analyses revealed similar chromosomal aberrations among matched primary tumor vs. metastases. Such intratumoral and intertumoral similarities were detected for chromosomal regions 8q22.3-q24.3, 13q12.13-q12.3 and 20q11.21-q13.33 (Fig. 2).
Discussion
Brain metastasis occurs as a late-stage phenomenon of CRC and is associated with an unfavorable prognosis. Although the incidence of brain metastasis is low (2.3%), most patients do not survive the first year after the diagnosis (Diep et al. 2006). In this study, we analyzed genetic aberrations in CRC, liver metastasis and brain metastasis in matched pairs to reveal specific aberrations at the transition to metastatic disease, especially into the brain. An advantage of matched pairs is to reduce intertumoral heterogeneity, which is well known in colorectal cancer (Sylvester and Vakiani 2015).
We could confirm many previously described chromosomal aberrations for primary CRC as well as liver and brain metastases, whereby most of chromosomal imbalances were detected for brain metastases. Also recently presented data of Golas et al. figured out significant differences in the total number of detected aberrations, with most imbalances identified in brain metastases (34th GfH annual meeting, Abstract ID. 183, unpublished data). Gain for chromosomal region 13q11-q34 obviously represents an early event, since it was present in primary CRC and all metastatic sites (Golas et al. 2022). Accordingly, this aberration had been described at the transition from adenoma to primary CRC (Hermsen et al. 2002).
Gains of 7, 8q and 20q as well as losses of 8p and 18q (DCC/SMAD4) were conserved from the primary CRC to the metastatic sites. In the literature, loss of 8p had not been more frequently detected in liver metastases compared to primary CRC by Knösel et al. (Knösel et al. 2004). Conversely, gain of 8q could play a role in CRC, since this region harbors genes which are altered in CRC (e.g. UBR5, KLF10, EIF3H) or which are associated with the metastatic process (e.g. RRM2B, NOV, RAD21) (Muñoz-Bellvis et al. 2012). Gutenberg et al. had identified 20q as the most frequently occurring copy number change in 11 patients with brain metastases (Gutenberg et al. 2010). Additionally, this gain had often been detected in CRC primary tumors and been described as an early event in CRC tumor progress (Golas et al. 2022).
Some chromosomal region aberrations, like gain of 10q24.1-q26.3 (primary CRC and brain metastasis in patient 4) have not yet been previously published in analyses of CRC. Within chromosomal region 10q24.1, the FRAT1 gene (frequently rearranged in advanced T-Cell lymphomas 1) is localized. The gene is involved in the process of carcinogenesis in CRC by the Wnt/β-catenin pathway. According to Zhu et al., FRAT1 regulates the proliferation in colon cancer cells and constitutes a potential target for the treatment of CRC (Zhu et al. 2016).
Homologous recombination as a result of repairing a double-strand break, has been proposed as a possible cause for segmental UPD or cn-LOH formation in cancer, and thus may constitute a further genetic mechanism involved in tumorigenesis (Teh et al. 2005). Cn-LOHs may be a mechanism for inactivating tumor suppressor genes (Torabi et al. 2015).
In a recent study by Nguyen et al., the results indicate an early occurrence of CNVs during tumor development of CRC. These suggest that the more frequent detection of cn-LOH aberrations in brain metastases could be a result of a subsequent somatic reduplication/repair mechanism after previously acquired CNVs during tumor genesis (Golas et al. 2022; Nguyen et al. 2022). Only in one primary tumor, we detected cn-LOH at the chromosomal region 3p21.31-p21.1, which harbors the tumor suppressor gene von Hippel-Lindau (VHL) (Torabi et al. 2015). Torabi et al. had detected a UPD in the region 5q of a primary tumor, which then may lead to functional inactivation of the APC gene in colon carcinoma (Torabi et al. 2019). Loss of chromosome 5q (including APC gene) has been described for the CRC (Takayama et al. 2006).Within these region, we detected a loss in only one of the four primary CRCs, but aberrations (loss or cn-LOHs) in all four brain metastases.
Similar aberrations were detected on chromosome 18 (primary tumor and liver metastasis: loss, brain: loss or cn-LOH). The losses of chromosome 18 are consistent with pre-described aberrations for primary tumors as well as liver metastases (Diep et al. 2006; Gutenberg et al. 2010). Cn-LOH on chromosome 18 has not been previously disclosed in CRC-based brain metastases. Among the previously undescribed detected cn-LOH regions, the aberration within the region 19p13.3-q13.43 (including CEA gene) seems to be interesting. A few studies discussed the association of an increased carcinoembryonic antigen (CEA) level and brain metastases, whereas Bryani et al. did not found this relationship. In our study, the CEA gene, affected by a cn-LOH aberration, could have an impact on the incidence of CRC-based brain metastases. Further analyses on this gene may further elucidate the importance of CEA in context of brain metastases development.
Applying genome-wide SNP array analyses, we identified most of the cn-LOH aberrations in brain metastases. For the chromosomal region 17p13.3-p11.1, a cn-LOH aberration was detected in two patients and a loss in the other two patients (Fig. 2). Loss of 17p had been described as the most common numerical change in brain metastases (Gutenberg et al. 2010), containing the tumor suppressor gene TP53.
The cancer genes located within the cn-LOH regions of the brain metastases are involved in different interesting pathways. For instance, the transcriptional regulation of the tumor suppressor gene RUNX3 is described for colorectal cancer (Weisenberger et al. 2006). The detected cancer genes NOTCH1 and RUNX1 could possibly influence the RUNX3 signaling pathway and could be meaningful for metastases formation (Gao et al. 2010; Nishina et al. 2011; Whittle et al. 2015). Furthermore, these genes are also described in context with brain tumors (Hai et al. 2018; Steponaitis et al. 2019; Zhao et al. 2019).
However, for primary CRC, many genetic studies had revealed a high grade of heterogeneity within the tumor, with most frequent losses at chromosomal regions 1p, 5q, 8p, 17p, 18p, 18q, 20p and 20q. These losses were confirmed in all four patients of the present study at 5q, 8p, 17p, 18p, 18q and 20p (Diep et al. 2006; Jasmine et al. 2012; Jones et al. 2005; Zarzour et al. 2015).
Besides the detected heterogeneity, chromosomal regions with a similar pattern of chromosomal imbalances between primary tumor and the associated metastases were confirmed for some chromosomal aberrations, e.g., gain on 8q, 13q and 20q. According to Gutenberg et al. (2010), gain of chromosomal regions 13q and 20q had been the most frequent aberrations in both primary tumor and brain metastases (Gutenberg et al. 2010). Diep et al. had found gains of 13q and 20q in both primary tumor and the corresponding liver metastases (Diep et al. 2004). Gains of 13q and 20q are significant molecular events in adenoma-carcinoma progression as well as metastasis formation. These aberrations seem to be important prognostic markers in CRC with a significantly worse outcome for rectal carcinoma (Maharaj et al. 2022).
In the present study, sample materials of a small cohort including primary tumors with liver metastases and brain metastases were analyzed. Therefore, statistical statements are limited. In follow-up studies, a larger cohort would be useful to verify the detected results. Rectal cancer seems to be associated with an increased risk of BM (in comparison to colon cancer) (Christensen et al. 2016). Therefore, a comparative genetic analysis using colon cancer and rectal cancer could be of special interest.
Conclusions
Employing SNP array analyses on primary colorectal cancer and matched metachronous liver and brain metastases, we detected undescribed chromosomal aberrations in both, primary tumor as well as corresponding metastases. Despite considerable heterogeneity, comprehensive analyses of CRC and matched metastases revealed regions with intratumoral (e.g., gain 13q12.13-q12.3) and intertumoral homogeneity (e.g., gain for 8q22.3-q24.3). Furthermore, the SNP array data revealed a high number of cn-LOH aberrations in brain metastases, which suggests cn-LOHs to be involved when CRC cells develop into the cerebral seeding phenotype. Therefore, further analyses are needed to get more information on the increased occurrence of cn-LOHs in brain metastases.
Data availability
The datasets generated and/ or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CRC:
-
Colorectal carcinoma
- CIN:
-
Chromosomal instability
- MSI:
-
Microsatellite instability
- CIMP:
-
CpG island methylator phenotype
- CNV:
-
Copy Number Variation
- CGH:
-
Comparative genomic hybridization
- SNP:
-
Single nucleotide polymorphism
- cn-LOH:
-
Copy-neutral loss of heterozygosity
- UPD:
-
Uniparental disomy
- LM:
-
Liver metastasis
- BM:
-
Brain metastasis
References
Bacolod MD, Barany F (2011) Molecular profiling of colon tumors: the search for clinically relevant biomarkers of progression, prognosis, therapeutics, and predisposition. Ann Surg Oncol 18:3694–3700. https://doi.org/10.1245/s10434-011-1615-5
Beroukhim R, Lin M, Park Y, Hao K, Zhao X, Garraway LA, Fox EA, Hochberg EP, Mellinghoff IK, Hofer MD, Descazeaud A, Rubin MA, Meyerson M, Wong WH, Sellers WR, Li C (2006) Inferring loss-of-heterozygosity from unpaired tumors using high-density oligonucleotide SNP arrays. PLoS Comput Biol 2:e41. https://doi.org/10.1371/journal.pcbi.0020041
Cardoso J, Boer J, Morreau H, Fodde R (2007) Expression and genomic profiling of colorectal cancer. Biochim Biophys Acta 1775:103–137. https://doi.org/10.1016/j.bbcan.2006.08.004
Christensen TD, Spindler K-LG, Palshof JA, Nielsen DL (2016) Systematic review: brain metastases from colorectal cancer–Incidence and patient characteristics. BMC Cancer 16:260. https://doi.org/10.1186/s12885-016-2290-5
Damiens K, Ayoub JPM, Lemieux B, Aubin F, Saliba W, Campeau MP, Tehfe M (2012) Clinical features and course of brain metastases in colorectal cancer: an experience from a single institution. Curr Oncol 19:254–258. https://doi.org/10.3747/co.19.1048
Diep CB, Teixeira MR, Thorstensen L, Wiig JN, Eknaes M, Nesland JM, Giercksky K-E, Johansson B, Lothe RA (2004) Genome characteristics of primary carcinomas, local recurrences, carcinomatoses, and liver metastases from colorectal cancer patients. Mol Cancer 3:6. https://doi.org/10.1186/1476-4598-3-6
Diep CB, Kleivi K, Ribeiro FR, Teixeira MR, Lindgjaerde OC, Lothe RA (2006) The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer 45:31–41. https://doi.org/10.1002/gcc.20261
Dressler L, Bortolomeazzi M, Keddar MR, Misetic H, Sartini G, Acha-Sagredo A, Montorsi L, Wijewardhane N, Repana D, Nulsen J, Goldman J, Pollitt M, Davis P, Strange A, Ambrose K, Ciccarelli FD (2022) Comparative assessment of genes driving cancer and somatic evolution in non-cancer tissues: an update of the Network of Cancer Genes (NCG) resource. Genome Biol 23:35. https://doi.org/10.1186/s13059-022-02607-z
Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61:759–767. https://doi.org/10.1016/0092-8674(90)90186-i
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386. https://doi.org/10.1002/ijc.29210
Gao J, Chen Y, Wu K-C, Liu J, Zhao Y-Q, Pan Y-L, Du R, Zheng G-R, Xiong Y-M, Xu H-L, Fan D-M (2010) RUNX3 directly interacts with intracellular domain of Notch1 and suppresses Notch signaling in hepatocellular carcinoma cells. Exp Cell Res 316:149–157. https://doi.org/10.1016/j.yexcr.2009.09.025
Golas MM, Gunawan B, Cakir M, Cameron S, Enders C, Liersch T, Füzesi L, Sander B (2022) Evolutionary patterns of chromosomal instability and mismatch repair deficiency in proximal and distal colorectal cancer. Colorectal Dis 24:157–176. https://doi.org/10.1111/codi.15946
Grady WM, Carethers JM (2008) Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135:1079–1099. https://doi.org/10.1053/j.gastro.2008.07.076
Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, de Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S (2015) The consensus molecular subtypes of colorectal cancer. Nat Med 21:1350–1356. https://doi.org/10.1038/nm.3967
Gutenberg A, Gerdes JS, Jung K, Sander B, Gunawan B, Bock HC, Liersch T, Brück W, Rohde V, Füzesi L (2010) High chromosomal instability in brain metastases of colorectal carcinoma. Cancer Genet Cytogenet 198:47–51. https://doi.org/10.1016/j.cancergencyto.2009.12.006
Hai L, Zhang C, Li T, Zhou X, Liu B, Li S, Zhu M, Lin Y, Yu S, Zhang K, Ren B, Ming H, Huang Y, Chen L, Zhao P, Zhou H, Jiang T, Yang X (2018) Notch1 is a prognostic factor that is distinctly activated in the classical and proneural subtype of glioblastoma and that promotes glioma cell survival via the NF-κB(p65) pathway. Cell Death Dis 9:158. https://doi.org/10.1038/s41419-017-0119-z
Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, Roemen G, Arends J-W, Williams R, Giaretti W, de Goeij A, Meijer G (2002) Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology 123:1109–1119. https://doi.org/10.1053/gast.2002.36051
Jasmine F, Rahaman R, Dodsworth C, Roy S, Paul R, Raza M, Paul-Brutus R, Kamal M, Ahsan H, Kibriya MG (2012) A genome-wide study of cytogenetic changes in colorectal cancer using SNP microarrays: opportunities for future personalized treatment. PLoS ONE 7:e31968. https://doi.org/10.1371/journal.pone.0031968
Jones AM, Douglas EJ, Halford SE, Fiegler H, Gorman PA, Roylance RR, Carter NP, Tomlinson IPM (2005) Array-CGH analysis of microsatellite-stable, near-diploid bowel cancers and comparison with other types of colorectal carcinoma. Oncogene 24:118–129. https://doi.org/10.1038/sj.onc.1208194
Knösel T, Petersen S, Schwabe H, Schlüns K, Stein U, Schlag PM, Dietel M, Petersen I (2002) Incidence of chromosomal imbalances in advanced colorectal carcinomas and their metastases. Virchows Arch 440:187–194. https://doi.org/10.1007/s004280100493
Knösel T, Schlüns K, Stein U, Schwabe H, Schlag PM, Dietel M, Petersen I (2004) Chromosomal alterations during lymphatic and liver metastasis formation of colorectal cancer1. Neoplasia 6:23–28
Knosel T, Schluns K, Dietel M, Petersen I (2005) Chromosomal alterations in lung metastases of colorectal carcinomas: associations with tissue specific tumor dissemination. Clin Exp Metastasis 22:533–538. https://doi.org/10.1007/s10585-005-5239-7
Liu J, Zeng W, Huang C, Wang J, Yang D, Ma D (2018) Predictive and prognostic implications of mutation profiling and microsatellite instability status in patients with metastatic colorectal carcinoma. Gastroenterol Res Pract. https://doi.org/10.1155/2018/4585802
Maharaj S, Sharaf R, Redman RA, Rojan A (2022) Chromosome 20q and 13q gain in metastatic colorectal cancer: prognostic significance and genomic correlates. JCO 40:e15553–e15553. https://doi.org/10.1200/JCO.2022.40.16_suppl.e15553
Mauri G, Sartore-Bianchi A, Russo A-G, Marsoni S, Bardelli A, Siena S (2019) Early-onset colorectal cancer in young individuals. Mol Oncol 13:109–131. https://doi.org/10.1002/1878-0261.12417
Mitra D, Clark JW, Shih HA, Oh KS, Brastianos PK, Wo JY, Strickland MR, Curry WT, Parikh AR, Corcoran RB, Ryan DP, Iafrate AJ, Borger DR, Lennerz JK, Hong TS (2019) Enrichment of HER2 amplification in brain metastases from primary gastrointestinal malignancies. Oncologist 24:193–201. https://doi.org/10.1634/theoncologist.2018-0152
Muñoz-Bellvis L, Fontanillo C, González-González M, Garcia E, Iglesias M, Esteban C, Gutierrez ML, Abad MM, Bengoechea O, de Las Rivas J, Orfao A, Sayagués JM (2012) Unique genetic profile of sporadic colorectal cancer liver metastasis versus primary tumors as defined by high-density single-nucleotide polymorphism arrays. Mod Pathol 25:590–601. https://doi.org/10.1038/modpathol.2011.195
Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, Walch H, Chatila WK, Madupuri R, Kundra R, Bielski CM, Mastrogiacomo B, Donoghue MT, Boire A, Chandarlapaty S, Ganesh K, Harding JJ, Iacobuzio-Donahue CA, Razavi P, Reznik E, Rudin CM, Zamarin D, Abida W, Abou-Alfa GK, Aghajanian C, Cercek A, Chi P, Feldman D, Ho AL, Iyer G, Janjigian YY, Morris M, Motzer RJ, O’Reilly EM, Postow MA, Raj NP, Riely GJ, Robson ME, Rosenberg JE, Safonov A, Shoushtari AN, Tap W, Teo MY, Varghese AM, Voss M, Yaeger R, Zauderer MG, Abu-Rustum N, Garcia-Aguilar J, Bochner B, Hakimi A, Jarnagin WR, Jones DR, Molena D, Morris L, Rios-Doria E, Russo P, Singer S, Strong VE, Chakravarty D, Ellenson LH, Gopalan A, Reis-Filho JS, Weigelt B, Ladanyi M, Gonen M, Shah SP, Massague J, Gao J, Zehir A, Berger MF, Solit DB, Bakhoum SF, Sanchez-Vega F, Schultz N (2022) Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 185:563-575.e11. https://doi.org/10.1016/j.cell.2022.01.003
Nishina S-I, Shiraha H, Nakanishi Y, Tanaka S, Matsubara M, Takaoka N, Uemura M, Horiguchi S, Kataoka J, Iwamuro M, Yagi T, Yamamoto K (2011) Restored expression of the tumor suppressor gene RUNX3 reduces cancer stem cells in hepatocellular carcinoma by suppressing Jagged1-notch signaling. Oncol Rep 26:523–531. https://doi.org/10.3892/or.2011.1336
Pino MS, Chung DC (2010) The chromosomal instability pathway in colon cancer. Gastroenterology 138:2059–2072. https://doi.org/10.1053/j.gastro.2009.12.065
Sefrioui D, Vermeulin T, Blanchard F, Chapusot C, Beaussire L, Armengol-Debeir L, Sesboué R, Gangloff A, Hebbar M, Copin M-C, Houivet E, Schwarz L, Clatot F, Tuech J-J, Bénichou J, Martin L, Bouvier A-M, Sabourin J-C, Sarafan-Vasseur N, Frébourg T, Lepage C, Michel P, Di Fiore F (2017) Copy number variations in DCC/18q and ERBB2/17q are associated with disease-free survival in microsatellite stable colon cancer. Int J Cancer 140:1653–1661. https://doi.org/10.1002/ijc.30584
Steponaitis G, Kazlauskas A, Vaitkienė P, Deltuva VP, Mikuciunas M, Skiriutė D (2019) Oncosuppressive role of RUNX3 in human astrocytomas. J Oncol 2019:1232434. https://doi.org/10.1155/2019/1232434
Sylvester BE, Vakiani E (2015) Tumor evolution and intratumor heterogeneity in colorectal carcinoma: insights from comparative genomic profiling of primary tumors and matched metastases. J Gastrointest Oncol 6:668–675. https://doi.org/10.3978/j.issn.2078-6891.2015.083
Takayama T, Miyanishi K, Hayashi T, Sato Y, Niitsu Y (2006) Colorectal cancer: genetics of development and metastasis. J Gastroenterol 41:185–192. https://doi.org/10.1007/s00535-006-1801-6
Teh M-T, Blaydon D, Chaplin T, Foot NJ, Skoulakis S, Raghavan M, Harwood CA, Proby CM, Philpott MP, Young BD, Kelsell DP (2005) Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res 65:8597–8603. https://doi.org/10.1158/0008-5472.CAN-05-0842
Torabi K, Miró R, Fernández-Jiménez N, Quintanilla I, Ramos L, Prat E, Del Rey J, Pujol N, Killian JK, Meltzer PS, Fernández PL, Ried T, Lozano JJ, Camps J, Ponsa I (2015) Patterns of somatic uniparental disomy identify novel tumor suppressor genes in colorectal cancer. Carcinogenesis 36:1103–1110. https://doi.org/10.1093/carcin/bgv115
Torabi K, Erola P, Alvarez-Mora MI, Díaz-Gay M, Ferrer Q, Castells A, Castellví-Bel S, Milà M, Lozano JJ, Miró R, Ried T, Ponsa I, Camps J (2019) Quantitative analysis of somatically acquired and constitutive uniparental disomy in gastrointestinal cancers. Int J Cancer 144:513–524. https://doi.org/10.1002/ijc.31936
Vatandoust S, Price TJ, Karapetis CS (2015) Colorectal cancer: metastases to a single organ. World J Gastroenterol 21:11767–11776. https://doi.org/10.3748/wjg.v21.i41.11767
Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, Benson AB, Hamilton SR (2001) Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med 344:1196–1206. https://doi.org/10.1056/NEJM200104193441603
Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38:787–793. https://doi.org/10.1038/ng1834
Weiss L, Grundmann E, Torhorst J, Hartveit F, Moberg I, Eder M, Fenoglio-Preiser CM, Napier J, Horne CH, Lopez MJ (1986) Haematogenous metastatic patterns in colonic carcinoma: an analysis of 1541 necropsies. J Pathol 150:195–203. https://doi.org/10.1002/path.1711500308
Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, Wood LD, Goggins M, Hruban RH, Chang AE, Calses P, Thorsen SM, Hingorani SR (2015) RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell 161:1345–1360. https://doi.org/10.1016/j.cell.2015.04.048
Zarzour P, Boelen L, Luciani F, Beck D, Sakthianandeswaren A, Mouradov D, Sieber OM, Hawkins NJ, Hesson LB, Ward RL, Wong JWH (2015) Single nucleotide polymorphism array profiling identifies distinct chromosomal aberration patterns across colorectal adenomas and carcinomas. Genes Chromosom Cancer 54:303–314. https://doi.org/10.1002/gcc.22243
Zhao K, Cui X, Wang Q, Fang C, Tan Y, Wang Y, Yi K, Yang C, You H, Shang R, Wang J, Kang C (2019) RUNX1 contributes to the mesenchymal subtype of glioblastoma in a TGFβ pathway-dependent manner. Cell Death Dis 10:877. https://doi.org/10.1038/s41419-019-2108-x
Zhu K, Guo J, Wang H, Yu W (2016) FRAT1 expression regulates proliferation in colon cancer cells. Oncol Lett 12:4761–4766. https://doi.org/10.3892/ol.2016.5300
Žilina O, Koltšina M, Raid R, Kurg A, Tõnisson N, Salumets A (2015) Somatic mosaicism for copy-neutral loss of heterozygosity and DNA copy number variations in the human genome. BMC Genom 16:703. https://doi.org/10.1186/s12864-015-1916-3
Acknowledgements
The authors thank the Department of Pathology at the Leipzig University Medical Center, Institute of Pathology at University Hospital Bochum and the Institute of Pathology Weißenfels for their transfer of the sample material as well as for the performance of the pathological investigations.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: HH. Formal Analysis: V-PB. Investigation: MR, MW, V-PB. Project Administration: HH. Resources: CS, CF. Supervision: HH. Visualization: LH, CS, V-PB. Writing – Original Draft Preparation: HH, CS, MW, V-PB. Writing – Review & Editing: HH, RK, UN.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the University Leipzig (Az.: 005/17-ek).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brandt, VP., Holland, H., Wallenborn, M. et al. SNP array genomic analysis of matched pairs of brain and liver metastases in primary colorectal cancer. J Cancer Res Clin Oncol 149, 18173–18183 (2023). https://doi.org/10.1007/s00432-023-05505-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05505-4