Abstract
Background
Colorectal cancer (CRC) is one of the most common causes of cancer-related deaths in the Western world, and despite the fact that metastases are usually the ultimate cause of deaths, the knowledge of the genetics of advanced stages of this disease is limited. In order to identify potential genetic abnormalities underlying the development of local and distant metastases in CRC patients, we have, by comparative genomic hybridization, compared the DNA copy number profiles of 10 primary carcinomas, 14 local recurrences, 7 peritoneal carcinomatoses, and 42 liver metastases from 61 CRC patients.
Results
The median number of aberrations among the primary carcinomas, local recurrences, carcinomatoses, and liver metastases was 10, 6, 13, and 14, respectively. Several genetic imbalances, such as gains of 7, 8q, 13q, and 20, and losses of 4q, 8p, 17p, and 18, were common in all groups. In contrast, gains of 5p and 12p were more common in the carcinomatoses than in other stages of the disease. With hierarchical cluster analysis, liver metastases could be divided into two main subgroups according to clusters of chromosome changes.
Conclusions
Each stage of CRC progression is characterized by a particular genetic profile, and both carcinomatoses and liver metastases are more genetically complex than local recurrences and primary carcinomas. This is the first genome profiling of local recurrences and carcinomatoses, and gains of 5p and 12p seem to be particularly important for the spread of the CRC cells within the peritoneal cavity.
Similar content being viewed by others
Background
More than a decade ago, a genetic model for the adenoma–carcinoma sequence in the large bowel was presented by Fearon and Vogelstein [1]. It is now widely accepted that colorectal cancer (CRC) arises through the accumulation of genetic and epigenetic changes [2]. The order as well as the number of events are important in the process that transform normal cells into neoplastic precursors and subsequently into malignant tumors, which may further metastasize [3]. Inactivation of tumorsupressor genes, APC and TP53, as well as components of mismatch repair system, is commonly found in colorectal tumors. A recent study suggest alternative molecular pathways for colorectal carcinomas based on the observation that APC, KRAS2, and TP53 are all frequently mutated but rarely in the same tumor [4]. Twelve to fifteen percent of all primary colorectal carcinomas display microsatellite instability [5–8], a result of defect mismatch repair [9]. The majority of colorectal carcinomas, however, harbor numerous aberrations at the chromosome level, and chromosomal instability seems to be pronounced in these tumors [2]. This type of instability may be caused by varying mechanisms, including telomer dysfunction, defect DNA double-strand break repair and disturbances during chromosome segregation [10].
The genomes of hundreds of primary colorectal carcinomas have now been studied by conventional chromosomal banding technique as well as by comparative genomic hybridization (CGH) [11–28]. These studies have revealed a nonrandom pattern of genomic abnormalities in primary carcinomas, including frequent gain of material from chromosomes and chromosome arms 7, 8q, 13, and 20, and losses from 4, 8p, 14, 17p, and 18q. However, in spite of the fact that metastases are usually the ultimate cause of death in CRC patients [29], the cytogenetic changes that characterize and presumably drive the advanced stages of this disease have been poorly described [reviewed in [25, 30]]. We have recently analyzed 17 CRC liver metastases, combining chromosome banding with CGH, and found that the former technique, in contrast to CGH profiles, did not always detect the cytogenetically abnormal clones [30]. The use of CGH have provided profiles of various series of liver metastases [reviewed in [30]]. To our knowledge the genome aberrations in local recurrences and peritoneal carcinomatoses from CRC patient have not previously been described. In order to identify genetic changes underlying the development of local and distant metastases, we have compared the genomes of primary CRC with those of local recurrences, peritoneal carcinomatoses, and liver metastases using a molecular cytogenetic approach.
Results
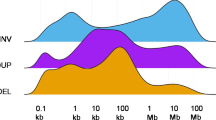
The complete CGH profiles are presented in the supplementary Table 1 (additional data file 1). All tumors, except one primary carcinoma, three local recurrences and one liver metastasis, exhibited DNA copy number changes, and the overall copy number profiles for each tumor stage are illustrated in Fig. 1. The number of imbalances per case ranged from 0 to 28 (median, 11). Although all chromosomes were involved, the distribution of the imbalances was clearly nonrandom. The most common copy number changes, found in more than 20% of each tumor group, were gains of 7, 8q, 13q, and 20, and losses of 4q, 8p, 17p, and 18, but however, frequency variations were observed among primary tumors, local recurrences, and liver metastases (Fig. 1). Twenty tumors showed amplifications (Table 1, supplementary data, additional data file 1) in one to nine discrete regions: chromosome arms 13q (ten cases), 20q (nine cases), 8q (eight cases), 20p (six cases), 5p (one case), and chromosome X (two cases) and 7 (one case). The frequency of amplification did not differ significantly among the different tumor groups.
Graphic comparison of the overall genomic gains (A) and losses (B) detected by CGH in different CRC stages. For each case, the presence or absence of imbalance in every chromosome band (from 1p36 to Xq28) was computed in a spreadsheet. The total number of imbalances detected in every band was then used to prepare the graphic comparison. All chromosome arms were involved in imbalances. (A) Loss of 18q was one of the most frequent aberrations seen in all groups. In addition to 18q loss, primary carcinomas and liver metastases often showed loss of 8p. Additionally, liver metastases and carcinomatoses frequently harbored loss of 4q. (B) Gain of 8q and 20q were common in all tumor types. In addition, carcinomatoses often contained gains of 5p, 7p, 9q, 12, and 13q, whereas liver metastases often showed gains of 7p, and 13q. * Multiple samples from a single tumor (case no. 53R1 and 53R2; 64C1 and 64C2; 21L1 and 21L2; 42L1 and 42L2; 52L1 and 52L2; 76L1, 76L2, and 76L3) were counted as one sample.
The number of imbalances per case for primary carcinomas, local recurrences, carcinomatoses, and liver metastases ranged from 0 to 16 (median, 10), 0 to 17 (median, 6), 1 to 20 (median, 13), and 0 to 28 (median, 14), respectively. Increased copy numbers of chromosome arms 5p and 12p were significantly more frequent in the carcinomatoses than in the other lesions, except the association for 5p between carcinomatoses and primary carcinomas (Table 1).
A hierarchical cluster analysis was performed for the colorectal tumors based on the gains and losses from all chromosome arms (Fig. 2). No chromosome changes seemed to be tumor-stage specific. Furthermore, a separate cluster analysis for the 37 liver metastases is illustrated in Fig. 3. The majority of the liver metastases harbored changes at 7, 8p, 13q, 17p, 18, and 20 (pink tree). The top dendogram indicates that the liver metastases can be subdivided into two main groups (red and brown trees) according to the chromosome changes, one of these subgroup is characterized by additional changes at 4q, 5q, 6p, 8q, 16 (blue tree).
Hierarchical clustering of tumors from different stages of colorectal cancer. The hierarchical clustering is based on chromosome aberrations of all chromosome arms. Chromosome arms 13p, 14p, 15p, 21p, 22p, and chromosome Y, due to high content of heterochromatin, were excluded from the cluster analysis. The chromosome arms are given in the right dendogram (gains in green and losses in blue). Each tumor sample is depicted and coded (white – primary carcinomas; yellow – local recurrence; green – peritoneal carcinomatoses; red – liver metastases) at the top of the dendogram. Each row represents the alterations from a separate chromosome arm over all tumor samples, and each column represents all changes in each tumor.
Hierarchical clustering of liver metastases from colorectal cancer patients. The hierarchical clustering is based on chromosome aberrations of all chromosome arms. Chromosome arms 13p, 14p, 15p, 21p, 22p, and chromosome Y, due to high content of heterochromatin, were excluded from the cluster analysis. The chromosome arms are given in the right dendogram (gains in green and losses in blue). Each tumor sample is depicted at the top of the dendogram. Each row represents alterations from a separate chromosome arm over all tumor samples, and each column represents all changes in each tumor.
In four of the five patients from whom both primary carcinomas and liver metastases were analyzed, the liver metastases had more aberrations than the corresponding primary carcinomas (Table 1, supplementary data, additional data file 1). Gains of 8q (four cases) and 20q (three cases), and losses of 8p (three cases) and 18q (three cases) were seen in both the primary carcinomas and metastatic samples, whereas gain of 7p and loss of 15q were more common in the liver metastases than in the primary carcinomas (7p: in 5 metastases versus 2 primary carcinomas; 15q: in 4 metastases versus 1 primary carcinoma).
Discussion
The present study is the first CGH study of peritoneal carcinomatoses and local recurrences from CRC patients and includes one of the largest series of liver metastases analyzed. Our data provide clues to the understanding of the genetic basis of the advanced disease stages. Local recurrences are re-growth of tumor cells at the site of the primary carcinomas. We found that the median number of alterations in these tumors was lower than in the primary carcinomas. In a heterogeneous primary tumor, regrowth to a local recurrency might originate from genetically less complex cells in the primary tumor. This observation should be interpreted with caution since the analyzed primary carcinomas and local recurrences were not from the same patients, and in fact the ranges of aberrations were large for both tumor groups.
Peritoneal carcinomatosis reflects the regional spread of cancer cells within the peritoneal cavity. Although carcinomatosis is not always associated with widespread visceral or extra-peritoneal disease, it almost always reflects an incurable disease [31, 32]. The present comparisons of the distribution and frequency of specific chromosomal imbalances among the different tumor stages revealed significant differences between carcinomatoses and each of the other stages of the disease regarding gains at 5p and 12p (Table 1). The frequencies of 5p gains were not significantly different between primary carcinomas and carcinomatoses (Table 1), is best explained by small sample sets. However, these chromosome aberrations are indeed quite rare in primary colorectal carcinomas as evaluated from previous studies, and among 670 cases the average frequencies for 5p and 12p gains were less than 10% each [11–23, 28, 33–40]. Summarized, the present study and the previous data, lead us to speculate that genes located at these chromosome arms are involved in development of peritoneal carcinomatoses. Although gain of 5p12-p14 was confirmed in both samples from the same patient (Fig. 4), a smallest region of overlapping gain can not determined from one case only.
Gains of 5p and 12p in carcinomatoses. Gains of 5p (samples 17C, 64C1, and 64C2) and 12p (samples 36C, 64C1, and 62C2) were clearly detected by CGH. The central line (red) in the CGH profile shows the average fluorescence ratio along the chromosome, and the flanking curves (brown) represent the 95% confidence interval. The red and the green lines represent the cut-off values, 0.83 and 1.17, respectively.
Our findings agree well with previous CGH studies of primary colorectal carcinomas and liver metastases, as the type and the frequency of copy number changes are comparable [11–23, 28, 30, 33–42]. The increasing number of aberrations found in distant metastases compared to primary carcinomas is also in line with a previous allelotype study in which we found a higher fractional allelic imbalance in liver metastases than in primary carcinomas and local recurrences [43].
Some observation from the results obtained from the five primary carcinomas and their corresponding liver metastases are worth mentioning. Gain of 7p and loss of 15q were more frequent in the latter, and interestingly the three primary carcinomas that each showed one of these aberrations had synchronous liver metastases. When the present cases and those of previous reports were combined [11–23, 28, 41, 42], statistical significant frequency differences between these two stages were found (7p: 216/670 versus 74/147, P < 0.001; 15q: 114/670 versus 39/147, P < 0.001), indicating that 7p gains and 15q losses may be involved in the development of distant metastases.
Losses involving chromosomes 17 and 18 were seen in 19 (31%) and 42 (69%), respectively, of the cases in the present study. These changes have been suggested to be late and important events in the adenoma-carcinoma sequence [1], as they have been shown to be more frequent in carcinomas than in adenomas [24, 44, 45]. Deletions of 17p and 18q usually reflect inactivation of TP53 [1], DCC [46], MADH2, and MADH4 [46, 47]. Changes at chromosome 8, usually loss of 8p and gain of 8q, are important late events in colorectal carcinogenesis, and occur more often in carcinomas and metastases than in adenomas [15, 24, 44]. Deletions of 8p, 17p, and 18q and inactivation of their potential target genes are associated with short survival [48, 49], again supporting their important role in the disease progression.
The present study showed that gains of 13q and 20q are also frequent in the late stages of CRC (73% and 89%, respectively). Increased copy number of chromosome arm 13q, often reflecting trisomy of chromosome 13, is also found as the sole aberration in colorectal adenomas [25], suggesting this to be an early event in colorectal tumorigenesis. Gain of 20q is an aberration frequently seen in carcinomas and less commonly in adenomas [15]. In an assessment of the clonal composition of colorectal adenomas using FISH analyses with centromere probes, we found that 7, 13, and 20 were present with >2 signals in at least 1/3 of the adenomas [50]. This and the present results reveal a major cluster of changes in all advanced stages (containing gains of 7, 13q, 20q, and losses of 8p, 17p and 18q), one may therefore speculate that gains of 7, 13q, and 20q precede the losses of 8p, 17p, 18q, and all aberrations seem to be, in most cases, part of the clone within the primary tumor with the ability to metastasize, locally and peripheral.
Conclusions
A number of genetic imbalances common to all tumor groups were demonstrated, as well as consistent genetic differences among the different stages. Whereas local recurrences were genetically less complex than primary colorectal carcinomas, liver metastases and peritoneal carcinomatoses usually have more DNA copy number changes than the lesions from which they originated. Gains of 5p and 12p seem to be particularly important for the spread of CRC cells within the peritoneal cavity.
Material and methods
Patients and tumor samples
Ten primary carcinomas, 14 local recurrences, 7 peritoneal carcinomatoses, and 42 liver metastases from 61 CRC patients admitted to the Norwegian Radium Hospital (Oslo, Norway) and to the Lund University Hospital (Lund, Sweden), were included in the present study. Both primary carcinomas and liver metastases were obtained from each of five patients. Among these, three (76, 93, 136) occurred synchronously and two (3, 112) metachronously.
From each frozen tumor sample, a five μm section was stained with hematoxylin and eosin and evaluated by a pathologist (J. M. N.). The tumor area was identified, and dissected to enrich the fraction of tumor cells before DNA extraction, which was performed using standard phenol and chloroform extraction followed by ethanol precipitation [51] (nucleic acid extractor, Model 340A, Applied Biosystems, Foster City, CA).
CGH analysis
The CGH method initially described by Kallioniemi et al. [52] was used with modifications previously described by Kraggerud et al. [53]. Briefly, test (tumor) and reference (peripheral blood lymphocytes from 4 healthy females or 4 healthy males) DNA were labeled in nick-translation reactions with a mixture of two fluorochrome-conjugated nucleotides (FITC-12-dCTP and FITC-12-dUTP for tumor DNA, and Texas Red-6-dCTP and Texas Red-6-dUTP for reference DNA, New England Nuclear, Boston, MA). The same amounts of labeled tumor and reference DNA (1 μg each) were mixed with 20 μg of unlabeled Cot-1 DNA (Life Technologies, Rockville, MD), ethanol-precipitated, dried, and dissolved in hybridization buffer (Vysis, Downers Grove, IL). Normal metaphase cells were obtained from lymphocyte culture from healthy donors. After denaturing, the DNA was hybridized to normal, denatured metaphase spreads and incubated in a humidified chamber for 2–3 days at 37°C. Finally, the slides were washed and mounted in an antifade solution with DAPI (Vector Laboratories, Burlingame, CA). Good-quality metaphase spreads were selected. Three images – FITC (green) and Texas Red (red) hybridization signals and DAPI counterstain – were sequentially captured with a Cohu 4900 CCD (12 bits gray scale) camera, using an automated filter wheel coupled to a Zeiss Axioplan fluorescence microscope (Zeiss, Oberkochen, Germany), and a CytoVision system (Applied Imaging, Newcastle, UK). Chromosomes were identified based on their inverted DAPI banding, and fluorescence ratio profiles (green to red fluorescence) were calculated for each chromosome using data from at least eight representative copies of each chromosome (range 8 – 22). Average ratio profiles with 95% confidence intervals were generated for each tumor. The Y chromosome was not evaluated. Due to the high quality DNA extracted from frozen tissue, the threshold values 1.17 and 0.83 were used to score gain and loss of DNA sequences, respectively. This threshold allows the detection of one chromosome copy number change present in at least 50% of the cells in a sample from a triploid tumor. Amplification was defined as a ratio equal to or above 2.0, corresponding to the detection of at least 6 and 9 chromosomes in 50% of the cells in a diploid and triploid tumor, respectively. All scorings were performed independently by two of the authors (C. B. D. and R. A. L.) with few interobserver differences; these were resolved after joint reevaluation. A negative (normal versus normal) and a positive (the colon cell line LoVo with known copy number changes) control was included in every set of experiments. The description of the CGH copy number changes followed the guidelines suggested in the International System for Human Cytogenetic Nomenclature [54]. In order to distinguish between a CGH profile that was abnormal throughout the chromosome from one that returns to normal within the terminal band, we used "ter" in the former and the respective band number in the latter when writing the copy number changes. CGH profiles for 17 of the liver metastases have previously been published [30].
Statistical analyzes
For hierarchical clustering the average-linkage method was used with Pearson's correlation similarity measure. The cluster analyses and the drawing of the dendogram were performed with J-Express Pro [55]. For comparisons of different groups, the two-sided Fisher exact test was used, and all the statistical analyzes were performed with the SPSS© software (SPSS, Chicago, IL).
Abbreviations
- CGH:
-
comparative genomic hybridization
- CRC:
-
colorectal cancer.
References
Fearon ER, Vogelstein B: A genetic model for colorectal tumorigenesis. Cell. 1990, 61: 759-767.
Lengauer C, Kinzler KW, Vogelstein B: Genetic instability in colorectal cancers. Nature. 1997, 386: 623-627.
Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell. 1996, 87: 159-170.
Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR: Mutations in APC, Kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002, 99: 9433-9438.
Aaltonen LA, Pennanen S, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR: Clues to the pathogenesis of familial colorectal cancer. Science. 1993, 260: 812-816.
Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M: Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993, 363: 558-561.
Lothe RA, Peltomäki P, Meling GI, Aaltonen LA, Nystrom-Lahti M, Pylkkanen L, Heimdal K, Andersen TI, Møller P, Rognum TO: Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993, 53: 5849-5852.
Thibodeau SN, Bren G, Schaid DJ: Microsatellite instability in cancer of the proximal colon. Science. 1993, 260: 816-819.
Fishel R, Kolodner RD: Identification of mismatch repair genes and their role in the development of cancer. Curr Opin Genet Dev. 1995, 5: 382-395.
Albertson DG, Collins C, McCormick F, Gray JW: Chromosome aberrations in solid tumors. Nat Genet. 2003, 34: 369-376.
Al-Mulla F, Keith WN, Pickford IR, Going JJ, Birnie GD: Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer. 1999, 24: 306-314.
Paredes-Zaglul A, Kang JJ, Essig YP, Mao W, Irby R, Wloch M, Yeatman TJ: Analysis of colorectal cancer by comparative genomic hybridization: evidence for induction of the metastatic phenotype by loss of tumor suppressor genes. Clin Cancer Res. 1998, 4: 879-886.
De Angelis PM, Clausen OP, Schjolberg A, Stokke T: Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br J Cancer. 1999, 80: 526-535.
De Angelis PM, Stokke T, Beigi M, Mjaland O, Clausen OP: Prognostic significance of recurrent chromosomal aberrations detected by comparative genomic hybridization in sporadic colorectal cancer. Int J Colorectal Dis. 2001, 16: 38-45.
Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G: Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996, 15: 234-245.
Aust DE, Willenbucher RF, Terdiman JP, Ferrell LD, Chang CG, Moore DH, Molinaro-Clark A, Baretton GB, Loehrs U, Waldman FM: Chromosomal alterations in ulcerative colitis-related and sporadic colorectal cancers by comparative genomic hybridization. Hum Pathol. 2000, 31: 109-114.
Nakao K, Shibusawa M, Tsunoda A, Yoshizawa H, Murakami M, Kusano M, Uesugi N, Sasaki K: Genetic changes in primary colorectal cancer by comparative genomic hybridization. Surg Today. 1998, 28: 567-569.
Nakao K, Shibusawa M, Ishihara A, Yoshizawa H, Tsunoda A, Kusano M, Kurose A, Makita T, Sasaki K: Genetic changes in colorectal carcinoma tumors with liver metastases analyzed by comparative genomic hybridization and DNA ploidy. Cancer. 2001, 91: 721-726.
Schlegel J, Stumm G, Scherthan H, Bocker T, Zirngibl H, Ruschoff J, Hofstadter F: Comparative genomic in situ hybridization of colon carcinomas with replication error. Cancer Res. 1995, 55: 6002-6005.
Aragane H, Sakakura C, Nakanishi M, Yasuoka R, Fujita Y, Taniguchi H, Hagiwara A, Yamaguchi T, Abe T, Inazawa J, Yamagishi H: Chromosomal aberrations in colorectal cancers and liver metastases analyzed by comparative genomic hybridization. Int J Cancer. 2001, 94: 623-629.
Meijer GA, Hermsen MA, Baak JP, van Diest PJ, Meuwissen SG, Belien JA, Hoovers JM, Joenje H, Snijders PJ, Walboomers JM: Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol. 1998, 51: 901-909.
Rooney PH, Murray GI, Stevenson DA, Haites NE, Cassidy J, McLeod HL: Comparative genomic hybridization and chromosomal instability in solid tumours. Br J Cancer. 1999, 80: 862-873.
Knösel T, Petersen S, Schwabe H, Schluns K, Stein U, Schlag PM, Dietel M, Petersen I: Incidence of chromosomal imbalances in advanced colorectal carcinomas and their metastases. Virchows Arch. 2002, 440: 187-194.
Bardi G, Sukhikh T, Pandis N, Fenger C, Kronborg O, Heim S: Karyotypic characterization of colorectal adenocarcinomas. Genes Chromosomes Cancer. 1995, 12: 97-109.
Mitelman F, Johansson B, Mertens F: Mitelman database of chromosome aberrations in cancer. http://cgap nci nih gov/Chromosomes/Mitelman. 2003
Mertens F, Johansson B, Höglund M, Mitelman F: Chromosomal imbalance maps of malignant solid tumors: a cytogenetic survey of 3185 neoplasms. Cancer Res. 1997, 57: 2765-2780.
Muleris M, Salmon RJ, Dutrillaux B: Cytogenetics of colorectal adenocarcinomas. Cancer Genet Cytogenet. 1990, 46: 143-156.
Ghadimi BM, Grade M, Liersch T, Langer C, Siemer A, Fuzesi L, Becker H: Gain of chromosome 8q23-24 is a predictive marker for lymph node positivity in colorectal cancer. Clin Cancer Res. 2003, 9: 1808-1814.
Registry Norwegian Cancer: Cancer in Norway. http://www kreftregisteret no/. 2001
Diep CB, Parada LA, Teixeira MR, Eknæs M, Nesland JM, Johansson B, Lothe RA: Genetic profiling of colorectal cancer liver metastases by combined comparative genomic hybridization and G-banding analysis. Genes Chromosomes Cancer. 2003, 36: 189-197.
Culliford AT, Brooks AD, Sharma S, Saltz LB, Schwartz GK, O'Reilly EM, Ilson DH, Kemeny NE, Kelsen DP, Guillem JG, Wong WD, Cohen AM, Paty PB: Surgical debulking and intraperitoneal chemotherapy for established peritoneal metastases from colon and appendix cancer. Ann Surg Oncol. 2001, 8: 787-795.
Sadeghi B, Arvieux C, Glehen O, Beaujard AC, Rivoire M, Baulieux J, Fontaumard E, Brachet A, Caillot JL, Faure JL, Porcheron J, Peix JL, Francois Y, Vignal J, Gilly FN: Peritoneal carcinomatosis from non-gynecologic malignancies: results of the EVOCAPE 1 multicentric prospective study. Cancer. 2000, 88: 358-363.
Alcock HE, Stephenson TJ, Royds JA, Hammond DW: Analysis of colorectal tumor progression by microdissection and comparative genomic hybridization. Genes Chromosomes Cancer. 2003, 37: 369-380.
Georgiades IB, Curtis LJ, Morris RM, Bird CC, Wyllie AH: Heterogeneity studies identify a subset of sporadic colorectal cancers without evidence for chromosomal or microsatellite instability. Oncogene. 1999, 18: 7933-7940.
He QJ, Zeng WF, Sham JS, Xie D, Yang XW, Lin HL, Zhan WH, Lin F, Zeng SD, Nie D, Ma LF, Li CJ, Lu S, Guan XY: Recurrent genetic alterations in 26 colorectal carcinomas and 21 adenomas from Chinese patients. Cancer Genet Cytogenet. 2003, 144: 112-118.
Leung SY, Yuen ST, Chan TL, Chan AS, Ho JW, Kwan K, Fan YW, Hung KN, Chung LP, Wyllie AH: Chromosomal instability and p53 inactivation are required for genesis of glioblastoma but not for colorectal cancer in patients with germline mismatch repair gene mutation. Oncogene. 2000, 19: 4079-4083.
Chan TL, Curtis LC, Leung SY, Farrington SM, Ho JW, Chan AS, Lam PW, Tse CW, Dunlop MG, Wyllie AH, Yuen ST: Early-onset colorectal cancer with stable microsatellite DNA and near-diploid chromosomes. Oncogene. 2001, 20: 4871-4876.
Richter H, Slezak P, Walch A, Werner M, Braselmann H, Jaramillo E, Ost A, Hirata I, Takahama K, Zitzelsberger H: Distinct chromosomal imbalances in nonpolypoid and polypoid colorectal adenomas indicate different genetic pathways in the development of colorectal neoplasms. Am J Pathol. 2003, 163: 287-294.
Leslie A, Carey FA, Pratt NR, Steele RJ: The colorectal adenoma-carcinoma sequence. Br J Surg. 2002, 89: 845-860.
Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, Roemen G, Arends JW, Williams R, Giaretti W, De Goeij A, Meijer G: Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002, 123: 1109-1119.
Korn WM, Yasutake T, Kuo WL, Warren RS, Collins C, Tomita M, Gray JW, Waldman FM: Chromosome arm 20q gains and other genomic alterations in colorectal cancer metastatic to liver, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Genes Chromosomes Cancer. 1999, 25: 82-90.
Platzer P, Upender MB, Wilson K, Willis J, Lutterbaugh JD, Nosrati A, Willson JK, Mack D, Ried T, Markowitz SD: Silence of chromosomal amplifications in colon cancer. Cancer Res. 2002, 62: 1134-1138.
Thorstensen L, Qvist H, Nesland JM, Giercksky KE, Lothe RA: Allelotype profiles of local recurrences and distant metastases from colorectal-cancer patients. Int J Cancer. 1996, 69: 452-456.
Griffin CA, Lazar S, Hamilton SR, Giardiello FM, Long P, Krush AJ, Booker SV: Cytogenetic analysis of intestinal polyps in polyposis syndromes: comparison with sporadic colorectal adenomas. Cancer Genet Cytogenet. 1993, 67: 14-20.
Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL: Genetic alterations during colorectal-tumor development. N Engl J Med. 1988, 319: 525-532.
Cho KR, Fearon ER: DCC: linking tumor suppressor genes and altered cell surface interactions in cancer?. Curr Opin Genet Dev. 1995, 5: 72-78.
Arends JW: Molecular interactions in the Vogelstein model of colorectal carcinoma. J Pathol. 2000, 190: 412-416.
McLeod HL, Murray GI: Tumour markers of prognosis in colorectal cancer. Br J Cancer. 1999, 79: 191-203.
Halling KC, French AJ, McDonnell SK, Burgart LJ, Schaid DJ, Peterson BJ, Moon-Tasson L, Mahoney MR, Sargent DJ, O'Connell MJ, Witzig TE, Farr G.H., Jr, Goldberg RM, Thibodeau SN: Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999, 91: 1295-1303.
Bomme L, Lothe RA, Bardi G, Fenger C, Kronborg O, Heim S: Assessments of clonal composition of colorectal adenomas by FISH analysis of chromosomes 1, 7, 13 and 20. Int J Cancer. 2001, 92: 816-823.
Kunkel LM, Smith KD, Boyer SH, Borgaonkar DS, Wachtel SS, Miller OJ, Breg WR, Jones HW, Rary JM: Analysis of human Y-chromosome-specific reiterated DNA in chromosome variants. Proc Natl Acad Sci USA. 1977, 74: 1245-1249.
Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992, 258: 818-821.
Kraggerud SM, Szymanska J, Abeler VM, Kaern J, Eknæs M, Heim S, Teixeira MR, Trope CG, Peltomäki P, Lothe RA: DNA copy number changes in malignant ovarian germ cell tumors. Cancer Res. 2000, 60: 3025-3030.
, : An International System for Human Cytogenetic Nomenclature. 1995, Basel, Karger, Mitelman F.
Dysvik B, Jonassen I: J-Express: exploring gene expression data using Java. Bioinformatics. 2001, 17: 369-370.
Acknowledgements
The technical assistance of Merete Hektoen for DNA extraction is gratefully acknowledged. This work was supported by a PhD grant (CBD) from the Norwegian Foundation for Health and Rehabilitation (CBD), as well as grants from the Norwegian Cancer Society (LT, RAL), and the Swedish Cancer Society (BJ).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
CBD, LT, and ME carried out the experimental work. CBD interpreted the result, performed bioinformatics and statistical analyses and drafted the manuscript. MRT participated in the evaluation of the cytogenetic data and in the manuscript preparation. JNW, K-EG, and BJ were responsible for referring the patients, collecting tissue specimens and clinical information. JMN determined the histological diagnoses. RAL conceived the study, was responsible for its design and coordination, participated in the evaluation of the data and in the manuscript preparation. All authors read and approved the final version.
Electronic supplementary material
12943_2003_57_MOESM1_ESM.doc
Additional data file 1: CGH copy number changes of different tumor stages in colorectal cancer. Complete CGH profiles of 10 primary carcinomas, 14 local recurrences, 7 peritoneal carcinomatoses, and 42 liver metastases from colorectal cancer patients. (DOC 103 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Diep, C.B., Teixeira, M.R., Thorstensen, L. et al. Genome characteristics of primary carcinomas, local recurrences, carcinomatoses, and liver metastases from colorectal cancer patients. Mol Cancer 3, 6 (2004). https://doi.org/10.1186/1476-4598-3-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-4598-3-6