Abstract
The expression of Neuritin-1 (NRN1), a neurotrophic factor crucial for neurodevelopment and synaptic plasticity, is enhanced by the Brain Derived Neurotrophic Factor (BDNF). Although the receptor of NRN1 remains unclear, it is suggested that NRN1’s activation of the insulin receptor (IR) pathway promotes the transcription of the calcium voltage-gated channel subunit alpha1 C (CACNA1C). These three genes have been independently associated with schizophrenia (SZ) risk, symptomatology, and brain differences. However, research on how they synergistically modulate these phenotypes is scarce. We aimed to study whether the genetic epistasis between these genes affects the risk and clinical presentation of the disorder via its effect on brain structure. First, we tested the epistatic effect of NRN1 and BDNF or CACNA1C on (i) the risk for SZ, (ii) clinical symptoms severity and functionality (onset, PANSS, CGI and GAF), and (iii) brain cortical structure (thickness, surface area and volume measures estimated using FreeSurfer) in a sample of 86 SZ patients and 89 healthy subjects. Second, we explored whether those brain clusters influenced by epistatic effects mediate the clinical profiles. Although we did not find a direct epistatic impact on the risk, our data unveiled significant effects on the disorder’s clinical presentation. Specifically, the NRN1-rs10484320 x BDNF-rs6265 interplay influenced PANSS general psychopathology, and the NRN1-rs4960155 x CACNA1C-rs1006737 interaction affected GAF scores. Moreover, several interactions between NRN1 SNPs and BDNF-rs6265 significantly influenced the surface area and cortical volume of the frontal, parietal, and temporal brain regions within patients. The NRN1-rs10484320 x BDNF-rs6265 epistasis in the left lateral orbitofrontal cortex fully mediated the effect on PANSS general psychopathology. Our study not only adds clinical significance to the well-described molecular relationship between NRN1 and BDNF but also underscores the utility of deconstructing SZ into biologically validated brain-imaging markers to explore their mediation role in the path from genetics to complex clinical manifestation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Schizophrenia (SZ) is a severe psychiatric disorder with a substantial burden affecting 21 million people worldwide (Charlson et al. 2018). It has a strong genetic component that is reflected in its high heritability (h2 = 65–79%) (Hilker et al. 2018; Sullivan et al. 2003). Moreover, genome-wide association studies (GWAS) have provided solid evidence of its polygenicity by describing a hundred genetic variants with additive effects (Pardiñas et al. 2018; Trubetskoy et al. 2022). Interestingly, due to the convergence of those variants in molecular networks related to synaptic plasticity, it has been proposed as a key pathophysiological mechanism in SZ (Hall and Bray 2022). However, the identified risk variants together only account for a proportion (h2SNP = 24%) of the variance in liability of the phenotype (Trubetskoy et al. 2022).
It has been suggested that such missing heritability could arise, among other factors, from nonlinear molecular interactions, which are not typically explored in classical GWAS models (Zuk et al. 2012). In fact, gene-gene interactions between two or more loci have been extensively studied in animal and cellular models, suggesting that epistatic networks might represent an essential molecular mechanism involved in the modulation of complex traits (Mackay and Moore 2014; Özsoy et al. 2021). When the collective effect of large-scale genetic interactions is considered in humans, the association with complex traits, such as SZ, becomes more robust and new variants appear (Woo et al. 2017). Therefore, considering that genetic interactions mainly occur between genes involved in the same molecular pathways (Roguev et al. 2008), inspecting epistatic effects among those involved in synaptic plasticity in SZ may add relevant data on how genetic factors lead to the emergence of the disorder.
Considering the phenotypic complexity of SZ as reflected in its diverse outcomes, neuroimaging genetics approaches provide a neurobiological context for studying how genetic variants act to confer an increased risk for the disorder (van der Meer and Kaufmann 2022). Notably, SZ-related genes identified by GWAS are highly expressed in brain regions with structural differences in SZ (Ji et al. 2021) and have been associated with symptomatology (Legge et al. 2021; Sengupta et al. 2017). This indicates a partial genetic overlap within the axis connecting the brain and symptoms, with specific genes delineated as potential contributors to cerebral alterations that sustain distinct manifestations. Indeed, some studies have shown that genetic effects on clinical phenotypes are mediated by specific brain regions and functions in mental disorders (Sudre et al. 2020). Nevertheless, few cases in the literature explore the impact of epistasis on brain phenotypes (Callicott et al. 2013; Guardiola-Ripoll et al. 2022; Tecelão et al. 2019; Xu et al. 2018) and its mediation effect on clinical symptoms in SZ. These studies have highlighted the methodological challenges in exploring epistasis while effectively unveiling non-independent effects between those genes that go unnoticed when examining main effects alone. Therefore, such approaches reveal the important role of gene interactions in the architecture of common human diseases while helping to connect the statistical perspective with the complex dynamics of biological systems (Phillips 2008).
One of the genes intimately implicated in synaptic plasticity processes is Neuritin-1 (NRN1, 6p25.1), which encodes for a neurotrophin that is highly expressed in the hippocampus, the cerebral cortex and the cerebellum (Naeve et al. 1997). Its expression is experience-dependent (Harwell et al. 2005; Nedivi et al. 1996) and regulated by Ca2+ influx via the N-methyl-D-aspartate (NMDA) receptors (Fujino et al. 2003). Additionally, it has been described that another neuropeptide, the Brain-Derived Neurotrophic Factor (BDNF, 11p13), modulates NRN1 expression. This gene is highly expressed in the same cerebral regions as NRN1 (Esvald et al. 2023) and also in an experience-dependent manner (Tongiorgi 2008). As neurotrophins, both genes exert multiple functions in the developing brain, being involved, for example, in enhancing neurite and dendritic growth, stabilising active synapses, improving synaptic maturation, increasing neuronal migration and regulating apoptosis of proliferative neurons, but are also involved in regulating the neuronal plasticity in the adult brain (Sasi et al. 2017; Yao et al. 2018). In the synaptic cleft, BDNF binds tyrosine kinase receptor B (TrkB) activating the transcription factor CREB (cAMP response element-binding protein) that attaches, among others, to the endogenous NRN1 promoter in vivo (Finkbeiner et al. 1997; Fujino et al. 2003) fostering functional and structural neuronal changes and leading to the consolidation of long-term synaptic plasticity (Kaldun and Sprecher 2019). Besides, animal-based models have described the direct relationship between BDNF and NRN1 expression, showing that the intraventricular injection or the intrahippocampal infusion of BDNF into neonatal rat pups results in the up-regulation of NRN1 expression in vivo (Wibrand et al. 2006).
Current research looking for potential receptors for NRN1 suggests a role in regulating synaptic excitability through the activation of the insulin receptor (IR) and downstream pathway, which trigger the transcription and trafficking of L-type voltage-gated calcium channel (L-VGCC) subunits to the membrane of cortical neurons, specifically Cav1.3 and Cav1.2 (Lu et al. 2017; J.-J. Yao et al. 2012; Zhao et al. 2018). The latter is encoded by humans’ calcium voltage-gated channel subunit alpha1 C gene (CACNA1C, 12p13.33). Notably, L-VGCC channels constitute the most abundant calcium channels in the human brain, accounting for approximately 90% (Striessnig et al. 2014). These channels are located at the post-synapsis, specifically at the neurons’ soma and dendritic spines and shaft (Jenkins et al. 2010). There, they facilitate Ca2+ influx in response to membrane depolarisation, which acts as a cellular messenger that triggers diverse cellular responses, including the strengthening of short- and long-term synaptic plasticity and the promotion of activity-dependent gene expression in a process called excitation-transcription coupling (Ma et al. 2011).
The genetic association of BDNF and CACNA1C genes has been widely reported in psychiatric disorders, and NRN1, while far less explored, has been identified as an interesting candidate gene, involved in age at onset, general cognitive abilities and brain activity in SZ (Almodóvar-Payá et al. 2022; Chandler et al. 2010; Fatjó-Vilas et al. 2016). Regarding the role of BDNF in SZ, the rs6265 polymorphism has been associated not only with the risk for the disorder (Kheirollahi et al. 2016; Rosa et al. 2006) but also with a range of clinical features, including the age of onset, symptoms, therapeutic responsiveness, neurocognitive function and brain morphology and activity (see review by Notaras et al. 2005). About CACNA1C variability, the rs1006737 polymorphism has been repeatedly associated with the risk for SZ both through GWAS and meta-analysis (Trubetskoy et al. 2022; Zhu et al. 2019) and with disrupted cognitive performance (Novaes de Oliveira Roldan et al., 2023) and altered brain structure and function in subjects with SZ (Gurung and Prata 2015). However, whether epistatic effects among these genes are involved in the aetiology of SZ remains largely unexplored. Two preceding studies have suggested that NRN1 x BDNF-rs6265 epistasis is associated with the susceptibility for SZ-spectrum disorders and modulates depressive symptoms in healthy subjects (HS) (Fatjó-Vilas et al. 2016; Prats et al. 2017). In summary, the currently available molecular and genetic association data point towards the potential synergistic role of NRN1, BDNF and CACNA1C in the risk of SZ and the modulation of the disorder’s presentation.
Consequently, we hypothesised that genetic epistasis between NRN1, BDNF and CACNA1C would be associated with the clinical manifestations of the disorder and that such effects would be mediated by their modulation of brain cortical structure. To test this hypothesis, first, we explored the impact of polymorphic variability along the NRN1 gene in combination with BDNF (rs6265) or CACNA1C (rs1006737) on (i) the risk for SZ; (ii) the clinical severity and functionality plus age at onset; and, (iii) the neuroanatomical cortical measures (thickness (CT), surface area (CSA) and volume (CV)). Second, we investigated whether those brain clusters under a significant epistatic effect might be mediating the clinical outcomes of the patients.
Methods
Sample
The sample consisted of a case-control dataset of 175 individuals: 86 healthy subjects (HS) and 89 subjects diagnosed with SZ. All participants provided a biological sample for genotypic analyses and underwent a comprehensive clinical evaluation and a magnetic resonance imaging (MRI) session, detailed in the subsequent sections. The subjects with SZ were recruited from Germanes Hospitalaries psychiatric hospitals in the Barcelona area (Hospital Benito Menni and Hospital Sant Rafael), and the healthy controls were recruited from the same area.
All participants were of European origin with ages between 18 and 65 years, had an Intelligence Quotient (IQ) > 75 according to the Wechsler Adult Intelligence Scale III (WAIS-III) (Wechsler 1997) and were right-handed. Experienced psychiatrists evaluated patients using the Structured Clinical Interview for DSM Disorders (SCID) (First et al. 2002) and met the DSM-IV-TR criteria for SZ. The HS had no personal history of mental disorders or treatment. All participants met the same exclusion criteria, which included a major medical illness affecting brain function, neurological conditions, a history of head trauma with loss of consciousness and a history of drug abuse or dependence.
Ethical approval
was obtained from the research ethics committee, and all participants provided written consent after being informed about the study procedures and implications. All procedures were carried out according to the Declaration of Helsinki.
Clinical assessment
The patients with SZ underwent a clinical evaluation including (i) the age at onset of the first episode, based on the first appearance of psychotic symptoms and established by experienced psychiatrists through clinical information derived from case notes and information provided by the patient and close relatives; (ii) symptom severity, evaluated with the Positive and Negative Symptoms Scale (PANSS) (Kay et al. 1987); (iii) the Global Assessment of Functioning (GAF) (Endicott et al. 1976); and, (iv) the Clinical Global Impression scale (CGI) (Table 1).
Genotyping
Genomic DNA was obtained for all individuals from buccal mucosa through cotton swabs and extracted using ATP Genomic DNA Mini Kit Tissue (Teknokroma Analítica, S.A., Sant Cugat del Vallès, Barcelona, Spain) or peripheral blood cells by punction and extracted using Realpure SSS Kit for DNA Extraction (Durviz, S.L.U, Valencia, Spain). We genotyped: (i) eleven single nucleotide polymorphisms (SNP) at the Neuritin1 gene (NRN1, 6p25.1), (ii) the SNP rs6265 (also known as Val66Met; GRCh38 position: 27,658,369) at the Brain-Derived Neurotrophic Factor gene (BDNF, 11p13) for which the T allele encodes for the amino acid methionine (Met) and the C allele encodes for valine (Val), and (iii) the SNP rs1006737 (GRCh38 position: 2,236,129) at calcium voltage-gated channel subunit alpha1 gene (CACNA1C, 12p13.33). The genotyping was carried out at the National Genotyping Center (CeGen) of the Network Platform of Biomolecular and Bioinformatics Resources (PRB3) of the Carlos III Health Institute (ISCIII). The genotyping call rate was 97% and, as shown in Table 2, the minor allele frequencies were comparable to the ones described for the EUR population in the 1,000 Genomes Project, and the genotype frequencies were in Hardy-Weinberg equilibrium (PLINK v.1.07) (Purcell et al. 2007).
MRI data acquisition, processing, and analyses
The neuroimaging protocol was conducted at the Hospital Sant Joan de Déu using a 1.5T GE Sigma MRI scanner. High-resolution structural-T1 imaging was obtained using the following acquisition parameters: matrix size = 512 × 512; 180 contiguous axial slices; voxel-resolution 0.47 × 0.47 × 1 mm3; echo (TE), repetition (TR) and inversion (TI) times, (TE / TR / TI) = 3.93 / 2000 / 710 ms, respectively; flip angle of 15°. This scan was used to estimate the three cortical measures: cortical thickness (CT), cortical surface area (CSA) and cortical volume (CV).
Structural MRI data were analysed with the FreeSurfer (http://surfer.nmr.mgh.harvard.edu/). Briefly, the pre-processing included the removal of non-brain tissue, automated Talairach transformation, tessellation of the grey and white matter boundaries and surface deformation (Fischl et al. 2004), after which individual images were normalised to a common stereotaxic space. Some deformation procedures were performed in the data analysis pipeline, including surface inflation and registration to a spherical atlas. This method uses both intensity and continuity information from the entire three-dimensional images in the segmentation and deformation procedures to produce vertex-wise representations of CT, CSA, and CV.
All subjects included in this study passed the standardised quality-control protocols from the ENIGMA consortium (http://enigma.ini.usc.edu/protocols/imaging-protocols) that have previously been applied in large-scale multi-centre studies (Hibar et al. 2018).
Statistical analyses
Epistasis models between NRN1 and its interactors (BDNF and CACNA1C) on SZ phenotypes
Taking into account previously reported association data, and to maximise the power of the detected gene-gene interactions, all the analyses were carried out grouping the minor and the heterozygous genotypes for all the NRN1 SNPs, for the BDNF-rs6265 (Met-allele carriers and Val/Val homozygotes) and CACNA1C-rs1006737 (A-allele carriers and GG homozygotes; Supplementary Table S1 and S2).
First, the epistatic effects on the risk for the disorder were studied using two-way interaction factors in logistic regression models (adjusted by sex). Second, we used linear regression models to test the gene-gene effects on the clinical data within subjects with SZ (adjusted by age and sex). We only considered those interactions significant after the Bonferroni correction (0.05 / 11 = 0.0045, based on the number of SNPs analysed). Statistical power calculations were performed using version 1.0.2 of the ‘genpwr’ R package, considering a minor allele frequency (MAF) range of 0.17 to 0.49 and a power of 0.80. In the case of epistasis analyses on the risk (case-control logistic regression), we had a detectable odds ratio ≥ 1.85. For the epistasis analyses on the clinical measures (linear regressions within patients), we had a potentially detectable regression coefficient ≥ 1.76. Third, the impact of genetic interactions on brain measurements was explored within subjects with SZ using the Qdec graphical user interface that implements the general linear models. Specifically, our epistasis analyses were based on the two-factor, two-level ANOVA implemented in the FreeSurfer software, with the two genes (NRN1 and the interactor, BDNF or CACNA1C) as groups and three covariates (sex, age, and intracranial volume (ICV)). We used a Monte Carlo Null-Z simulation with a z-value threshold of 2.3 (equivalent to p < 0.005, two tails) to correct for multiple comparisons. In these analyses, according to the Bonferroni threshold mentioned before, only those clusters with cluster-wise p-threshold that met that criterion were considered. Anatomical locations of the significant regions were determined using the surface atlas included in the FreeSurfer software. Fourth, the values for each significant cluster were imported to SPSS to explore their association with clinical measures using linear regression models (adjusted by age and sex), for graphical purposes and further moderated mediation analyses.
Moderated mediation analyses
A moderated mediation model included those epistatic models significantly associated with clinical profiles and neuroanatomical measures (Fig. 1). This approach allowed us to investigate whether those brain clusters where we detected significant epistasis between NRN1 and BDNF or CACNA1C might mediate the effect detected on clinical profiles. We built a moderated mediation analysis with NRN1 as the predictor variable (X), clinical features as the response variable (Y), BDNF-rs6265 or CACNA1C-rs1006737 as the moderator variable (Mo), and the brain measures as the mediating variable (Me). We used the R package “mediation” (Tingley et al. 2014). This tests, first, if the effect of X on the Me differs conditional to Mo (model 1: Me = β0 + β1X + β2Mo + β3XMo + ε), and second, if Me and the interaction between X and Mo are significantly associated with Y (model 2: Y = β0 + β1Me + β2X + β3Mo + β4XMo + ε). Lastly, it allows the calculation of the indirect effect (the index of moderation mediation), which is quantified as the product of the regression coefficient for the X and Mo interaction in model 1 and the regression coefficient of Me in model 2 (βindirect = β3XMo * β1Me). Once the indirect effect is calculated, the confidence interval and the significance levels for the entire indirect effect are estimated based on the bootstrap method with 10,000 resampling iterations. The conceptual and statistical diagrams of the model are shown in Fig. 1.
Schematic representation of the different moderated mediation models used. (A) Conceptual diagrams representing the used model. The grey arrows indicate the indirect effect of X on Y through Me and modulated by Mo. (B) Statistical diagram depicting the different equations used in the moderated mediation model. Two equations are used to estimate the index of moderation mediation (IMM): first, the effect of X (a1) and Mo (a2) variables and their interaction (a3) on Me (model 1: Me = β0 + β1X + β2Mo + β3XMo + ε); second, the effect of X (c’1), Mo (c’2), their interaction (c’3) and Me (b1) on Y (model 2: Y = β0 + β1Me + β2X + β3Mo + β4XMo + ε). The different letters represent the coefficients of each one of these effects. Then, the index of moderated mediation was calculated as the product of the coefficients a3*b1
Results
Sample characteristics
Table 1 shows the main sociodemographic and clinical data of the sample. As shown, subjects with SZ and HS presented sex differences; then, this variable was used as a covariable in the analyses exploring the epistatic effect on the risk.
Epistatic effects on the risk for SZ
There were no NRN1 x BDNF-rs6265 nor NRN1 x CACNA1C-rs1006737 epistatic effects on the risk for the disorder (Supplementary Table S1 and S1).
Epistatic effects on SZ clinical measures
We detected a significant two-order gene-gene interaction between NRN1-rs10484320 and BDNF-rs6265 on PANSS general psychopathology. As shown in Table 3, while the main effects of NRN1-rs10484320 and BDNF-rs6265 were not detected, the interaction of both was significant. Adding the interaction term significantly improved the model’s overall fit (Δ-R2 = 0.098, p-value of the change = 0.003). As illustrated in Fig. 2A, patients presented inverse patterns in PANSS general psychopathology scores as a function of both NRN1 and BDNF genotypes. Remarkably, the significance of this effect is amplified with the inclusion of treatment, as chlorpromazine (CPZ) equivalents in mg/day, as a covariate in the model (β = -0.651, SE = 4.154, p = 0.001, model Adj-R2 = 0.099).
We also found a significant NRN1-rs4960155 x CACNA1C-rs1006737 epistasis effect on GAF scores. As indicated in Table 3, no main genotypic effects were detected, whereas their interaction was significant. When the interaction term was included, the model’s overall fit significantly improved (Δ-R2 = 0.112, p-value of the change = 0.002). As shown in Fig. 2B, patients presented inverse patterns in GAF scores as a function of both NRN1 and CACNA1C genotypes. This effect did not substantially change when CPZ was added as a covariable in the model (β = 0.582, SE = 6.832, p = 0.003, model Adj-R2 = 0.102).
Bar plots showing the significant epistatic effects detected on clinical measures within SZ subjects. Each bar represents the scores’ marginal mean (± 2 standard error) for each epistatic group. A) NRN1-rs10484320 x BDNF-rs6265 interaction on Positive and Negative Symptoms Scale (PANSS) general psychopathology. B) NRN1-rs4960155 x CACNA1C-rs1006737 interaction on Global Assessment of Functioning (GAF)
Epistatic effects on SZ brain cortical measures
Regarding the interaction between NRN1 SNPs and BDNF-rs6265, we detected many significant effects affecting the CSA of frontal, parietal and temporal regions. We also found that some epistasis effects regulated the CV of frontal regions (Table 4; Fig. 3).
Concerning the interaction between NRN1 SNPs and CACNA1C-rs1006737, we detected significant effects modulating the CSA of temporal regions and the CV of frontal regions (Table 4; Fig. 4).
All the results remained significant after incorporating CPZ as a covariate in the model (data not shown).
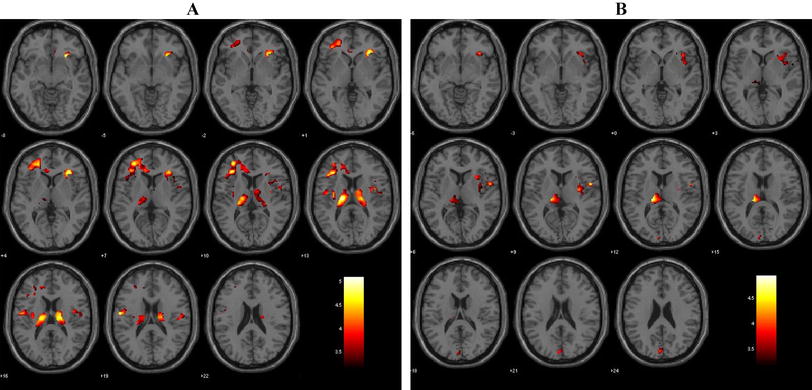
Brain regions where significant epistatic effects between NRN1 and BDNF were detected in the whole-brain FreeSurfer analyses. We represented significant gene-gene interactions related to cortical surface area (CSA) (A) and cortical volume (CV) (B) on the pial surface, where a specific colour distinguishes each cluster, and both lateral and medial views are presented for each hemisphere (LH: left hemisphere; RH: right hemisphere). The bar plots depict the marginal mean scores (± 2 standard error) for each epistatic group
Brain regions where significant epistatic effects between NRN1 and CACNA1C were detected in the whole-brain FreeSurfer analyses. We represented significant gene-gene interactions related to cortical surface area (CSA) (A) and cortical volume (CV) (B) on the pial surface, where each cluster is distinguished by a specific colour, and both lateral and medial views are presented for each hemisphere (LH: left hemisphere; RH: right hemisphere). The bar plots depict the marginal mean scores (± 2 standard error) for each epistatic group
Cortical measures effects on clinical features
We found a significant association between the cluster comprising the left lateral orbitofrontal cortex (L-LOFC) (cluster 8) and PANSS subscales and total scores (Table 5). As illustrated in Fig. 5A, B and C, greater volumes of L-LOFC were associated with elevated PANSS scores, which are indicative of more severe symptoms. These effects ramined significant when CPZ was incorporated as a covariate in the model (Positive β = 0.322, SE = 0.003, p = 0.004, model Adj-R2 = 0.063; Negative β = 0.269, SE = 0.004, p = 0.014, model Adj-R2 = 0.105; General Psychopathology β = 0.281, SE = 0.005, p = 0.011, model Adj-R2 = 0.059; Total scores β = 0.340, SE = 0.009, p = 0.001, model Adj-R2 = 0.116). We also observed a significant correlation between the inferior temporal region (cluster 11) and GAF scores (Table 5). As depicted in Fig. 5D, greater volumes of the inferior temporal cortex were associated with lower GAF values, which are indicative of worse functioning. This effect did not change when CPZ was added as a covariate in the model (β = -0.243, SE = 0.006, p = 0.024, model Adj-R2 = 0.082).
Scatter plots showing the significant correlations between brain clusters and clinical measures within subjects with SZ. For both clinical and morphometric measures, the reported values signify estimated marginal means. These means have been adjusted for potential confounding factors, including age and sex. In the case of brain measures, the adjustments also encompass intracranial volume. PANSS: positive and negative syndrome scale; GP: general psychopathology; GAF: Global Assessment Functioning; CV: cortical volume; CSA: cortical surface area; L-LOFC: left lateral orbitofrontal cortex
Brain structure as a mediator of the epistatic effects on clinical features in SZ
Given the NRN1-rs10484320 x BDNF-rs6265 epistatic effect on both PANSS general psychopathology (Table 3; Fig. 2A) and the CV of the L-LOFC (Table 4; Fig. 3, cluster 8), we explored whether this brain cluster mediated the detected epistatic effect on psychopathology. As shown in Table 6 and schematised in Fig. 6, after extracting the mean values of the L-LOFC cluster for each individual, we confirmed that NRN1-rs10484320 x BDNF-rs6265 epistasis significantly accounted for variation of the L-LOFC volume. However, when the NRN1-rs10484320 x BDNF-rs6265 interaction and the L-LOFC volume were included in the same model to predict PANSS general psychopathology, the mediator was significantly associated, but the interaction did not retain its significance. The significant index of moderated mediation indicates that the influence of NRN1-rs10484320 x BDNF-rs6265 on PANSS general psychopathology is entirely mediated by its impact on L-LOFC volume. This effect remained significant when CPZ was added as a covariate in the model (IMM = -25.839; SE = 13.942; Bootstrap 95% CI [-59.225 – -5.108], p = 0.024)
Discussion
Based on data coming from several animal and cellular studies that highlight the molecular links between NRN1, BDNF and CACNA1C genes (Fujino et al. 2003; Wibrand et al. 2006), this study explores the neurobiological pathways through which the epistasis between these synaptic plasticity-related genes may be associated with the clinical presentation of SZ. By combining genetic, neuroimaging and clinical data, the main findings show the epistasis between NRN1 and its interactors (BDNF and CACNA1C) on SZ clinical features and indicate that this joint genetic background may exert its effect through the impact on neuroanatomical measures.
Regarding our analyses exploring the epistatic effect on the risk for SZ, we did not detect any significant association. While the individual effect of NRN1, BDNF and CACNA1C on the risk for SZ is reported in different studies, there is little evidence showing that NRN1 role seems to be not independent of BDNF (Fatjó-Vilas et al. 2016; Prats et al. 2017), and no previous study has explored the epistasis with CACNA1C. Thus, the counterpart to the novelty in our study is the challenge in comparability, as there is only one prior study (Fatjó-Vilas et al. 2016) that described a significant interaction effect between NRN1-rs9379002 and BDNF-rs6265 on a broader clinical spectrum, including both SZ and bipolar disorder. These discrepancies could be due to differences in diagnosis and sample size. Nonetheless, the current results do not indicate a direct epistatic effect on the risk of SZ, but reveal significant epistatic effects relevant to the clinical presentation of the disorder.
In this sense, we found a significant interaction between NRN1-rs10484320 and BDNF-rs6265 impacting PANSS general psychopathology, which encompasses symptoms like anxiety, guilt, tension, depression, and disorientation (Kay et al. 1987). Albeit earlier studies noted the negative impact of BDNF-rs6265 ValVal genotype on various PANSS subscales (Chang et al. 2009; Numata et al. 2006; Zhai et al. 2013), our data revealed that NRN1-rs10484320 modifies this effect. We observed more severe symptoms in patients carrying the NRN1 T allele and the BDNF ValVal genotype or the NRN1 CC genotype and the BDNF Met allele than in those carrying the opposite genotypic combinations. We also identified a significant interaction between NRN1-rs4960155 and CACNA1C-rs1006737 affecting GAF scores, summarising personal, social, and psychological functioning (Endicott et al. 1976). Again, previous research has established the CACNA1C-rs1006737 A allele as a risk factor for SZ through GWAS and meta-analytic approaches (Liu et al. 2020; Trubetskoy et al. 2022) and has demonstrated its detrimental effect on longitudinal GAF scores and recovery after psychotic episodes (Heilbronner et al. 2015). Our data adds to such evidence by revealing that NRN1-rs4960155 modulates the CACNA1C effects. Patients with the NRN1 TT genotype and the A allele for CACNA1C and those with the NRN1 GG genotype but carrying the C allele for CACNA1C exhibited poorer functioning. It is worth mentioning that both NRN1 SNPs, rs10484320 and rs4960155, have been previously associated with the risk of SZ-spectrum disorders and have been found to influence IQ scores among these individuals (Chandler et al. 2010; Fatjó-Vilas et al. 2016).
Nevertheless, taking into account the clinical heterogeneity of SZ and its complex aetiology, expecting a direct influence of genetic background on the disorder’s clinical presentation seems unrealistic; instead, genetic networks might modulate a lower-level trait, which, in turn, sustains the manifestation of symptoms (Glahn et al. 2010; Meijer et al. 2021). Therefore, following other previous studies (Kirschner et al. 2020; Miranda et al. 2019; Sudre et al. 2020), we have proposed that deconstructing SZ into biologically validated and stable trait markers, such as brain structural measures and investigating their role in mediating symptomatology could help to fill the gap in the path from synaptic plasticity genetic variability to the complex and heterogeneous clinical presentation of the disorder.
In this regard, we discovered that NRN1-rs2208870 and several upstream variants (rs10484320, rs4960155, rs9379002, rs1475157) interact with BDNF-rs6265 to modulate CSA and CV among individuals with SZ. Additionally, we identified that NRN1-rs1475157 and NRN1-12333117 interact with CACNA1C-rs1006737, modulating the CSA and CV, respectively, among individuals with SZ. These findings highlight the role of epistatic interactions involving NRN1, BDNF, and CACNA1C in contributing to the strong genetic underpinnings of CSA, which is estimated to have a heritability of 91% (Eyler et al. 2012). The critical roles of these genes in neural development (Sasi et al. 2017; Yao et al. 2018) also support its inclusion in the pool of genetic factors influencing CSA, which is driven by regulatory elements active during prenatal cortical development (Grasby et al. 2020).
Those brain regions significantly modulated by epistasis effects, mainly frontal regions as well as some parietal and temporal regions, have been previously reported to present CSA and CV reductions in patients with SZ (Erp et al. 2018; Madre et al. 2020; Rimol et al. 2012). Voxel-based morphometry meta-analyses have also evidenced CV reductions, especially in frontotemporal regions (Bora et al. 2011; Honea et al. 2005). Though our analysis was confined to patients with SZ patients, our findings indicate that these specific epistatic combinations distinctly impact brain structure in this group. Thus, it is plausible that these genetic interactions might also play a role in the molecular mechanisms contributing to the previously mentioned differences in cortical structure between patients and HS.
From all the epistasis effects on brain structure, we highlight the NRN1-rs10484320 x BDNF-rs6265 interaction effect on the L-LOFC volume since these genetic variants also jointly modulate PANSS general psychopathology. Our moderated mediation model was designed to deep into the relationship between these significant associations and confirmed the mediation effect of the L-LOFC in the relationship between the NRN1-rs10484320 x BDNF-rs6265 epistasis and the PANSS general psychopathology scores.
However, contrary to the expected, the same patients who presented the lowest PANSS general psychopathology scores, those carriers of the CC genotype for NRN1-rs10484320 and the ValVal genotype for BDNF-rs6265, were the ones with the smaller left L-LOFC volume. The L-LOFC, as part of a functional network including the medial prefrontal cortex (Öngür and Price 2000), plays multiple roles, such as the integration of multiple sensory information, modulation of visceral reactions, and participation in learning, prediction, and decision-making for emotional and reward-related behaviours (Kringelbach and Rolls 2004; Rolls 2004). Findings in SZ regarding structural alterations of the OFC are controversial. While some studies have reported volume reductions (Madre et al. 2020; Rimol et al. 2012), others have described increased volume of the left OFC (Lacerda et al. 2007). These incongruities are not clarified by exploring the relationship with symptoms, as both volume increase and decrease have been associated with SZ symptomatology (Baaré et al. 1999; Gur et al. 2000; Koutsouleris et al. 2008; Lacerda et al. 2007; Nakamura et al. 2008). Interestingly, proteomic studies have linked N-methyl-D-aspartate (NMDA) receptors hypofunction and disruption of calcium homeostasis with OFC volumetric alterations in SZ (Nascimento and Martins-de-Souza 2015; Velásquez et al. 2019). At the clinical level, decreasing glutamatergic neurotransmission through NMDA receptor antagonists generates and worsens psychotic traits, suggesting its significance as a major pathway for symptom development in SZ (Lewis and González-Burgos, 2007). In fact, NRN1 expression in the cortex is regulated by Ca2+ signalling via the NMDA receptor (Fujino et al. 2003), and NMDA receptor-mediated neurotransmission and plasticity are particularly affected by BDNF-rs6265 genotype within the hippocampus and infralimbic medial prefrontal cortex (Ninan et al. 2010; Pattwell et al. 2012). A significant pathway in the development of positive and negative symptoms may result from the reduced activity of glutamate, mainly mediated through the NMDA receptor (Lewis and González-Burgos, 2007). Therefore, both neurotrophic factors might play a role in the molecular pathways underlying the volumetric differences of the L-LOFC in SZ subjects that are behaviourally reflected in general psychopathology. Thus, our findings offer insight into the previously debated results, providing evidence that these genetic factors may contribute to volumetric variability among individuals, which, in turn, may underlie the emergence of general psychopathology during lifespan.
To understand how those genetic variants modulate psychopathology through their impact on brain structure, the functional consequences of those polymorphisms must be considered. On the one hand, animal and cell-based models have demonstrated that the BDNF-rs6265 Met variant affects both its intracellular distribution and activity-dependent secretion (Chen et al. 2005, 2006; Chiaruttini et al. 2009; Egan et al. 2003). On the other hand, data from the GTEx Project identifies BDNF-rs6265 and NRN1-rs10484320 variants as expression quantitative trait loci (eQTLs) affecting the expression of both genes in the brain context but does not indicate an epistatic effect. However, it is essential to note that statistical interaction between polymorphisms does not necessarily imply a direct impact on gene expression; instead, the identified epistasis could reflect the influence of these polymorphisms on intermediate pathways.
Finally, we acknowledge several limitations in our study, with the primary concern being the sample size. To mitigate potential overfitting, we calculated the smallest detectable effect in our sample for both risk assessment and clinical associations and applied the Bonferroni threshold to select significant epistasis. Concerning power estimation related to brain structure analyses it is discouraged due to the inherent properties of whole-brain approaches. Additionally, power calculations using vertices inside a significant cluster may create circularity because these vertices have already been selected for having highly different values (Vul et al. 2009). Despite these challenges, we employed various methodological strategies to avoid type I errors. Firstly, we selected polymorphic variants with known associations with SZ and described functional interactions. Secondly, we applied the cluster-wise correction method to correct for multiple comparisons and selected only those clusters that met the Bonferroni criterion. Simultaneously, to enhance the robustness of our findings and avoid potential pitfalls, we connected epistatic effects on symptomatology and brain structure through a mediation exploration. These approaches allowed for the concurrent detection of the interaction effect between NRN1-rs10484320 and BDNF-rs6265 on SZ PANSS general psychopathology and L-LOFC volume while describing that this effect on the clinical manifestation is fully mediated by brain structure. However, we must acknowledge the potential for type II errors that may have impeded the detection of epistatic effects in other regions. Another limitation is that our study did not directly investigate the molecular mechanisms underlying the genetic interactions that contribute to the clinical presentation of SZ. It is important to note that our analyses focus on identifying statistical relationships, which do not necessarily imply direct biological connections. However, the biological plausibility of our findings is supported by prior cellular and animal models describing functional interactions related to synaptic plasticity between the studied genes. Nevertheless, further studies are necessary to validate the statistical and biological impact of genetic interactions on brain structure and symptoms. Lastly, while the homogeneity of our sample concerning ethnicity and demographic variables minimises potential errors, it also restricts the generalizability of our results. Upcoming research should strive for larger samples with more diverse representations to enhance the external validity of our findings.
Conclusions
In conclusion, our study adds clinical significance to the well-described molecular relationship between NRN1 and its molecular interactors, BDNF and CACNA1C, since we provide the first evidence of their epistatic impact on PANSS general psychopathology through its effect on L-LOFC volume specifically within subjects with SZ.
Data availability
No datasets were generated or analysed during the current study.
References
Almodóvar-Payá C, Guardiola-Ripoll M, Giralt-López M, Gallego C, Salgado-Pineda P, Miret S, Salvador R, Muñoz MJ, Lázaro L, Guerrero-Pedraza A, Parellada M, Carrión MI, Cuesta MJ, Maristany T, Sarró S, Fañanás L, Callado LF, Arias B, Pomarol-Clotet E, Fatjó-Vilas M (2022) NRN1 gene as a potential marker of early-onset Schizophrenia: evidence from genetic and neuroimaging approaches. Int J Mol Sci 23(13) Article 13. https://doi.org/10.3390/ijms23137456
Baaré WFC, Pol HEH, Hijman R, Mali WPT, Viergever MA, Kahn RS (1999) Volumetric analysis of frontal lobe regions in schizophrenia: relation to cognitive function and symptomatology. Biol Psychiatry 45(12):1597–1605. https://doi.org/10.1016/S0006-3223(98)00266-2
Bora E, Fornito A, Radua J, Walterfang M, Seal M, Wood SJ, Yücel M, Velakoulis D, Pantelis C (2011) Neuroanatomical abnormalities in schizophrenia: a multimodal voxelwise meta-analysis and meta-regression analysis. Schizophr Res 127(1):46–57. https://doi.org/10.1016/j.schres.2010.12.020
Callicott JH, Feighery EL, Mattay VS, White MG, Chen Q, Baranger DAA, Berman KF, Lu B, Song H, Ming G, Weinberger DR (2013) DISC1 and SLC12A2 interaction affects human hippocampal function and connectivity. J Clin Investig 123(7):2961–2964. https://doi.org/10.1172/JCI67510
Chandler D, Dragović M, Cooper M, Badcock JC, Mullin BH, Faulkner D, Wilson SG, Hallmayer J, Howell S, Rock D, Palmer LJ, Kalaydjieva L, Jablensky A (2010) Impact of Neuritin 1 (NRN1) polymorphisms on fluid intelligence in schizophrenia. Am J Med Genet Part B: Neuropsychiatric Genet 153B(2):428–437. https://doi.org/10.1002/ajmg.b.30996
Chang H-A, Lu R-B, Shy M-J, Chang C-C, Lee M-S, Huang S-Y (2009) Brain-derived neurotrophic factor Val66Met Polymorphism: Association with psychopathological symptoms of Schizophrenia? J Neuropsychiatry Clin Neurosci 21(1):30–37. https://doi.org/10.1176/jnp.2009.21.1.30
Charlson FJ, Ferrari AJ, Santomauro DF, Diminic S, Stockings E, Scott JG, McGrath JJ, Whiteford HA (2018) Global Epidemiology and burden of Schizophrenia: findings from the global burden of Disease Study 2016. Schizophr Bull 44(6):1195–1203. https://doi.org/10.1093/schbul/sby058
Chen Z-Y, Ieraci A, Teng H, Dall H, Meng C-X, Herrera DG, Nykjaer A, Hempstead BL, Lee FS (2005) Sortilin Controls Intracellular sorting of brain-derived neurotrophic factor to the regulated secretory pathway. J Neurosci 25(26):6156–6166. https://doi.org/10.1523/JNEUROSCI.1017-05.2005
Chen Z-Y, Jing D, Bath KG, Ieraci A, Khan T, Siao C-J, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS (2006) Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science 314(5796):140–143. https://doi.org/10.1126/science.1129663
Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E (2009) Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci USA 106(38):16481–16486. https://doi.org/10.1073/pnas.0902833106
de Novaes AC, Fernandes Júnior LCC, de Oliveira CEC, Nunes SOV (2023) Impact of ZNF804A rs1344706 or CACNA1C rs1006737 polymorphisms on cognition in patients with severe mental disorders: a systematic review and meta-analysis. World J Biol Psychiatry 24(3):195–208. https://doi.org/10.1080/15622975.2022.2097308
Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR (2003) The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112(2):257–269. https://doi.org/10.1016/S0092-8674(03)00035-7
Endicott J, Spitzer RL, Fleiss JL, Cohen J (1976) The Global Assessment Scale: A Procedure for Measuring overall severity of Psychiatric Disturbance. Arch Gen Psychiatry 33(6):766–771. https://doi.org/10.1001/archpsyc.1976.01770060086012
Esvald E-E, Tuvikene J, Kiir CS, Avarlaid A, Tamberg L, Sirp A, Shubina A, Cabrera-Cabrera F, Pihlak A, Koppel I, Palm K, Timmusk T (2023) Revisiting the expression of BDNF and its receptors in mammalian development. Front Mol Neurosci 16. https://www.frontiersin.org/articles/https://doi.org/10.3389/fnmol.2023.1182499
Eyler LT, Chen C-H, Panizzon MS, Fennema-Notestine C, Neale MC, Jak A, Jernigan TL, Fischl B, Franz CE, Lyons MJ, Grant M, Prom-Wormley E, Seidman LJ, Tsuang MT, Fiecas MJA, Dale AM, Kremen WS (2012) A comparison of heritability maps of cortical surface area and thickness and the influence of Adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res Hum Genetics: Official J Int Soc Twin Stud 15(3):304–314. https://doi.org/10.1017/thg.2012.3
Fatjó-Vilas M, Prats C, Pomarol-Clotet E, Lázaro L, Moreno C, González-Ortega I, Lera-Miguel S, Miret S, Muñoz MJ, Ibáñez I, Campanera S, Giralt-López M, Cuesta MJ, Peralta V, Ortet G, Parellada M, González-Pinto A, McKenna PJ, Fañanás L (2016) Involvement of NRN1 gene in schizophrenia-spectrum and bipolar disorders and its impact on age at onset and cognitive functioning. World J Biol Psychiatry 17(2):129–139. https://doi.org/10.3109/15622975.2015.1093658
Finkbeiner S, Tavazoie SF, Maloratsky A, Jacobs KM, Harris KM, Greenberg ME (1997) CREB: a major mediator of neuronal neurotrophin responses. Neuron 19(5):1031–1047. https://doi.org/10.1016/S0896-6273(00)80395-5
First M, Spitzer R, Gibbon M, Williams J (2002) Structured clinical interview for DSM-IV-TR Axis I Disorders, Research Version, Non-patient Edition. En (SCID-I/P)
Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM (2004) Automatically parcellating the human cerebral cortex. Cereb Cortex 14(1):11–22. https://doi.org/10.1093/cercor/bhg087
Fujino T, Lee W-CA, Nedivi E (2003) Regulation of cpg15 by signaling pathways that mediate synaptic plasticity. Mol Cell Neurosci 24(3):538–554. https://doi.org/10.1016/S1044-7431(03)00230-6
Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL, Laird AR, Smith SM, Beckmann CF, Fox PT, Blangero J (2010) Genetic control over the resting brain. Proceedings of the National Academy of Sciences, 107(3), 1223–1228. https://doi.org/10.1073/pnas.0909969107
Grasby KL, Jahanshad N, Painter JN, Colodro-Conde L, Bralten J, Hibar DP, Lind PA, Pizzagalli F, Ching CRK, McMahon MAB, Shatokhina N, Zsembik LCP, Thomopoulos SI, Zhu AH, Strike LT, Agartz I, Alhusaini S, Almeida MAA, Alnæs D, ENHANCING NEUROIMAGING GENETICS THROUGH META-ANALYSIS CONSORTIUM (ENIGMA)—GENETICS WORKING GROUP (2020) The genetic architecture of the human cerebral cortex. Science 367(6484):eaay6690. https://doi.org/10.1126/science.aay6690
Guardiola-Ripoll M, Almodóvar-Payá C, Lubeiro A, Sotero A, Salvador R, Fuentes-Claramonte P, Salgado-Pineda P, Papiol S, Ortiz-Gil J, Gomar JJ, Guerrero-Pedraza A, Sarró S, Maristany T, Molina V, Pomarol-Clotet E, Fatjó-Vilas M (2022) A functional neuroimaging association study on the interplay between two schizophrenia genome-wide associated genes (CACNA1C and ZNF804A). Eur Arch Psychiatry Clin NeuroSci 272(7):1229–1239. https://doi.org/10.1007/s00406-022-01447-z
Gur RE, Cowell PE, Latshaw A, Turetsky BI, Grossman RI, Arnold SE, Bilker WB, Gur RC (2000) Reduced dorsal and Orbital Prefrontal Gray Matter Volumes in Schizophrenia. Arch Gen Psychiatry 57(8):761–768. https://doi.org/10.1001/archpsyc.57.8.761
Gurung R, Prata DP (2015) What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol Med 45(12):2461–2480. https://doi.org/10.1017/S0033291715000537
Hall J, Bray NJ (2022) Schizophrenia Genomics: convergence on synaptic development, adult synaptic plasticity, or both? Biol Psychiatry 91(8):709–717. https://doi.org/10.1016/j.biopsych.2021.10.018
Harwell C, Burbach B, Svoboda K, Nedivi E (2005) Regulation of cpg15 expression during single whisker experience in the Barrel cortex of adult mice. J Neurobiol 65(1):85–96. https://doi.org/10.1002/neu.20176
Heilbronner U, Malzahn D, Strohmaier J, Maier S, Frank J, Treutlein J, Mühleisen TW, Forstner AJ, Witt SH, Cichon S, Falkai P, Nöthen MM, Rietschel M, Schulze TG (2015) A common risk variant in CACNA1C supports a sex-dependent effect on longitudinal functioning and functional recovery from episodes of schizophrenia-spectrum but not bipolar disorder. Eur Neuropsychopharmacol 25(12):2262–2270. https://doi.org/10.1016/j.euroneuro.2015.09.012
Hibar DP, Westlye LT, Doan NT, Jahanshad N, Cheung JW, Ching CRK, Versace A, Bilderbeck AC, Uhlmann A, Mwangi B, Krämer B, Overs B, Hartberg CB, Abé C, Dima D, Grotegerd D, Sprooten E, Bøen E, Jimenez E, Andreassen OA (2018) Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA bipolar disorder Working Group. Mol Psychiatry 23(4) Article 4. https://doi.org/10.1038/mp.2017.73
Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, Nordentoft M, Glenthøj B (2018) Heritability of Schizophrenia and Schizophrenia Spectrum based on the nationwide Danish twin Register. Biol Psychiatry 83(6):492–498. https://doi.org/10.1016/j.biopsych.2017.08.017
Honea R, Crow TJ, Passingham D, Mackay CE (2005) Regional deficits in Brain volume in Schizophrenia: a Meta-analysis of Voxel-based Morphometry studies. Am J Psychiatry 162(12):2233–2245. https://doi.org/10.1176/appi.ajp.162.12.2233
Jenkins MA, Christel CJ, Jiao Y, Abiria S, Kim KY, Usachev YM, Obermair GJ, Colbran RJ, Lee A (2010) Ca2+-Dependent facilitation of Cav1.3 Ca2 + channels by Densin and Ca2+/Calmodulin-Dependent protein kinase II. J Neurosci 30(15):5125–5135. https://doi.org/10.1523/JNEUROSCI.4367-09.2010
Ji Y, Zhang X, Wang Z, Qin W, Liu H, Xue K, Tang J, Xu Q, Zhu D, Liu F, Yu C (2021) Genes associated with gray matter volume alterations in schizophrenia. NeuroImage 225:117526. https://doi.org/10.1016/j.neuroimage.2020.117526
Kaldun JC, Sprecher SG (2019) Initiated by CREB: Resolving Gene Regulatory Programs in Learning and Memory. BioEssays 41(8):1900045. https://doi.org/10.1002/bies.201900045
Kay SR, Fiszbein A, Opler LA (1987) The positive and negative syndrome scale (PANSS) for Schizophrenia. Schizophr Bull 13(2):261–276. https://doi.org/10.1093/schbul/13.2.261
Kheirollahi M, Kazemi E, Ashouri S (2016) Brain-derived neurotrophic factor gene Val66Met polymorphism and risk of Schizophrenia: a Meta-analysis of case–control studies. Cell Mol Neurobiol 36(1):1–10. https://doi.org/10.1007/s10571-015-0229-z
Kirschner M, Shafiei G, Markello RD, Makowski C, Talpalaru A, Hodzic-Santor B, Devenyi GA, Paquola C, Bernhardt BC, Lepage M, Chakravarty MM, Dagher A, Mišić B (2020) Latent clinical-anatomical dimensions of Schizophrenia. Schizophr Bull 46(6):1426–1438. https://doi.org/10.1093/schbul/sbaa097
Koutsouleris N, Gaser C, Jäger M, Bottlender R, Frodl T, Holzinger S, Schmitt GJE, Zetzsche T, Burgermeister B, Scheuerecker J, Born C, Reiser M, Möller H-J, Meisenzahl EM (2008) Structural correlates of psychopathological symptom dimensions in schizophrenia: a voxel-based morphometric study. NeuroImage 39(4):1600–1612. https://doi.org/10.1016/j.neuroimage.2007.10.029
Kringelbach ML, Rolls ET (2004) The functional neuroanatomy of the human orbitofrontal cortex: evidence from neuroimaging and neuropsychology. Prog Neurobiol 72(5):341–372. https://doi.org/10.1016/j.pneurobio.2004.03.006
Lacerda ALT, Hardan AY, Yorbik O, Vemulapalli M, Prasad KM, Keshavan MS (2007) Morphology of the orbitofrontal cortex in first-episode schizophrenia: relationship with negative symptomatology. Prog Neuropsychopharmacol Biol Psychiatry 31(2):510–516. https://doi.org/10.1016/j.pnpbp.2006.11.022
Legge SE, Cardno AG, Allardyce J, Dennison C, Hubbard L, Pardiñas AF, Richards A, Rees E, Di Florio A, Escott-Price V, Zammit S, Holmans P, Owen MJ, O’Donovan MC, Walters JTR (2021) Associations between Schizophrenia Polygenic liability, Symptom dimensions, and cognitive ability in Schizophrenia. JAMA Psychiatry 78(10):1143–1151. https://doi.org/10.1001/jamapsychiatry.2021.1961
Liu Y, Wu X, Xia X, Yao J, Wang B (2020) The genome-wide supported CACNA1C gene polymorphisms and the risk of schizophrenia: an updated meta-analysis. BMC Med Genet 21(1):159. https://doi.org/10.1186/s12881-020-01084-0
Lu J-M, Liu D-D, Li Z-Y, Ling C, Mei Y-A (2017) Neuritin enhances synaptic transmission in Medial Prefrontal Cortex in mice by increasing CaV3.3 surface expression. Cereb Cortex 27(7):3842–3855. https://doi.org/10.1093/cercor/bhx082
Mackay TF, Moore JH (2014) Why epistasis is important for tackling complex human disease genetics. Genome Med 6(6):42. https://doi.org/10.1186/gm561
Madre M, Canales-Rodríguez EJ, Fuentes-Claramonte P, Alonso-Lana S, Salgado-Pineda P, Guerrero-Pedraza A, Moro N, Bosque C, Gomar JJ, Ortíz-Gil J, Goikolea JM, Bonnin CM, Vieta E, Sarró S, Maristany T, McKenna PJ, Salvador R, Pomarol-Clotet E (2020) Structural abnormality in schizophrenia versus bipolar disorder: a whole brain cortical thickness, surface area, volume and gyrification analyses. NeuroImage: Clin 25:102131. https://doi.org/10.1016/j.nicl.2019.102131
Ma H, Groth RD, Wheeler DG, Barrett CF, Tsien RW (2011) Excitation–transcription coupling in sympathetic neurons and the molecular mechanism of its initiation. Neurosci Res 70(1):2–8. https://doi.org/10.1016/j.neures.2011.02.004
Meijer M, Keo A, van Leeuwen JMC, Dzyubachyk O, Meijer OC, Vinkers CH, Mahfouz A (2021) Molecular characterisation of the stress network in individuals at risk for schizophrenia. Neurobiol Stress 14:100307. https://doi.org/10.1016/j.ynstr.2021.100307
Miranda GG, Rodrigue KM, Kennedy KM (2019) Fronto-parietal cortical thickness mediates the effect of COMT Val158Met polymorphism on age-associated executive function. Neurobiol Aging 73:104–114. https://doi.org/10.1016/j.neurobiolaging.2018.08.027
Naeve GS, Ramakrishnan M, Kramer R, Hevroni D, Citri Y, Theill LE (1997) Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc Natl Acad Sci USA 94(6):2648–2653
Nakamura M, Nestor PG, Levitt JJ, Cohen AS, Kawashima T, Shenton ME, McCarley RW (2008) Orbitofrontal volume deficit in schizophrenia and thought disorder. Brain 131(Pt 1):180–195. https://doi.org/10.1093/brain/awm265
Nascimento JM, Martins-de-Souza D (2015) The proteome of schizophrenia. Npj Schizophrenia 1(1). https://doi.org/10.1038/npjschz.2014.3
Nedivi E, Fieldust S, Theill LE, Hevron D (1996) A set of genes expressed in response to light in the adult cerebral cortex and regulated during development. Proc Natl Acad Sci USA 93(5):2048–2053
Öngür D, Price JL (2000) The Organization of Networks within the Orbital and Medial Prefrontal cortex of rats, monkeys and humans. Cereb Cortex 10(3):206–219. https://doi.org/10.1093/cercor/10.3.206
Ninan I, Bath KG, Dagar K, Perez-Castro R, Plummer MR, Lee FS, Chao MV (2010) The BDNF Val66Met polymorphism impairs NMDA receptor-dependent synaptic plasticity in the Hippocampus. J Neurosci 30(26):8866–8870. https://doi.org/10.1523/JNEUROSCI.1405-10.2010
Notaras M, Hill R, van den Buuse M (2005) A role for the BDNF gene Val66Met polymorphism in schizophrenia? A comprehensive review. Neurosci Biobehav Rev 51:15–30. https://doi.org/10.1016/j.neubiorev.2014.12.016
Numata S, Ueno S, Iga J, Yamauchi K, Hongwei S, Ohta K, Kinouchi S, Shibuya-Tayoshi S, Tayoshi S, Aono M, Kameoka N, Sumitani S, Tomotake M, Kaneda Y, Taniguchi T, Ishimoto Y, Ohmori T (2006) Brain-derived neurotrophic factor (BDNF) Val66Met polymorphism in schizophrenia is associated with age at onset and symptoms. Neurosci Lett 401(1):1–5. https://doi.org/10.1016/j.neulet.2006.02.054
Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, Hubbard L, Lynham A, Mantripragada K, Rees E, MacCabe JH, McCarroll SA, Baune BT, Breen G, Walters JTR (2018) Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50(3) Article 3. https://doi.org/10.1038/s41588-018-0059-2
Pattwell SS, Bath KG, Perez-Castro R, Lee FS, Chao MV, Ninan I (2012) The BDNF Val66Met polymorphism impairs synaptic transmission and plasticity in the Infralimbic Medial Prefrontal Cortex. J Neurosci 32(7):2410–2421. https://doi.org/10.1523/JNEUROSCI.5205-11.2012
Phillips PC (2008) Epistasis–the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9(11):855–867. https://doi.org/10.1038/nrg2452
Prats C, Arias B, Ortet G, Ibáñez MI, Moya J, Pomarol-Clotet E, Fañanás L, Fatjó-Vilas M (2017) Role of neurotrophins in depressive symptoms and executive function: Association analysis of NRN1 gene and its interaction with BDNF gene in a non-clinical sample. J Affect Disord 211:92–98. https://doi.org/10.1016/j.jad.2016.11.017
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a Tool Set for whole-genome Association and Population-based linkage analyses. Am J Hum Genet 81(3):559–575
Rimol LM, Nesvåg R, Hagler DJ, Bergmann Ø, Fennema-Notestine C, Hartberg CB, Haukvik UK, Lange E, Pung CJ, Server A, Melle I, Andreassen OA, Agartz I, Dale AM (2012) Cortical volume, Surface Area, and thickness in Schizophrenia and Bipolar Disorder. Biol Psychiatry 71(6):552–560. https://doi.org/10.1016/j.biopsych.2011.11.026
Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park H-O, Hayles J, Hoe K-L, Kim D-U, Ideker T, Grewal SI, Weissman JS, Krogan NJ (2008) Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322(5900):405–410. https://doi.org/10.1126/science.1162609
Rolls ET (2004) The functions of the orbitofrontal cortex. Brain Cogn 55(1):11–29. https://doi.org/10.1016/S0278-2626(03)00277-X
Rosa A, Cuesta MJ, Fatjó-Vilas M, Peralta V, Zarzuela A, Fañanás L (2006) The Val66Met polymorphism of the brain-derived neurotrophic factor gene is associated with risk for psychosis: evidence from a family-based association study. Am J Med Genet Part B: Neuropsychiatric Genet 141B(2):135–138. https://doi.org/10.1002/ajmg.b.30266
Sasi M, Vignoli B, Canossa M, Blum R (2017) Neurobiology of local and intercellular BDNF signaling. Pflügers Archiv - Eur J Physiol 469(5):593–610. https://doi.org/10.1007/s00424-017-1964-4
Sengupta SM, MacDonald K, Fathalli F, Yim A, Lepage M, Iyer S, Malla A, Joober R (2017) Polygenic risk score associated with specific symptom dimensions in first-episode psychosis. Schizophr Res 184:116–121. https://doi.org/10.1016/j.schres.2016.11.039
Striessnig J, Pinggera A, Kaur G, Bock G, Tuluc P (2014) L-type Ca2 + channels in heart and brain. Wiley Interdisciplinary Reviews: Membrane Transp Signal 3(2):15–38. https://doi.org/10.1002/wmts.102
Sudre G, Frederick J, Sharp W, Ishii-Takahashi A, Mangalmurti A, Choudhury S, Shaw P (2020) Mapping associations between polygenic risks for childhood neuropsychiatric disorders, symptoms of attention deficit hyperactivity disorder, cognition, and the brain. Mol Psychiatry 25(10). https://doi.org/10.1038/s41380-019-0350-3. Article 10
Sullivan PF, Kendler KS, Neale MC (2003) Schizophrenia as a Complex Trait: evidence from a Meta-analysis of Twin studies. Arch Gen Psychiatry 60(12):1187. https://doi.org/10.1001/archpsyc.60.12.1187
Tecelão D, Mendes A, Martins D, Fu C, Chaddock CA, Picchioni MM, McDonald C, Kalidindi S, Murray R, Prata DP (2019) The effect of psychosis associated CACNA1C, and its epistasis with ZNF804A, on brain function. Genes Brain Behav 18(4):e12510. https://doi.org/10.1111/gbb.12510
Tingley D, Yamamoto T, Hirose K, Keele L, Imai K (2014) Mediation: R Package for Causal Mediation Analysis. J Stat Softw 59:1–38. https://doi.org/10.18637/jss.v059.i05
Tongiorgi E (2008) Activity-dependent expression of brain-derived neurotrophic factor in dendrites: facts and open questions. Neurosci Res 61(4):335–346. https://doi.org/10.1016/j.neures.2008.04.013
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, Bryois J, Chen C-Y, Dennison CA, Hall LS, Lam M, Watanabe K, Frei O, Ge T, Harwood JC, Koopmans F, Magnusson S, Richards AL, Sidorenko J, O’Donovan MC (2022) Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604(7906) Article 7906. https://doi.org/10.1038/s41586-022-04434-5
van der Meer D, Kaufmann T (2022) Mapping the genetic architecture of cortical morphology through neuroimaging: progress and perspectives. Translational Psychiatry 12(1). https://doi.org/10.1038/s41398-022-02193-5. Article 1
van Erp TGM, Walton E, Hibar DP, Schmaal L, Jiang W, Glahn DC, Pearlson GD, Yao N, Fukunaga M, Hashimoto R, Okada N, Yamamori H, Bustillo JR, Clark VP, Agartz I, Mueller BA, Cahn W, de Zwarte SMC, Pol HEH, Turner JA (2018) Cortical brain abnormalities in 4474 individuals with Schizophrenia and 5098 control subjects via the Enhancing Neuro Imaging Genetics through Meta Analysis (ENIGMA) Consortium. Biol Psychiatry 84(9):644–654. https://doi.org/10.1016/j.biopsych.2018.04.023
Velásquez E, Martins-de-Souza D, Velásquez I, Carneiro GRA, Schmitt A, Falkai P, Domont GB, Nogueira FCS (2019) Quantitative subcellular proteomics of the Orbitofrontal Cortex of Schizophrenia Patients. J Proteome Res 18(12):4240–4253. https://doi.org/10.1021/acs.jproteome.9b00398
Vul E, Harris C, Winkielman P, Pashler H (2009) Voodoo correlations in social neuroscience. Perspect Psychol Sci 4(3):274–290
Wechsler D (1997) Wechsler Adult Intelligence Scale (3rd ed.)
Wibrand K, Messaoudi E, Håvik B, Steenslid V, Løvlie R, Steen VM, Bramham CR (2006) Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur J Neurosci 23(6):1501–1511. https://doi.org/10.1111/j.1460-9568.2006.04687.x
Woo HJ, Yu C, Kumar K, Reifman J (2017) Large-scale interaction effects reveal missing heritability in schizophrenia, bipolar disorder and posttraumatic stress disorder. Translational Psychiatry 7(4) Article 4. https://doi.org/10.1038/tp.2017.61
Xu Q, Xiong Y, Yuan C, Liu F, Zhao F, Shen J, Qin W, Yu C (2018) ZNF804A rs1344706 interacts with COMT rs4680 to affect prefrontal volume in healthy adults. Brain Imaging Behav 12(1):13–19. https://doi.org/10.1007/s11682-016-9671-x
Yao J-J, Gao X-F, Chow C-W, Zhan X-Q, Hu C-L, Mei Y-A (2012) Neuritin activates insulin receptor pathway to Up-regulate Kv4.2-mediated transient outward K + current in rat cerebellar granule neurons *. J Biol Chem 287(49):41534–41545. https://doi.org/10.1074/jbc.M112.390260
Yao J, Zhao Q, Lu J, Mei Y (2018) Functions and the related signaling pathways of the neurotrophic factor neuritin. Acta Pharmacol Sin 39(9):1414–1420. https://doi.org/10.1038/aps.2017.197
Zhai J, Yu Q, Chen M, Gao Y, Zhang Q, Li J, Wang K, Ji F, Su Z, Li W, Li X, Qiao J (2013) Association of the brain-derived neurotrophic factor gene G196A rs6265 polymorphisms and the cognitive function and clinical symptoms of schizophrenia. Int J Clin Exp Pathol 6(8):1617–1623
Zhao Q-R, Lu J-M, Li Z-Y, Mei Y-A (2018) Neuritin promotes neurite and spine growth in rat cerebellar granule cells via L-type calcium channel-mediated calcium influx. J Neurochem 147(1):40–57. https://doi.org/10.1111/jnc.14535
Zhu D, Yin J, Liang C, Luo X, Lv D, Dai Z, Xiong S, Fu J, Li Y, Lin J, Lin Z, Wang Y, Ma G (2019) CACNA1C (rs1006737) may be a susceptibility gene for schizophrenia: an updated meta-analysis. Brain Behav 9(6):e01292. https://doi.org/10.1002/brb3.1292
Özsoy ED, Yılmaz M, Patlar B, Emecen G, Durmaz E, Magwire MM, Zhou S, Huang W, Anholt RRH, Mackay TFC (2021) Epistasis for head morphology in Drosophila melanogaster. G3: Genes|Genomes|Genetics, 11. 10jkab285. https://doi.org/10.1093/g3journal/jkab285
Zuk O, Hechter E, Sunyaev SR, Lander ES (2012) The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci 109(4):1193–1198. https://doi.org/10.1073/pnas.1119675109
Acknowledgements
We are grateful to all the participants, whose generosity made possible this study. This study received funding provided by: i) Fundación Alicia Koplowitz; ii) Acadèmia de les Ciències Mèdiques i de la Salut de Catalunya i de Balears (predoctoral contract to C A-P); iii) Instituto de Salud Carlos III through the project PI18/01535 and a Miguel Servet contract to M F-V (CP20/00072), co-funded by European Regional Development Fund (ERDF)/European Social Fund “Investing in your future”; iv) Comissionat per a Universitats i Recerca del DIUE of the Generalitat de Catalunya (Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR), 2021SGR1475). The funding sources played no role in the design of the study, the collection, analysis, or interpretation of data, or the decision to submit this manuscript for publication.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Author information
Authors and Affiliations
Contributions
Conceptualisation: CA-P and MF-V. Funding acquisition: MM, JS-V, CG, EP-C and MF-V. Supervision: MF-V. Resources: CA-P, MG-R, MG-L, MO-I, MM and JS-V, EP-C and MF-V. Methodology: CA-P, MG-R, EP-C and MF-V. Data curation: CA-P, MG-R, EJC-R. Formal analysis and investigation: CA-P, MG-R, MG-L, LFC, BA, CG, EJC-R and MF-V. Visualization: CA-P. Writing (original draft): CA-P, MG-R and MF-V. Writing (review and editing): all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Almodóvar-Payá, C., Guardiola-Ripoll, M., Giralt-López, M. et al. NRN1 epistasis with BDNF and CACNA1C: mediation effects on symptom severity through neuroanatomical changes in schizophrenia. Brain Struct Funct 229, 1299–1315 (2024). https://doi.org/10.1007/s00429-024-02793-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-024-02793-5