Abstract
In the spectrum of oncocytic renal neoplasms, a subset of tumors with high-grade-appearing histologic features harboring pathogenic mutations in mammalian target of rapamycin (mTOR) and hitherto clinical indolent behavior has been described. Three cases (2F,1 M) with histologically documented metastases (lymph node, skull, and liver) were retrieved and extensively investigated by immunohistochemistry, FISH, and next-generation sequencing. Tumors were composed of eosinophilic cells with prominent nucleoli (G3 by ISUP/WHO) arranged in solid to nested architecture. Additionally, there were larger cells with perinuclear cytoplasmic shrinkage and sparse basophilic Nissl-like granules, superficially resembling the so-called spider cells of cardiac rhabdomyomas. The renal tumors, including the skull and liver metastases, showed immunoexpression PAX8, CK8-18, and cathepsin-K, and negativity for vimentin. NGS identified mTOR genetic alterations in the three cases, including the skull and liver metastases. One patient was then treated with Everolimus (mTOR inhibitors) with clinical response (metastatic tumor shrinkage). We present a distinct renal tumor characterized by high-grade eosinophilic cells, cathepsin-K immunohistochemical expression, and harboring mTOR gene mutations demonstrating a malignant potential and showing responsiveness to mTOR inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last decade, our understanding of renal cell carcinoma (RCC) has vastly improved due to careful morphological evaluation of cases with correlation of data from high-throughput molecular profiling. This is particularly notable in oncocytic tumors where analysis of cases that were not easily classifiable as typical oncocytoma and chromophobe RCC has led to the establishment of additional clinicopathologically RCCs or a broadening of the spectrum within previously known RCC subtypes [1] including eosinophilic solid and cystic (ESC)-RCC [2, 3], succinate dehydrogenase (SDH)-deficient RCC [4], low-grade fumarate hydratase (FH)-deficient RCC [5], MiTF family translocation RCC [6], and TFEB-amplified RCC [7, 8].
Furthermore, increasing data regarding RCCs harboring mammalian target of rapamycin (mTOR) and tuberous sclerosis complex (TSC) mutations have been published. In detail, it has been known that alterations of the mTOR pathway are implicated in the pathogenesis of renal tumors arising in patients affected by the inherited tuberous sclerosis syndrome such as renal angiomyolipoma and related lesions [9, 10] and TSC-RCC [11, 12]. On the other hand, mutations of TSC1/TSC2 and mTOR genes have also been found in some sporadic renal cell neoplasms including RCC with leiomyomatous stroma [13], ESC-RCC [14,15,16], chromophobe RCC [17], epithelioid angiomyolipoma/pure epithelioid PEComa [10], low-grade oncocytic tumor (LOT) [18, 19], and eosinophilic vacuolated tumor (EVT), among others [20, 21]. Although an indolent clinical course has been accustomed to most of such tumors, recently, Tjota et al. reported the first case of an eosinophilic tumor harboring mTOR gene mutation with liver metastasis [22]. In this study, we describe the clinical, morphological, immunohistochemical, and molecular characteristics of three additional cases of eosinophilic RCC harboring mTOR gene mutations with histologically documented metastases and, in one case, the clinical response to targeted therapy.
Methods
Patients and samples
From our archives of unclassified oncocytic renal tumors, we identified four previously unreported high-grade eosinophilic renal cell tumors from three different patients. All of them were consult cases. All procedures performed in our study involving human participants received approval (Prog. 4136CESC) and were in accordance with the ethical standards of the institutional and/or national research committee and with the declaration of Helsinki. All patients gave their written informed consent to diagnostic procedures and treatment according to institutional rules for everyday clinical practice and experimental evaluations on archival tissue. All slides (28 slides for case 1, 14 slides for case 2, 9 slides for case 3) were reviewed by an experienced pathologist (GM). Samples of both primary tumor and metastases were available.
Immunohistochemistry
Sections from tissue blocks of primary and metastatic samples were immunohistochemically stained with the antibodies listed in Supplementary Table 1. All samples were processed using a sensitive Bond Polymer Refine detection system in an automated Bond immunohistochemistry instrument (Leica-Biosystems, Germany). The appropriate positive and negative controls were concurrently carried out. Labeling for each marker was recorded as the percentage of positive cells. P70S6 Kinase and ph4E-BP1 were performed in all samples using an automated Ventana Discovery system (Roche).
Fluorescence in situ hybridization (FISH)
FISH was carried out on primary and metastatic samples using dual-color break-apart TFE3 and TFEB probes (Cytotest, USA) and 1p36/1q25 probe, spectrum-orange/spectrum-green (Vysis) as previously described [23]. Scoring was performed by two experienced pathologists (AC and MB). At least 100 neoplastic non-overlapping nuclei were included in the scoring. To avoid false positive results due to nuclear truncation, cells with a single fluorescent signal were not evaluated.
Next-generation sequencing
DNA extraction
Sections were cut from all FFPE tissue blocks of primary and metastatic samples and manually microdissected to isolate a high percentage of neoplastic cells (> 50%). DNA was isolated using the GeneRead DNA FFPE kit (Qiagen, Hilden, Germany, http://www.qiagen.com Cat. n. 180,134). DNA quality and amount were assessed using NanoDrop and Qubit instruments (Thermo-Fisher Scientific) following the manufacturer’s instructions.
Library preparation and deep amplicon sequencing
We performed deep sequencing of the whole coding region and intron–exon junctions of 17 kidney-cancer-related genes with a custom panel created using the Ampliseq Designer pipeline (Thermo-Fisher Scientific) as previously described [3]. The genes included: TSC1, TSC2, MTOR, AKT1, PIK3CA, PTEN, SDHB, FH, VHL, SETD2, BAP1, PBRM1, MET, FLCN, SMARCA4, SMARCB1, and TCEB1.
Variant calling
Data from the PGM sequencing were initially processed using the Ion Torrent platform-specific software (Torrent Suite AD 5.6.4) to generate sequence reads, alignment of the reads on the reference genome Hg19, trim adapter sequences, filter, and remove poor signal-profile reads. The variant calling from the sequencing data was generated using the Variant Caller plugin.
To provide reliable somatic variant analysis we considered suitable only samples with more than 400,000 reads, an average coverage > × 500, and a coverage uniformity > 95%. We applied the following filters to the Variant Caller plugin: minimum allele frequency value of 2% and minimum phred quality score of 30. Variant annotation and copy number variation analysis were performed using the Ion Reporter 5.12 software (Thermo-Fisher Scientific).
Variant annotations were also assessed using the Ensembl Variant Effect Predictor pipeline of the Wellcome Trust Sanger Institute [24] as a second database check. Filtered variants were visually examined using the Integrative Genomic Viewer tool to taste their level of quality and to confirm the variant presence on both “ + ” and “ − ” strands. The clinical relevance (pathogenicity) of the annotated variants was assessed using the COSMIC database (Wellcome Sanger Institute), OncoKB database [25], ClinVar (NCBI), and LOVD (IARC).
Results
Clinical and pathological features
The clinical and pathological features are summarized in Table 1. Two patients were female and one male. The patients’ ages at diagnosis were 21, 58, and 49 years respectively. None of the three patients showed clinical stigmata of tuberous sclerosis. Two of them underwent radical nephrectomy and one partial nephrectomy; one patient had two tumors simultaneously affecting the left kidney. The tumors ranged in size from 3.1 to 9.5 cm, and were all solid and brownish in color. The original diagnosis made at the referral institution was unclassified RCC in case 1, oncocytoma in case 2, and chromophobe RCC in case 3.
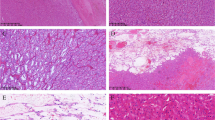
At light microscopy (Figs. 1 and 2), all the tumors were unencapsulated and composed of granular eosinophilic cells with cytoplasmatic vacuolization and nuclear atypia with prominent nucleoli (G3 by ISUP/WHO), mainly showing a solid/nested growth pattern. Additionally, in variable amount, there were larger cells with perinuclear cytoplasmic shrinkage and sparse basophilic Nissl-like granules, resembling the so-called spider cells of cardiac rhabdomyomas. Neoplastic cells were embedded within a fibrous and sometimes densely hyalinized stroma. Several thick-walled vessels were dispersed within the lesion along with peripherally entrapped normal renal tubules. Neither coagulative granular necrosis, mitoses, nor foamy macrophages were observed. In case 3, a neoplastic peritumoral vascular embolus was identified.
mTOR-mutated eosinophilic RCC. Low magnification demonstrates solid-nested growth (a). Large cells with perinuclear cytoplasmic shrinkage reminiscent of the so-called spider cells of cardiac rhabdomyomas were easily found in case 1 (b), or scattered in case 2 (c). Neoplastic vascular invasion was encountered in case 3 (d) (original magnification × 50 a, × 100 b and c, and × 200 d)
CT scan of case 2 revealed a skull mass (a) histologically characterized by large eosinophilic cells with round and enlarged nuclei (b). The neoplastic cells showed staining for PAX8 (c), cytokeratin 8–18 (d), but not for vimentin (e). The liver metastases of case 3 were histologically documented by biopsy (f) and were immunohistochemically positive for cathepsin-K (clone 3F9) (g) (original magnification × 50 f and g, × 100 b, d, and e, and × 200 c)
All the cases tested (Supplementary Table 2), expressed PAX8 along with immunolabelling for cathepsin-K (clone 3F9), whereas vimentin, and melanogenesis markers (HMB45 and Melan-A) were negative. Among the cytokeratins, strong and diffuse cytokeratin 8–18 expression was observed in all the neoplasms while in none of them significant staining of cytokeratin AE1/AE3 and cytokeratin 7 was found. All the cases were focally positive for CD117, expressed P70S6 Kinase and ph4E-BP1, and retained SDHB and FH. At FISH analysis all the cases showed 1p36/1q25 deletion (Fig. 3a); neither TFE3 nor TFEB rearrangements were identified.
One patient (case 1) had a synchronous small 9 mm nodal metastasis detected in one of seven regional lymph nodes and, to date, is alive without evidence of disease after 30 months. In another patient (case 2), a lesion was removed from the skull eleven years after surgical excision of the primary renal neoplasm. The skull lesion revealed overlapping histological and immunohistochemical findings with the kidney neoplasms and was therefore considered a metastasis. Finally, 3 years after surgery, two liver nodules were identified by imaging in the remaining patient (case 3); the largest of 3 cm as greater diameter was removed, whereas the other one was investigated by biopsy. Again, both liver lesions revealed overlapping histological and immunohistochemical findings with the renal tumor and were therefore considered metastases.
Next-generation sequencing results
Genetic alterations in the TSC or mTOR pathway were found in all the samples tested with next-generation sequencing (Table 2).
In case 1, a deletion in exon 30 of the mTOR gene (p.Tyr1450_Trp1456) was identified. This variant is reported both in COSMIC database (ref. COSM6972065) and OncoKB and occurs in the MTOR focal adhesion kinase targeting domain (FAT) domain, a key structural domain for the correct conformation of the catalytic pouch of the MTOR protein [26]. While its biological significance is still unknown, it has been identified as a statistically significant hotspot and is likely to be oncogenic. Moreover, it is also predicted to be pathogenic by the most common in-silico structural predictors (SIFT, POLYPHEN).
Case 2 showed pathogenic mTOR alterations in both primary renal tumors and the skull metastasis involving the exon 53 (p.Leu2427Arg—COSM2119114) (Fig. 3b).
Finally, case 3 showed pathogenic mTOR alterations in both the primary renal tumor and the liver metastases involving the exon 53 (p.Leu2427Gln—COSM1185313).
Both case 2 and case 3 variants involve the same aminoacidic residue of the MTOR Kinase Domain and are well known to be likely oncogenic [27].
No other pathogenetic mutation nor variant of unknown significance was identified in any of the primary tumor nor metastatic lesion. Furthermore, neither mTOR alterations nor TSC1/TSC2 gene mutation were found in the normal renal parenchyma.
Response to therapy
The patient of case 3 underwent radioembolization of the liver lesions. A new CT scan performed three months later documented the appearance of novel liver lesions (one lesion in the seventh hepatic segment 33 mm in long axis; 5 lesions in the fifth and sixth hepatic segments ranging from 17 to 47 mm in long axis; 1 lesion in the fourth hepatic segment 32 mm in long axis) and a pathological retrocaval lymphadenopathy (31 mm in short axis). Therefore, the patient started first-line systemic therapy with Pembrolizumab (200 mg intravenously, every 21 days) and Axitinib (5 mg orally twice a day). The best response achieved to this treatment was stable disease according RECIST 1.1, as shown by CT scans performed after 4, 8, and 12 months respectively. In the last examination, an osteolytic spot (22 mm in long axis) was identified at L2 so that the patient underwent external beam radiotherapy at this site (total dose: 20 Gy in 5 fractions). Fifteen months since the beginning of the systemic therapy, a new CT scan revealed further disease progression, with an increase in the size of all liver lesions, along with a new lesion in the eighth hepatic segment (6 mm in long axis) and new osteosclerotic spots in L5 and pelvic bone. Thus, shortly after first-line systemic treatment was permanently discontinued.
Based on the molecularly documented mTOR mutation (L2427Q, exon 53), the choice for second-line therapy was towards a combination of Lenvatinib (18 mg daily) and Everolimus (5 mg daily) in an off-label regimen. Within 4 months several treatment-related adverse events were observed including G2 hypertension, G2 fatigue, G2 diarrhea, and G2 mucositis. Hence, the dose of Lenvatinib was initially reduced to 14 mg daily and then to 10 mg daily. The first follow-up CT scan, after two months, showed reduction in size and vascularization of both all the target liver lesions and the retrocaval lymphadenopathy. Four months later, another CT scan revealed a further dimensional reduction of the liver lesions. Finally, 8 months after the beginning of the second-line therapy, the last CT scan available documented a numerical decrease in the liver lesions (with only two of them detectable to date) and an additional shrinkage of the retrocaval lymphadenopathy (< 10 mm in short axis) (Fig. 4). Currently, the patient is still on Lenvatinib (10 mg orally once a day) and Everolimus (5 mg orally once a day) treatment with fair tolerance.
Comparison between the baseline CT scan performed before the start of the first-line therapy with Pembrolizumab and Axitinib (January 2021), at the best response to the first-line therapy (May 2021), at the progression to the first-line therapy (April 2022), and after 9 months of the second-line therapy with Lenvatinib and Everolimus. Little to any tumor shrinkage was observed with Pembrolizumab and Axitinib. Conversely, following Lenvatinib and Everolimus, just a few hepatic lesions were detected, showing a decrease in size and vascularization. Disease stability for the osteolytic lesion in L2 was also recorded. All the images were taken in the arterial phase
Discussion
In this study, we reported four sporadic high-grade eosinophilic RCCs occurring in three patients with histologically documented metastases, characterized by the presence of “spider cells,” the immunohistochemical expression of cathepsin-K (clone 3F9), and mutations of mTOR gene.
In the last years, the differential diagnosis of eosinophilic tumors has become even more challenging for pathologists due to growing wide spectrum of tumors with oncocytic cells, usually characterized by non-aggressive behavior (Table 3) [1]. In this morphological scenario, cathepsin-K (clone 3F9) is a useful tool for differentiating renal oncocytoma, chromophobe RCC, and LOT which are negative for cathepsin-K (clone 3F9), from ESC-RCC, EVT, and mTOR-mutated eosinophilic RCC which are positive for this marker [28]. Whether the recognition of ESC-RCC is usually straightforward for uropathologists, EVT and mTOR-mutated eosinophilic RCC may show overlapping features, as highlighted by Tjota et al. [22]. However, EVT has prominent or extreme cytoplasmic vacuolation whereas in mTOR-mutated eosinophilic RCCs, we observed large cells with perinuclear cytoplasmic shrinkage resembling the so-called spider cells of cardiac rhabdomyomas occurring, interestingly, in tuberous sclerosis patients. This finding is morphologically reliable since similar elements are also focally reported by Tjota et al. as “somewhat rhabdoid appearance” [22].
Activating mTOR gene mutations are the genetic hallmarks of these high-grade eosinophilic RCCs. In accordance with the activation of the mTOR pathway, we observed an overexpression of two proteins downstream of the mTOR pathway, namely, the phosphorylated forms of 4EBP1 and S6K. Moreover, loss of chromosome 1 was identified in the four renal neoplasms and the skull and liver metastases, which represents the genomic location of the mTOR gene. The association of loss of chromosome 1 along with activating mTOR mutations has been also reported by Tjota et al. [22]. Being mTORC1 a dimer, it is possible to speculate that a heterodimer of wild-type and mTOR mutant proteins may not confer sufficient mTOR activation [29].
Recently, several renal tumors harboring mTOR gene mutations have been described (Table 4). In our comprehensive analysis of 57 neoplasms reported in the literature and our series, mTOR gene mutations have been observed in 16 EVTs, 24 LOTs, and 17 RCCs. The percentage of mTOR gene mutations in the molecularly tested cases is 31% of EVT (16 of 51 tumors) and 30.7% of LOT (24 of 78 tumors). Interestingly, the same mTOR gene mutation (L2427) has been detected in 11 of 16 (68.7%) EVTs, in 9 of 24 (37.5%) LOTs, and in 9 of 17 (53%) mTOR-mutated RCCs. The high prevalence of this hotspot genetic alteration, which falls in the catalytic subunit of the mTOR protein, may confer a selective advantage. Moreover, it has been demonstrated both in vitro and in vivo that cells harboring this mutation are highly sensitive to mTOR inhibitors [27, 30].
Differently from other eosinophilic tumors harboring mTOR mutations, the three patients presented in this study developed metastases. In case 1, the site of metastasis was a small lymph node of the renal sinus (i.e., still a loco-regional disease), which was removed together with the primary kidney lesion; in case 2, the metastasis was larger and occurred in the skull (i.e., a frankly metastatic disease) eleven years later the first diagnosis; finally, in case 3, multiple metastases in the liver were observed after 3 years from nephron-sparing surgery. Because of the amount of tumor available, we were able to test also the liver and skull metastases by immunohistochemistry and genetic analysis. We found morphological and immunohistochemical features (PAX8, cytokeratin 8–18, cathepsin-K clone 3F9 positivity, and vimentin negativity) overlapping with those observed in the primary renal tumors. The same mTOR gene mutation was identified in the liver metastasis and the primary renal tumor, as well as in the skull metastasis and the two renal tumors. A possible explanation of the latter finding is that one mass represented the intrarenal metastasis of the other tumor. Despite this possibility being well-known in other RCCs, it seems unlikely in this case since the skull metastasis occurred 11 years after the radical nephrectomy, suggesting instead a slow progression of the disease. Recently, Tjota et al. reported a similar case with liver metastasis harboring mTOR gene mutation (p.L2427K) and loss of chromosome 1 [22]. To date, the predictive value of mTOR gene mutations in metastatic RCC patients treated with mTOR inhibitors remains controversial, with studies suggesting such a predictive role [31], and others not [32]. A multicenter, histology-agnostic, single-arm prospective phase II trial of the mTORC1 inhibitor, Everolimus, in patients with solid tumors mainly harboring TSC1/TSC2 mutations ultimately failed, showing no association between these genomic alterations and response to targeted treatment in a broad spectrum of neoplasms, not including RCCs [33]. In our study, the clinical response to mTOR inhibitors observed in patient 3 might support the hypothesis that mTOR alterations could predict response to mTOR inhibitors, although our patient received a combination of one mTOR inhibitor, plus a multikinase inhibitor (mainly targeting VEGFRs), making it impossible to dissect the relative contribution of the two different classes of agents to the observed activity and efficacy.
In conclusion, herein, we present a distinct renal tumor characterized by high-grade eosinophilic cells, cathepsin-K (clone 3F9) immunohistochemical expression, and harboring mTOR gene mutation demonstrating a malignant potential and showing responsiveness to an mTOR inhibitor-containing combination. This latter observation encourages pathologists to investigate mTOR gene mutation in aggressive high-grade/cathepsin-K-positive eosinophilic RCC.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Amin MB, McKenney JK, Martignoni G, Campbell SC, Pal S, Tickoo SK (2022) Low grade oncocytic tumors of the kidney: a clinically relevant approach for the workup and accurate diagnosis. Mod Pathol 35:1306–1316. https://doi.org/10.1038/s41379-022-01108-5
Trpkov K, Hes O, Bonert M, Lopez JI, Bonsib SM, Nesi G, Comperat E, Sibony M, Berney DM, Martinek P, Bulimbasic S, Suster S, Sangoi A, Yilmaz A, Higgins JP, Zhou M, Gill AJ, Przybycin CG, Magi-Galluzzi C, McKenney JK (2016) Eosinophilic, solid, and cystic renal cell carcinoma: clinicopathologic study of 16 unique, sporadic neoplasms occurring in women. Am J Surg Pathol 40:60–71. https://doi.org/10.1097/PAS.0000000000000508
Munari E, Settanni G, Caliò A, Segala D, Lonardi S, Sandrini S, Vacca P, Tumino N, Marconi M, Brunelli M, Gobbo S, Netto GJ, Moretta L, Zamboni G, Martignoni G (2022) TSC loss is a clonal event in eosinophilic solid and cystic renal cell carcinoma: a multiregional tumor sampling study. Mod Pathol 35:376–385. https://doi.org/10.1038/s41379-021-00816-8
Gill AJ, Hes O, Papathomas T, Šedivcová M, Tan PH, Agaimy A, Andresen PA, Kedziora A, Clarkson A, Toon CW, Sioson L, Watson N, Chou A, Paik J, Clifton-Bligh RJ, Robinson BG, Benn DE, Hills K, Maclean F, Niemeijer ND, Vlatkovic L, Hartmann A, Corssmit EPM, van Leenders GJLH, Przybycin C, McKenney JK, Magi-Galluzzi C, Yilmaz A, Yu D, Nicoll KD, Yong JL, Sibony M, Yakirevich E, Fleming S, Chow CW, Miettinen M, Michal M, Trpkov K (2014) Succinate dehydrogenase (SDH)-deficient renal carcinoma: a morphologically distinct entity: a clinicopathologic series of 36 tumors from 27 patients. Am J Surg Pathol 38:1588–1602. https://doi.org/10.1097/PAS.0000000000000292
Smith SC, Sirohi D, Ohe C, McHugh JB, Hornick JL, Kalariya J, Karia S, Snape K, Hodgson SV, Cani AK, Hovelson D, Luthringer DJ, Martignoni G, Chen Y-B, Tomlins SA, Mehra R, Amin MB (2017) A distinctive, low-grade oncocytic fumarate hydratase-deficient renal cell carcinoma, morphologically reminiscent of succinate dehydrogenase-deficient renal cell carcinoma. Histopathology 71:42–52. https://doi.org/10.1111/his.13183
Caliò A, Segala D, Munari E, Brunelli M, Martignoni G (2019) MiT family translocation renal cell carcinoma: from the early descriptions to the current knowledge. Cancers (Basel) 11:1–12. https://doi.org/10.3390/cancers11081110
Argani P, Reuter VE, Zhang L, Sung Y-S, Ning Y, Epstein JI, Netto GJ, Antonescu CR (2016) TFEB-amplified renal cell carcinomas: an aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am J Surg Pathol 40:1484–1495. https://doi.org/10.1097/PAS.0000000000000720
Caliò A, Brunelli M, Segala D, Pedron S, Doglioni C, Argani P, Martignoni G (2019) VEGFA amplification/increased gene copy number and VEGFA mRNA expression in renal cell carcinoma with TFEB gene alterations. Mod Pathol 32:258–268. https://doi.org/10.1038/s41379-018-0128-1
Martignoni G, Pea M, Zampini C, Brunelli M, Segala D, Zamboni G, Bonetti F (2015) PEComas of the kidney and of the genitourinary tract. Semin Diagn Pathol 32:140–159. https://doi.org/10.1053/j.semdp.2015.02.006
Caliò A, Brunelli M, Segala D, Zamboni G, Bonetti F, Pea M, Martignoni G (2021) Angiomyolipoma of the kidney: from simple hamartoma to complex tumour. Pathology 53:129–140. https://doi.org/10.1016/j.pathol.2020.08.008
Guo J, Tretiakova MS, Troxell ML, Osunkoya AO, Fadare O, Sangoi AR, Shen SS, Lopez-Beltran A, Mehra R, Heider A, Higgins JP, Harik LR, Leroy X, Gill AJ, Trpkov K, Campbell SC, Przybycin C, Magi-Galluzzi C, McKenney JK (2014) Tuberous sclerosis-associated renal cell carcinoma: a clinicopathologic study of 57 separate carcinomas in 18 patients. Am J Surg Pathol 38:1457–1467. https://doi.org/10.1097/PAS.0000000000000248
Yang P, Cornejo KM, Sadow PM, Cheng L, Wang M, Xiao Y, Jiang Z, Oliva E, Jozwiak S, Nussbaum RL, Feldman AS, Paul E, Thiele EA, Yu JJ, Henske EP, Kwiatkowski DJ, Young RH, Wu C-L (2014) Renal cell carcinoma in tuberous sclerosis complex. Am J Surg Pathol 38:895–909. https://doi.org/10.1097/PAS.0000000000000237
Shah RB, Stohr BA, Tu ZJ, Gao Y, Przybycin CG, Nguyen J, Cox RM, Rashid-Kolvear F, Weindel MD, Farkas DH, Trpkov K, McKenney JK (2020) “Renal cell carcinoma with leiomyomatous stroma” harbor somatic mutations of TSC1, TSC2, MTOR, and/or ELOC (TCEB1): clinicopathologic and molecular characterization of 18 sporadic tumors supports a distinct entity. Am J Surg Pathol 44:571–581. https://doi.org/10.1097/PAS.0000000000001422
Parilla M, Kadri S, Patil SA, Ritterhouse L, Segal J, Henriksen KJ, Antic T (2018) Are sporadic eosinophilic solid and cystic renal cell carcinomas characterized by somatic tuberous sclerosis gene mutations? Am J Surg Pathol 42:911–917. https://doi.org/10.1097/PAS.0000000000001067
Palsgrove DN, Li Y, Pratilas CA, Lin M-T, Pallavajjalla A, Gocke C, De Marzo AM, Matoso A, Netto GJ, Epstein JI, Argani P (2018) Eosinophilic solid and cystic (ESC) renal cell carcinomas harbor tsc mutations: molecular analysis supports an expanding clinicopathologic spectrum. Am J Surg Pathol 42:1166–1181. https://doi.org/10.1097/PAS.0000000000001111
Mehra R, Vats P, Cao X, Su F, Lee ND, Lonigro R, Premkumar K, Trpkov K, McKenney JK, Dhanasekaran SM, Chinnaiyan AM (2018) Somatic bi-allelic loss of tsc genes in eosinophilic solid and cystic renal cell carcinoma. Eur Urol 74:483–486. https://doi.org/10.1016/j.eururo.2018.06.007
Roldan-Romero JM, Santos M, Lanillos J, Caleiras E, Anguera G, Maroto P, García-Donas J, de Velasco G, Martinez-Montes ÁM, Calsina B, Monteagudo M, Letón R, Leandro-García LJ, Montero-Conde C, Cascón A, Robledo M, Rodriguez-Antona C (2020) Molecular characterization of chromophobe renal cell carcinoma reveals mTOR pathway alterations in patients with poor outcome. Mod Pathol 33:2580–2590. https://doi.org/10.1038/s41379-020-0607-z
Morini A, Drossart T, Timsit M-O, Sibony M, Vasiliu V, Gimenez-Roqueplo A-P, Favier J, Badoual C, Mejean A, Burnichon N, Verkarre V (2022) Low-grade oncocytic renal tumor (LOT): mutations in mTOR pathway genes and low expression of FOXI1. Mod Pathol 35:352–360. https://doi.org/10.1038/s41379-021-00906-7
Kapur P, Gao M, Zhong H, Chintalapati S, Mitui M, Barnes SD, Zhou Q, Miyata J, Carrillo D, Malladi VS, Rakheja D, Pedrosa I, Xu L, Kinch L, Brugarolas J (2022) Germline and sporadic mTOR pathway mutations in low-grade oncocytic tumor of the kidney. Mod Pathol 35:333–343. https://doi.org/10.1038/s41379-021-00896-6
Chen Y-B, Mirsadraei L, Jayakumaran G, Al-Ahmadie HA, Fine SW, Gopalan A, Sirintrapun SJ, Tickoo SK, Reuter VE (2019) Somatic mutations of TSC2 or MTOR characterize a morphologically distinct subset of sporadic renal cell carcinoma with eosinophilic and vacuolated cytoplasm. Am J Surg Pathol 43:121–131. https://doi.org/10.1097/PAS.0000000000001170
Farcas M, Gatalica Z, Trpkov K, Swensen J, Zhou M, Alaghehbandan R, Williamson SR, Magi-Galluzzi C, Gill AJ, Tretiakova M, Lopez JI, Perez Montiel D, Sperga M, Comperat E, Brimo F, Yilmaz A, Siadat F, Sangoi A, Gao Y, Kuthi Ptákova NL, Pivovarcikova K, Rogala J, Agaimy A, Hartmann A, Fraune C, Rychly B, Hurnik P, Durcansky D, Bonert M, Gakis G, Michal M, Hora M, Hes O (2021) Mod Pathol 35(3):344–351. https://doi.org/10.1038/s41379-021-00923-6
Tjota MY, Segal J, Stadler WM, Antic T (2021) Eosinophilic renal cell carcinoma with isolated MTOR mutation metastatic to the liver: a novel case. Pathology 53:790–793. https://doi.org/10.1016/j.pathol.2020.10.013
Caliò A, Brunelli M, Segala D, Pedron S, Remo A, Ammendola S, Munari E, Pierconti F, Mosca A, Bollito E, Sidoni A, Fisogni S, Sacco C, Canu L, Sentinelli S, Fraccon AP, Fiorentino M, Scott C, Milella M, Porta C, Argani P, Martignoni G (2020) Comprehensive analysis of 34 MiT family translocation renal cell carcinomas and review of the literature: investigating prognostic markers and therapy targets. Pathology 52:297–309. https://doi.org/10.1016/j.pathol.2019.11.006
Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Charkhchi M, Cummins C, Da RinFioretto L, Davidson C, Dodiya K, El Houdaigui B, Fatima R, Gall A, Garcia Giron C, Grego T, Guijarro-Clarke C, Haggerty L, Hemrom A, Hourlier T, Izuogu OG, Juettemann T, Kaikala V, Kay M, Lavidas I, Le T, Lemos D, Gonzalez Martinez J, Marugán JC, Maurel T, McMahon AC, Mohanan S, Moore B, Muffato M, Oheh DN, Paraschas D, Parker A, Parton A, Prosovetskaia I, Sakthivel MP, Salam AIA, Schmitt BM, Schuilenburg H, Sheppard D, Steed E, Szpak M, Szuba M, Taylor K, Thormann A, Threadgold G, Walts B, Winterbottom A, Chakiachvili M, Chaubal A, De Silva N, Flint B, Frankish A, Hunt SE, IIsley GR, Langridge N, Loveland JE, Martin FJ, Mudge JM, Morales J, Perry E, Ruffier M, Tate J, Thybert D, Trevanion SJ, Cunningham F, Yates AD, Zerbino DR, Flicek P (2021) Ensembl. Nucleic Acids Res 49:D884–D891. https://doi.org/10.1093/nar/gkaa942
Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N (2017) OncoKB: a precision oncology knowledge base JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00011
Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP (2013) mTOR kinase structure, mechanism and regulation. Nature 497:217–223. https://doi.org/10.1038/nature12122
Chen Y-B, Xu J, Skanderup AJ, Dong Y, Brannon AR, Wang L, Won HH, Wang PI, Nanjangud GJ, Jungbluth AA, Li W, Ojeda V, Hakimi AA, Voss MH, Schultz N, Motzer RJ, Russo P, Cheng EH, Giancotti FG, Lee W, Berger MF, Tickoo SK, Reuter VE, Hsieh JJ (2016) Molecular analysis of aggressive renal cell carcinoma with unclassified histology reveals distinct subsets. Nat Commun 7:13131. https://doi.org/10.1038/ncomms13131
Caliò A, Brunelli M, Gobbo S, Argani P, Munari E, Netto G, Martignoni G (2021) Cathepsin K: a novel diagnostic and predictive biomarker for renal tumors. Cancers (Basel) 13(10):2441. https://doi.org/10.3390/cancers13102441
Kapur P, Gao M, Zhong H, Rakheja D, Cai Q, Pedrosa I, Margulis V, Xu L, Kinch L, Brugarolas J (2021) Eosinophilic vacuolated tumor of the kidney: a review of evolving concepts in this novel subtype with additional insights from a case with MTOR mutation and concomitant chromosome 1 loss. Adv Anat Pathol 28:251–257. https://doi.org/10.1097/PAP.0000000000000299
Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM (2014) A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov 4:554–563. https://doi.org/10.1158/2159-8290.CD-13-0929
Voss MH, Hakimi AA, Pham CG, Brannon AR, Chen Y-B, Cunha LF, Akin O, Liu H, Takeda S, Scott SN, Socci ND, Viale A, Schultz N, Sander C, Reuter VE, Russo P, Cheng EH, Motzer RJ, Berger MF, Hsieh JJ (2014) Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res 20:1955–1964. https://doi.org/10.1158/1078-0432.CCR-13-2345
Voss MH, Chen D, Reising A, Marker M, Shi J, Xu J, Ostrovnaya I, Seshan VE, Redzematovic A, Chen Y-B, Patel P, Han X, Hsieh JJ, Hakimi AA, Motzer RJ (2019) PTEN expression, not mutation status in TSC1, TSC2, or mTOR. Correlates with the outcome on everolimus in patients with renal cell carcinoma treated on the randomized RECORD-3 trial. Clin Cancer Res 25:506–514. https://doi.org/10.1158/1078-0432.CCR-18-1833
Adib E, Klonowska K, Giannikou K, Do KT, Pruitt-Thompson S, Bhushan K, Milstein MI, Hedglin J, Kargus KE, Sholl LM, Tsuji J, Hyman DM, Sisk A, Shapiro GI, Vargas HA, Harding JJ, Voss MH, Iyer G, Kwiatkowski DJ (2021) Phase II clinical trial of everolimus in a pan-cancer cohort of patients with mTOR pathway alterations. Clin Cancer Res 27:3845–3853. https://doi.org/10.1158/1078-0432.CCR-20-4548
Acknowledgements
The authors would like to thank Dr. Roldan-Romero JM for sharing some information regarding a few cases previously reported [17].
Funding
Open access funding provided by Università degli Studi di Verona within the CRUI-CARE Agreement. This study was funded by the “Bando di ricerca finalizzata 2021 (PC: GR-2021–12374462).”
Author information
Authors and Affiliations
Contributions
Conceptualization: A.C. and G.M.; methodology: A.C., S.M., G.S., and S.P.; formal analysis and investigation: A.C. and S.M.; writing—original draft preparation: A.C., S.M., and M.R.; writing—review and editing: S.G., L.S., E.M., M.B., L.M., A.P., S.F., F.P., M. R. R., A.M., and C.D.; supervision: M.B.A., C.P., and G.M.
Corresponding author
Ethics declarations
Ethics approval
All procedures involving human participants received institutional review board approval (Prog. 4136CESC) and were in accordance with the ethical standards of the institutional and/or national research committee and with the declaration of Helsinki.
Informed consent
All patients gave their written informed consent to diagnostic procedures and treatment according to institutional rules for everyday clinical practice.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Caliò, A., Marletta, S., Settanni, G. et al. mTOR eosinophilic renal cell carcinoma: a distinctive tumor characterized by mTOR mutation, loss of chromosome 1, cathepsin-K expression, and response to target therapy. Virchows Arch 483, 821–833 (2023). https://doi.org/10.1007/s00428-023-03688-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-023-03688-2