Abstract
Tropomyosin receptor kinase (TRK) inhibitors have been approved for metastatic solid tumors harboring NTRK fusions, but the detection of NTRK fusions is challenging. International guidelines recommend pan-TRK immunohistochemistry (IHC) screening followed by next generation sequencing (NGS) in tumor types with low prevalence of NTRK fusions, including metastatic colorectal cancer (mCRC). RNA-based NGS is preferred, but is expensive, time-consuming, and extracting good-quality RNA from FFPE tissue is challenging. Alternatives in daily clinical practice are warranted. We assessed the diagnostic performance of RNA-NGS, FFPE-targeted locus capture (FFPE-TLC), fluorescence in situ hybridization (FISH), and the 5′/3′ imbalance quantitative RT-PCR (qRT-PCR) after IHC screening in 268 patients with microsatellite-instability-high mCRC, the subgroup in which NTRK fusions are most prevalent (1–5%). A consensus result was determined after review of all assay results. In 16 IHC positive tumors, 10 NTRK fusions were detected. In 33 IHC negative samples, no additional transcribed NTRK fusions were found, underscoring the high sensitivity of IHC. Sensitivity of RNA-NGS, FFPE-TLC, FISH, and qRT-PCR was 90%, 90%, 78%, and 100%, respectively. Specificity was 100% for all assays. Robustness, defined as the percentage of samples that provided an interpretable result in the first run, was 100% for FFPE-TLC, yet more limited for RNA-NGS (85%), FISH (70%), and qRT-PCR (70%). Overall, we do not recommend FISH for the detection of NTRK fusions in mCRC due to its low sensitivity and limited robustness. We conclude that RNA-NGS, FFPE-TLC, and qRT-PCR are appropriate assays for NTRK fusion detection, after enrichment with pan-TRK IHC, in routine clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tropomyosin receptor kinase (TRK) inhibitors show durable responses in all tumor types harboring an NTRK fusion and are now a standard treatment option [1, 2]. Although NTRK fusions have been identified in all tumor types, their prevalence is typically low, complicating fusion detection in daily clinical practice. For instance, in metastatic colorectal cancer (mCRC), the prevalence of NTRK fusions is only 0.3% [3]. The vast majority of NTRK fusions are found in the subgroup of microsatellite-instability-high (MSI-H)/mismatch-repair-deficient (dMMR) tumors in which the prevalence is estimated to be 1–5% [4,5,6].
Implementation of NTRK fusion detection in routine care is hampered by the complexity of available testing strategies. International guidelines from both ASCO and ESMO recommend to use immunohistochemistry (IHC) as a screening assay followed by RNA-based next-generation sequencing (NGS) confirmation, which is considered the most sensitive test [7, 8]. However, RNA-based NGS is expensive, time-consuming, and may have decreased robustness. Specifically, extracting RNA from formalin-fixed, paraffin-embedded (FFPE) tissue is challenging due to crosslinking and fragmentation which decreases RNA quantity and quality [9, 10].
Ideally, molecular tests are implemented that are accurate, affordable, fast, and can be performed on archived FFPE tissue [11]. Alternative available tests for the confirmation of NTRK fusions include DNA-based NGS and fluorescence in situ hybridization (FISH) [12, 13]. More recently, the 5′/3′ imbalance quantitative RT-PCR (qRT-PCR) and FFPE-targeted locus capture (FFPE-TLC) have been added to the repertoire of gene fusion detection methods [14, 15]. Each assay has advantages and disadvantages, but head-to-head comparisons are lacking.
Like RNA-based NGS, DNA-based NGS can be used for multiplex and high-throughput analysis of gene fusions. DNA-based NGS has lower failure rates because DNA is more stable than RNA and is better preserved in FFPE tissues. However, the sensitivity for gene fusion detection of DNA-based NGS is limited by the large intronic regions with repetitive sequences, which are not covered by widely used targeted panels such as MSK-IMPACT and FoundationOneCDx [13]. FFPE-TLC, a novel DNA-based targeted NGS, can overcome this limitation of the standard DNA-based NGS. FFPE-TLC uses proximity ligation to obtain broad coverage on the fusion partners to detect fusion genes independent of the identification of the fusion breakpoint sequence. Recently, FFPE-TLC showed excellent diagnostic accuracy compared to FISH and DNA-NGS in detecting rearrangements in lymphoma FFPE samples [15].
FISH has a short turnaround time and is relatively inexpensive [16]. However, the fusion partner remains unknown, a positive test does not prove overexpression of the fusion product, and separate assays are needed to assess for rearrangements in NTRK1, NTRK2, and NTRK3.
The 5′/3′ imbalance quantitative RT-PCR (qRT-PCR), also known as Idylla GeneFusion Assay, combines reverse transcription quantitative real-time PCR (qRT-PCR) for the detection of known fusions and expression imbalance to detect fusions with unknown fusion partners, including NTRK. When a fusion is present, there will be an imbalance of the mRNA expression of the 3′ kinase domain, which is expressed because of its association with the novel fusion partner, and the original 5′ region which is not [14, 17]. This assay has a turnaround time of only three hours, is fully automated, and proves overexpression of the fusion protein. However, the fusion partner remains unknown and there is limited comprehensible quality or quantity control, while qRT-PCR faces the same challenge of using reduced quality RNA from FFPE tissue as RNA-NGS.
In order to find reliable alternatives for the detection of NTRK fusions that fits the need in routine care, we compare the diagnostic performance, defined as the robustness, sensitivity, and specificity, of RNA-based NGS, FFPE-TLC, FISH, and the qRT-PCR for the detection of NTRK fusions after pan-TRK IHC screening in MSI-H/dMMR mCRC.
Materials and methods
Patient selection and FFPE collection
All patients diagnosed with MSI-H/dMMR mCRC between 2015 and 2021 in the Netherlands were selected using the Netherlands Cancer Registry. All available formalin-fixed, paraffin-embedded (FFPE) tumor tissue blocks were retrieved using the Nationwide Network and Registry of Histo- and Cytopathology in the Netherlands (PALGA). Whenever possible, tissue of the primary tumor resection was used. If not available, tissue from a primary tumor biopsy or metastasis was used.
Immunohistochemistry screening
We used pan-TRK IHC as a screening test. The received tissue blocks were cut into 4-µm-thick slides and stained using a Ventana bench mark ultra autostainer using antigen retrieval for 24 min with CC1 (EDTA) and 32 min antibody exposure with a rabbit pan-TRK monoclonal antibody (mAb) (clone EPR17341, Abcam, Cambridge, MA) in a dilution of 1:500 according to standard procedures. Brain tissue was used as positive control.
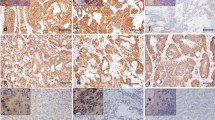
All slides were examined by two independent qualified pathologists (MML and LAAB) and scored for percentage of positive tumor cells, intensity, and staining pattern (cytoplasmic, nuclear, perinuclear, and/or membranous). In case of discordancy between the pathologists, the highest intensity score was used underlining the use as screening method. Positive staining was defined as ≥ 1% of tumor cells showing staining above background in any pattern and any intensity. Tumors that showed weak, non-specific granular cytoplasmic staining (Fig. 1B) were scored negative. All tissue samples with positive staining and 11 negative samples with weak, non-specific staining were included in the comparative analysis and analyzed with RNA-NGS, FFPE-TLC, FISH, and qRT-PCR.
Pan-Trk IHC, 40 × original magnification. (A) Negative staining, no additional testing was performed. (B) Weak, non-specific granular cytoplasmic staining (scored negative), no NTRK fusion was found. (C) Moderate cytoplasmic staining, no NTRK fusion was found. (D) Strong cytoplasmic + nuclear staining, LMNA::NTRK1 fusion was detected. (E) Strong cytoplasmic + perinuclear staining, LMNA::NTRK1 fusion was detected. (F) Strong cytoplasm + membranous staining, TPM3::NTRK1 was detected
In addition, a subset of pan-TRK negative cases was selected to examine the sensitivity of pan-TRK IHC. For this analysis, we selected the subgroup with the highest prevalence of NTRK fusions (MSI-H/dMMR, BRAF wild-type, and RAS wild-type) for FFPE-TLC analysis. Previously, we reported a NTRK fusion prevalence of 22% in this subgroup [18]. Transcription of NTRK fusions detected by FFPE-TLC was confirmed by RNA-NGS.
RNA-NGS
The Archer Fusionplex® Lung panel (ArcherDX, San Diego, USA) was used to assess NTRK1, NTRK2, and NTRK3 rearrangements (Supplementary Table S1). RNA was isolated using the Maxwell® RSC 48 instrument and the Maxwell® RSC RNA FFPE kit (Promega, Madison, USA) from four FFPE sections of 10 µm with a minimum of 10% tumor cell content. Isolated RNA was quantified using the Quantus™ Fluorometer and the QuantiFluor®RNA system (Promega, Madison, USA). A quantitative PCR (qPCR)-based method was used to determine the quality of the mRNA in each sample prior to the targeted library preparation. When a sample did not meet the quality criteria (Cq quantitative PCR should be < 30 cycles), the isolation and quality control was repeated using slides from a different tumor tissue block (if available). Libraries were prepared using the Archer FusionPlex reagent kit for Illumina (ArcherDX, San Diego, USA) according to manufacturer’s instructions. Results were analyzed on the ArcherDX website (https://analysis.archerdx.com/), version 6.2 or 7.0. Criteria for analysis were minimum of 200.00 RNA reads, unique RNA start sites per GSP2 control ≥ 10, on target deduplication ratio ≥ 3 and ≤ 10. Fusions must have > 3 SS unique start sites, > 5 breakpoint spanning reads, 10% fusion reads, and must be in frame. All results were examined by the same molecular biologist (WWL), who was blinded for the IHC result.
FFPE-targeted locus capture
A detailed description of FFPE-TLC was published before [15]. In brief, two to four FFPE sections of 10-µm thickness were deparaffinized to enable in situ DNA digestion by a restriction enzyme. After in situ ligation and overnight reverse crosslinking, standard protocols for library preparation and hybridization capture-based target enrichment were followed. Resulting libraries were paired-end sequenced on an Illumina platform. Two different capture panels were used to target NTRK1, NTRK2, NTRK3, and several other genes, including BRAF. For a subset of samples KRAS, NRAS, EGFR, and PIK3CA were also covered (Supplementary Table S2). One million on-target reads were aimed for 1 Mb region of interest.
After alignment of the raw sequencing data to the human genome, a computational pipeline PLIER was run to automatically detect NTRK1, NTRK2, and NTRK3 gene fusions. In brief, PLIER detects genomic intervals with significantly increased coverage of proximity ligation products per target locus and calculates a z-score by comparing its observed proximity score to the related expected proximity score. This PLIER pipeline has been optimized based on lymphoma cases that nearly always present as interchromosomal (trans) fusions, with fusion partners on a different chromosome. For CRC, several intrachromosomal (cis) fusions have been described, like for example LMNA::NTRK1, with both genes being located on chromosome 1. Therefore, visual inspection was performed in addition to PLIER to detect these intrachromosomal rearrangements. The scientists (ES, JFS, HF) involved in rearrangement calling were blinded for the results of the other assays.
Fluorescence in situ hybridization
FISH was performed on all pan-TRK positive cases [19]. NTRK1, NTRK2, and NTRK3 break-apart FISH probes (Z-2167, Z-2205, and Z-2206, ZytoVision GmbH, Bremerhaven, Germany) were used in a stepwise approach. First, the NTRK1 break-apart probe was used, as the NTRK1 gene is the most common NTRK fusion detected in colorectal cancer. Negative cases were subsequently analyzed using the NTRK3 break-apart probe. Lastly, FISH using the NTRK2 probe was performed for cases that were negative for both the NTRK1 and NTRK3 FISH.
For all tumor samples, a minimum of 100 non-overlapping nuclei were scored for their signals. FISH was considered positive for an NTRK fusion if ≥ 10% tumor cells showed a separation of red and green signals with a minimum of two signal diameters. With respect to single red signals: FISH was considered positive if ≥ 15% tumor cells showed a single red signal and equivocal if 10–15% of tumor cells showed a single red signal. FISH analysis was performed by qualified analysts and interpreted by a molecular biologist (WWL).
5′/3′ imbalance quantitative RT-PCR (qRT-PCR)
For the qRT-PCR we have used the Idylla GeneFusion Assay (Biocartis NV, Mechelen, Belgium). One 10-µm-thick slide was used according to the manufacturer’s instructions. A result was obtained for NTRK1, NTRK2, and NTRK3 individually. In case of errors or invalid test results, the assay was repeated once with another slide from the same tissue block. An invalid result for one out of three NTRK genes in the absence of a detected NTRK fusion was scored as invalid.
Comparison between assays
For all assays, the robustness was determined. Robustness was defined as the percentage of analyzed samples that provided an interpretable result in the first run. The assay results were all reviewed by two independent molecular biologists (WWL and RJAF), and their consensus decision regarding the presence of NTRK fusions per sample was used as truth set reference. Using this truth set reference, sensitivity and specificity were calculated for RNA-NGS, FFPE-TLC, FISH, and qRT-PCR. Subsequently, detected mutations in BRAFV600E and KRAS by RNA-NGS and FFPE-TLC were compared, and the level of agreement was evaluated.
Statistical analysis
Statistical analysis was performed using R Statistical Software (v4.0.1) and the epiR package. P-values of < 0.05 were considered statistically significant.
Results
We identified 383 patients diagnosed with MSI-H/dMMR mCRC between 2015 and 2021 in the Netherlands. Tumor tissue from 306 patients was collected, and 268 patients were included for the current analysis. The dropouts were due to administrative reasons (n = 77) or FFPE material did not contain sufficient cancer cells (n = 38) (Supplementary Fig. S1). Pan-TRK IHC was scored positive in 16 out of 268 (6%) patients (Fig. 1). All positive samples showed cytoplasmic staining and nine samples also showed nuclear, perinuclear, or membranous TRK expression (Supplementary Table S3). In general, there was homogenous pan-TRK staining with a median of 95% positive stained tumor cells (range: 10–100%) (Supplementary Table S3). Pan-TRK IHC negative samples often showed some weak, non-specific granular cytoplasmic staining (Fig. 1B). Patient characteristics of the NTRK positive patients have been published before [18]. No prior NTRK fusion tests were performed.
Robustness of assays
16 pan-TRK IHC positive and 11 negative tumor samples were included in the comparative analysis and analyzed with RNA-NGS, FFPE-TLC, FISH, and qRT-PCR (Fig. 2). Median age of the 27 FFPE tumor blocks that were analyzed was 43 months (range: 26–119 months), and median percentage of tumor cells in these tissues was 25% (range: < 10%–90%). For RNA-NGS, all samples passed the RNA quantity check but four samples did not generate an interpretable result. These samples had a median age of 65 months (range: 41–83 months) and contained a median tumor cell percentage of 40% (range: < 10%–90%) (Supplementary Table S3). One of these samples met the RNA quantity and quality check, but did not provide a result due to a high deduplication rate (119:1). The three other samples did not pass the RNA PCR quality test. For two out of four patients, a second FFPE tissue block was available and RNA-NGS was repeated successfully. Altogether, in 23 out of 27 samples, an interpretable result was generated in the first run, resulting in a robustness of 85% (95% CI: 66–96%) (Fig. 3). For FFPE-TLC, no repeat measurements were needed and robustness was 100% (95% CI: 87–100%). FISH analysis was successful for all tissue samples, but eight out of 27 samples were equivocal, resulting in a 70% (95% CI: 50–86%) robustness. For qRT-PCR, robustness was 70% (95% CI: 50–86%). Eight samples generated a non-interpretable result, including three technical errors related to the cartridge. Three of the invalid qRT-PCR samples also generated a non-interpretable result for RNA-NGS (Fig. 2). In three out of six samples, a valid result was generated after repeating the analysis, and two samples could not be repeated.
Comparison of NTRK fusion detection by IHC, RNA-NGS, FFPE-TLC, FISH, and qRT-PCR and the consensus result. For IHC, the assessment of both pathologists is shown. In case of discordancy between the pathologists, the highest intensity score was used underlining the use as screening method. The IHC of sample 019 is shown in Supplementary Fig. S2. Eleven pan-TRK IHC negative and 16 pan-TRK IHC positive tumors were analyzed. A total of 10 NTRK fusions were detected
NTRK fusion detection
An NTRK fusion was detected in 10 out of 16 (63%) pan-TRK IHC positive tumors according to the consensus results of all assays (Fig. 2). No NTRK fusions were detected in the 11 pan-TRK IHC negative samples with weak, non-specific staining. Nine tumors harbored an NTRK1 fusion with different fusion partners (4 × TPM3, 4 × LMNA, 1 × SFPQ), one tumor harbored an ETV6::NTRK3 fusion. All 10 tumor samples showed strong cytoplasmic TRK expression and nine of these patients also showed additional nuclear, perinuclear, or membranous TRK expression (Table 1).
RNA-NGS and FFPE-TLC detected nine NTRK fusions (Supplementary Table S4). Both assays missed a LMNA::NTRK1 fusion (samples 025 and 026). Sample 026 showed strong pan-TRK staining including nuclear staining. We repeated the RNA-NGS using a different FFPE tissue block, even though the sample past the RNA quantity and quality check, as the high deduplication ratio (130:1) that was found in the first run might indicate limited RNA quality. The repeated RNA-NGS did reveal the LMNA::NTRK1 fusion. The failure to detect the LMNA::NTRK1 fusion in sample 025 using FFPE-TLC remains unclear. FFPE-TLC was able to automatically detect the interchromosomal fusion ETV6::NTRK3 (Fig. 4A). The identified breakpoint position was in one of the large introns of NTRK3 and predicted an in-frame fusion product (Fig. 4B and C). The other eight intrachromosomal NTRK1 fusions had breakpoints between exons 8–12 and were detected after visual inspection (Fig. 4D).
Overview of FFPE-TLC results. (A) Genome-wide coverage of proximity ligation fragments containing NTRK3 sequences retrieved from sample 019. The rearranged region to the NTRK3 gene is identified by the clustered fragments around the ETV6 gene. (B) Butterfly plot confirming a true rearrangement within the targeted region. Proximity-ligation products between the target gene (NTRK3) and rearrangement partner (ETV6) are depicted in green. The transcriptional start site of both genes is indicated by the X mark. (C) Detailed overview of sequencing coverage on NTRK3 and ETV6. (D) Lollipop overview of breakpoint positions identified in NTRK1 and the corresponding fusion partner. The color of the dots indicates the 5′ fusion partner of NTRK1; TPM3 (blue), LMNA (yellow), SFPQ (red), and TPR (green). Lollipop numbers correspond with sample numbers
Using FISH, three tumors met the criteria of ≥ 10% tumor cells with break apart signals, including the ETV6::NTRK3 fusion. Furthermore, four NTRK1 fusions were detected by single red signals only, including three LMNA::NTRK1 fusions (Supplementary Table S5). Two LMNA::NTRK1 fusions and a TPM3::NTRK1 fusion were not detected by FISH.
All NTRK fusions were detected by qRT-PCR (Supplementary Table S6). Seven out of ten NTRK fusions were detected in the first run. The other three NTRK fusion-positive samples were detected after repeated qRT-PCR assays.
The overall agreement between the two molecular biologists for a consensus NTRK fusion score was 100% and is shown in the last column of Fig. 2. The sensitivity of RNA-NGS and FFPE-TLC was both 90% (95% CI: 55–100%). For qRT-PCR, sensitivity was 100% (95% CI: 69–100%). FISH had the lowest sensitivity: 78% (95% CI: 40–97%). In this comparative analysis of 27 samples, all tests showed 100% specificity set against the consensus decision.
Sensitivity of pan-TRK IHC using FFPE-TLC
To validate the excellent sensitivity (100%) of pan-TRK IHC in the 11 pan-TRK negative samples (Fig. 2), we additionally performed FFPE-TLC in the high NTRK fusions prevalent (22%) subgroup of mCRC patients with a MSI-H/dMMR, BRAF, and RAS wild-type tumor. Only pan-TRK IHC negative samples were selected, in which FFPE-TLC could be performed in 22 out of 23 samples. A TPR::NTRK1 fusion was detected by FFPE-TLC in sample 034 (Fig. 4D; Supplementary Fig. S3). This fusion was also detected by RNA-NGS in 100% of a small number of reads (n = 6) that showed intron fusions only, indicative that the fusion was not present in RNA reads, but only in DNA reads. The absence of RNA reads and the negative IHC suggests that the fusion was not transcribed. Therefore, it was concluded that FFPE-TLC identified a non-functional TPR::NTRK1 fusion. Accordingly, pan-TRK IHC identified all functional NTRK gene fusions.
Detection of other somatic alterations
A broader panel of genes was assessed, when performing RNA-NGS and FFPE-TLC for the detection of NTRK rearrangements (Supplementary Table S4). Concordance between RNA-NGS and FFPE-TLC was 100% for the detection of BRAFV600E and KRAS mutations. All tumors harboring an NTRK fusion (n = 10) were BRAF and KRAS wildtype.qRT-PCR and RNA-NGS also assess rearrangements in ALK, ROS1, and RET and detect MET exon 14 skipping, but these alterations were not detected in the tumors analyzed in this study.
Discussion
In this study, we show that FFPE-TLC and the qRT-PCR are reliable alternatives to the recommended RNA-NGS for the detection of NTRK fusions after IHC screening in mCRC, with a sensitivity of 90%, 100%, and 90%, respectively. Robustness for FFPE-TLC (100%) was good, reasonable for RNA-NGS (85%), but unsatisfactory for qRT-PCR (70%). Considering the high number of equivocal results and a limited sensitivity of 78%, we consider FISH analysis an inappropriate method for the detection of NTRK fusions in mCRC.
According to international guidelines, we used pan-TRK IHC as a screening tool to enrich for NTRK fusions [7, 8]. However, discriminating between weak, moderate, and strong TRK expression can be difficult and can lead to high inter-rater variability [3, 20]. To minimize variability, pan-TRK IHC was performed for all samples by the same optimized protocol, assessed by two experienced pathologists (Fig. 2, first column), and performed in a single cancer type (CRC). In this standardized and high-volume research setting, we found a 100% sensitivity and specificity for strong (3 +) TRK expression by IHC. However, in our opinion, this high sensitivity of pan-TRK IHC is unattainable in the routine diagnostic setting. This is reflected by the discordant pan-TRK IHC score in the NTRK3 fusion sample (Supplementary Fig. S2). A lower sensitivity for NTRK3 fusions by IHC has been described before [3]. Therefore, we recommend any above background positive pan-TRK IHC to be further analyzed by RNA or DNA based tests. For tumor (sub)types with a high prevalence of NTRK fusions, including MSI-H/dMMR, BRAF and RAS wildtype mCRC, upfront testing with RNA-NGS, FFPE-TLC, or qRT-PCR could be a reasonable approach.
Our results do not support FISH analysis for the detection of NTRK fusions in mCRC, which is in line with previous recommendations [7, 16]. Sensitivity of FISH was limited, possibly due to the occurrence of intrachromosomal NTRK1 fusions, including TPM3::NTRK1 and LMNA::NTRK1, which were most prevalent in our study. In addition, its usual advantages of being an inexpensive and easy assay are not apparent here, because three different break-apart probes are needed for NTRK1, NTRK2, and NTRK3, raising costs and complexity.
According to the results of this study, RNA-NGS, FFPE-TLC, and qRT-PCR can be used interchangeably in our opinion. Besides diagnostic performance, other factors are therefore important when deciding which assay to use, including costs, required input, turnaround time, availability, and whether or not simultaneous direct assessment of other somatic alterations is possible.qRT-PCR is an easy and fast assay to test for NTRK fusions with a turnaround time of only three hours and has a high diagnostic accuracy with an overall agreement of 100%, which is in line with previous reports [14, 17]. The most important limitation of qRT-PCR was the high rate of invalid test results. We found a robustness of 70% which is somewhat lower than reported in the aforementioned studies (88% and 89%, respectively). This may be explained by the use of older samples (median 43 months) with lower tumor percentages (median 25%) in our study. Increasing the tissue input seemed to increase the success rate.
RNA-NGS and FFPE-TLC are more expensive tests, but can be used more broadly by detecting other relevant mutations and rearrangements in one assay. In our study, RNA-NGS and FFPE-TLC performed equally well in terms of sensitivity. However, there are some important differences between these two assays. FFPE is the standard for preservation of tissue material in virtually all pathology labs worldwide. In FFPE tissue, DNA and RNA can be considerably damaged and fragmented, hindering DNA- and RNA-based NGS. For example, RNA-NGS could not detect an NTRK fusion in a tumor sample (case 026) that passed the RNA quality control. We hypothesized that the high deduplication ratio (130:1) was a sign of diminished RNA quality. Therefore, caution is advised for the interpretation of negative results in these instances. For FFPE-TLC, this fragmentation is irrelevant as FFPE-TLC is a proximity-ligation assay. On the other hand, transcription is not proven for fusions detected on DNA level by FFPE-TLC, emphasizing the importance of protein overexpression determined by IHC. Also, FFPE-TLC is not yet commercially available, so the perfect robustness needs to be confirmed in routine clinical practice. For broad implementation of FFPE-TLC, automated identification and characterization of rearrangements is needed. Therefore, further development of the computational pipeline PLIER is needed to automatically detect all intrachromosomal fusions. Nevertheless, the diagnostic performance and robustness could make FFPE-TLC a useful diagnostic tool for accurate detection of NTRK fusions.
This study provides a unique head-to-head comparison of available tests for the detection of NTRK fusions in a large population-based cohort of MSI-H/dMMR mCRC patients. Our study has several limitations. First, despite using this unbiased nationwide cohort, the number of detected NTRK fusions is low due to the rare occurrence of NTRK fusions in mCRC. Second, we used the consensus results as a reference standard with which we strived to find the ground truth. Because this reference standard is based on the assays that are being compared, sensitivity and specificity for these assays might be overestimated [21]. However, we think this epidemiological phenomenon might be limited in our study because two independent molecular biologists interpreted all results and were 100% concordant. Third, we did not perform a cost analysis which is an important factor for the adoption of an assay in daily practice. For determining total costs, not only materials and reagent cost should be taken into account but also hands-on-time costs, maintenance costs, and depreciation costs for all materials that are used.
We did not include whole genome sequencing (WGS) in our analysis because this is not feasible on FFPE material. However, WGS could be a solution for the identification of the diverse set of genetic biomarkers linked to the increasing number of targeted anticancer agents. Recently, the WIDE study has shown the feasibility of WGS for metastatic cancer in routine clinical practice [22]. However, costs are still very high, and the need for a repeat biopsy to obtain fresh-frozen tissue and the limited robustness (70%) are currently hurdles to overcome before WGS can be a suitable diagnostic tool in clinical practice.
To conclude, in this study, we compared pan-TRK IHC, RNA-NGS, FFPE-TLC, FISH, and qRT-PCR for the detection of NTRK fusions in dMMR/MSI mCRC, to find alternative diagnostic strategies for NTRK fusion detection that fit the need in routine care. We do not recommend FISH for screening or confirmation of NTRK fusions in mCRC due to its low sensitivity and robustness. Pan-TRK IHC is an appropriate screening tool for MSI-H/dMMR mCRC patients and NTRK fusions can be reliably confirmed by RNA-NGS, but also by FFPE-TLC and qRT-PCR.
Data availability
All results of our analysis are provided in supplementary files. Other data obtained during the current study are available from the corresponding author on reasonable request.
References
Hong DS, DuBois SG, Kummar S, Farago AF, Albert CM, Rohrberg KS et al (2020) Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol 21(4):531–540
Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF et al (2020) Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol 21(2):271–282
Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D et al (2020) NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol an Off J United States Can Acad Pathol Inc. 33(1):38–46
Chou A, Fraser T, Ahadi M, Fuchs T, Sioson L, Clarkson A et al (2020) NTRK gene rearrangements are highly enriched in MLH1/PMS2 deficient, BRAF wild-type colorectal carcinomas-a study of 4569 cases. Mod Pathol an Off J United States Can Acad Pathol Inc. 33(5):924–32
Cocco E, Benhamida J, Middha S, Zehir A, Mullaney K, Shia J et al (2019) Colorectal carcinomas containing hypermethylated MLH1 promoter and wild-type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res 79(6):1047–1053
Singh H, Li YY, Spurr LF, Shinagare AB, Abhyankar R, Reilly E et al (2021) Molecular characterization and therapeutic targeting of colorectal cancers harboring receptor tyrosine kinase fusions. Clin Cancer Res 27(6):1695–1705
Marchiò C, Scaltriti M, Ladanyi M, Iafrate AJ, Bibeau F, Dietel M et al (2019) ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol 30(9):1417–1427
Yoshino T, Pentheroudakis G, Mishima S, Overman MJ, Yeh K-H, Baba E et al (2020) JSCO-ESMO-ASCO-JSMO-TOS: international expert consensus recommendations for tumour-agnostic treatments in patients with solid tumours with microsatellite instability or NTRK fusions. Ann Oncol Off J Eur Soc Med Oncol 31(7):861–872
von Ahlfen S, Missel A, Bendrat K, Schlumpberger M (2007) Determinants of RNA quality from FFPE samples. PLoS One 2(12):e1261
Murphy DA, Ely HA, Shoemaker R, Boomer A, Culver BP, Hoskins I et al (2017) Detecting gene rearrangements in patient populations through a 2-step diagnostic test comprised of rapid IHC enrichment followed by sensitive next-generation sequencing. Appl Immunohistochem Mol Morphol AIMM 25(7):513–523
Donczo B, Guttman A (2018) Biomedical analysis of formalin-fixed, paraffin-embedded tissue samples: the holy grail for molecular diagnostics. J Pharm Biomed Anal 155:125–134
Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D et al (2017) Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol 41(11):1547–1551
Solomon JP, Hechtman JF (2019) Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res 79(13):3163–3168
Ukkola I, Nummela P, Kero M, Tammio H, Tuominen J, Kairisto V et al (2022) Gene fusions and oncogenic mutations in MLH1 deficient and BRAFV600E wild-type colorectal cancers. Virchows Arch [Internet] 480(4):807–17. https://doi.org/10.1007/s00428-022-03302-x
Allahyar A, Pieterse M, Swennenhuis J, Los-de Vries GT, Yilmaz M, Leguit R et al (2021) Robust detection of translocations in lymphoma FFPE samples using targeted locus capture-based sequencing. Nat Commun [Internet] 12(1):1–15. https://doi.org/10.1038/s41467-021-23695-8
Solomon JP, Benayed R, Hechtman JF, Ladanyi M (2019) Identifying patients with NTRK fusion cancer. Ann Oncol 30(Supplement 8):VIII16–22
Sorber L, Van Dorst B, Bellon E, Zwaenepoel K, Lambin S, De Winne K et al (2022) NTRK gene fusion detection in a pan-cancer setting using the Idylla GeneFusion Assay. J Mol Diagn 24(7):750–759
Schraa SJ, Laclé MM, Zwart K, Gort EH, Koopman M, De Leng WW et al Prevalence, treatment and survival of NTRK gene fusions in microsatellite instable metastatic colorectal cancer patientsle. Ann Oncol 33:S136–S196. https://doi.org/10.1016/annonc/annonc1048
Richardson SO, Huibers MMH, de Weger RA, de Leng WWJ, Hinrichs JWJ, Meijers RWJ et al (2019) One-fits-all pretreatment protocol facilitating fluorescence in situ hybridization on formalin-fixed paraffin-embedded, fresh frozen and cytological slides. Mol Cytogenet 12:27
Koopman B, Kuijpers CCHJ, Groen HJM, Timens W, Schuuring E, Willems SM et al (2022) Detection of NTRK fusions and TRK expression and performance of pan-TRK immunohistochemistry in routine diagnostics: results from a nationwide community-based cohort. Diagnostics (Basel) 12(3):668
Dendukuri N, Schiller I, de Groot J, Libman M, Moons K, Reitsma J et al (2018) Concerns about composite reference standards in diagnostic research. BMJ 360:j5779
Samsom KG, Schipper LJ, Roepman P, Bosch LJ, Lalezari F, Klompenhouwer EG et al (2022) Feasibility of whole-genome sequencing-based tumor diagnostics in routine pathology practice. J Pathol 258(2):179–188
Acknowledgements
The authors would like to thank PALGA, the nationwide network and registry of histo- and cytopathology in the Netherlands, for providing histopathological data and for their help in the collection of FFPE tissue. The authors also thank the registration team of the Netherlands Comprehensive Cancer Organization (IKNL) for the collection of data for the Netherlands Cancer Registry.
Funding
This work was supported by Bayer B.V. The funders had no role in the design and conduct of the study. Cergentis B.V. performed the FFPE-TLC analysis free of charge and Biocartis Group N.V. provided cartridges free of charge. Biocartis had no role in the design and conduct of the study.
Author information
Authors and Affiliations
Contributions
S.J.S, G.R.V., and G.M.B. performed study concept and design; S.J.S., E.S., M.M.L., J.F.S., L.A.A.B., and W.W.L. performed collection and assembly of data; S.J.S., E.S., R.J.F., H.F., W.W.L., G.R.V., and G.M.B. performed data analysis and interpretation; S.J.S, E.S., G.R.V., and G.M.B. wrote the paper; all authors performed final approval of the paper, are accountable for all aspects of the work, confirm that they had full access to all the data in the study, and accept responsibility to submit for publication.
Corresponding author
Ethics declarations
Competing interests
S.J. Schraa reported grants from Personal Genome Diagnostics (PGDx) outside the submitted work. E. Stelloo, J.F. Swennenhuis, and H. Feitsma are employees of Cergentis BV, the company that invented and owns the FFPE-TLC technology. R.J.A.F. reported public private partnership consortia grants in collaboration with Cergentis BV, Personal Genome Diagnostics, Delfi Diagnostics, MERCK BV, outside the submitted work. In addition, R.J.A.F. has several patents pending. M. Koopman reported institutional scientific grants from Bayer, Bristol Myers Squibb, Merck, Personal Genome Diagnostics (PGDx), Pierre Fabre, Roche, Sirtex and Servier, has an advisory role for Nordic Farma, Merck-Serono, Pierre Fabre, and Servier outside the submitted work and is principal investigator from the international cohort study PROMETCO with Servier as sponsor. W.W. de Leng reported grants from Roche, BMS, and Pfizer and was involved in advisory boards for BMS, Janssen, and Novartis outside the submitted work. G.R. Vink reported grants from BMS, Merck, Servier, Personal Genome Diagnostics (PGDx), Bayer, Sirtex, Pierre Fabre, Lilly, and Delfi Diagnostics outside the submitted work. G.M. Bol reports institutional scientific grants from Bayer, Pierre Fabre, and Terumo. No other disclosures were reported.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schraa, S.J., Stelloo, E., Laclé, M.M. et al. Comparison of NTRK fusion detection methods in microsatellite-instability-high metastatic colorectal cancer. Virchows Arch 482, 983–992 (2023). https://doi.org/10.1007/s00428-023-03538-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-023-03538-1