Abstract
The current study assessed the performance of the fully automated RT-PCR-based Idylla™ GeneFusion Assay, which simultaneously covers the advanced non-small cell lung carcinoma (aNSCLC) actionable ALK, ROS1, RET, and MET exon 14 rearrangements, in a routine clinical setting involving 12 European clinical centers. The Idylla™ GeneFusion Assay detects fusions using fusion-specific as well as expression imbalance detection, the latter enabling detection of uncommon fusions not covered by fusion-specific assays. In total, 326 archival aNSCLC formalin-fixed paraffin-embedded (FFPE) samples were included of which 44% were resected specimen, 46% tissue biopsies, and 9% cytological specimen. With a total of 179 biomarker-positive cases (i.e., 85 ALK, 33 ROS1, 20 RET fusions and 41 MET exon 14 skipping), this is one of the largest fusion-positive datasets ever tested. The results of the Idylla™ GeneFusion Assay were compared with earlier results of routine reference technologies including fluorescence in situ hybridization, immunohistochemistry, reverse-transcription polymerase chain reaction, and next-generation sequencing, establishing a high sensitivity/specificity of 96.1%/99.6% for ALK, 96.7%/99.0% for ROS1, 100%/99.3% for RET fusion, and 92.5%/99.6% for MET exon 14 skipping, and a low failure rate (0.9%). The Idylla™ GeneFusion Assay was found to be a reliable, sensitive, and specific tool for routine detection of ALK, ROS1, RET fusions and MET exon 14 skipping. Given its short turnaround time of about 3 h, it is a time-efficient upfront screening tool in FFPE samples, supporting rapid clinical decision making. Moreover, expression-imbalance-based detection of potentially novel fusions may be easily verified with other routine technologies without delaying treatment initiation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-small cell lung carcinoma (NSCLC) therapy has been transformed by the identification of actionable oncogenic driver mutations, of which ALK, ROS1, RET fusions and MET exon 14 skipping represent interesting drug targets [1]. ALK [2], ROS1 [3], and RET [4] are tyrosine kinases, and fusion of their kinase domains with the amino-terminal portions of a variety of protein partners leads to disturbance of downstream signaling cascades, resulting in uncontrolled cell proliferation and promotion of survival. MET exon 14 skipping on the other hand is an intragenic rearrangement of the MET gene resulting in sustained MET activation, ultimately leading to cell proliferation and tumor growth [5]. The prevalence of ALK fusion events in NSCLC is 4–7%, for ROS1 prevalence is 1–3%, and for RET it is 1–2% [6,7,8], while MET exon 14 skipping has a prevalence of 3% [8]. Therefore, the combined frequency of these gene rearrangements is about the same as the occurrence of EGFR mutations in an European advanced NSCLC population [9].
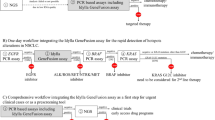
To determine eligibility to therapy targeting ALK, ROS1, RET, or MET alterations [10,11,12,13], NSCLC treatment guidelines recommend testing for ALK, ROS1, RET fusions and MET exon 14 skipping [14, 15]. Kinase fusions are usually detected using single-test methods like immunohistochemistry (IHC) for ALK fusions [15,16,17], and fluorescence in situ hybridization (FISH), which is the gold standard for detection of ALK and ROS1 fusions and to a lesser extent RET fusions [18, 19], or multiplex test methods like reverse-transcription polymerase chain reaction (RT-PCR) and next-generation sequencing (NGS) [20] as valuable alternatives to ALK, ROS1, and RET FISH and as the most effective detection methods for MET exon 14 skipping.
The CE-IVD Idylla™ GeneFusion Assay (Biocartis, Mechelen, Belgium) is an RNA-based fully automated RT-PCR assay, designed to concurrently detect presence of ALK, ROS1, RET fusions and MET exon 14 skipping in formalin-fixed, paraffin-embedded (FFPE) tumor tissue sections. This is realized by combining (i) specific detection of ALK, ROS1, RET fusions and MET exon 14 skipping and (ii) analysis of ALK, ROS1, and RET expression imbalance. The latter measures the difference between 3′ and 5′ gene expression levels of ALK, ROS1, or RET, which is indicative for the presence of a fusion, however, should always be confirmed with an alternative gene fusion test method [21]. The research which use only (RUO) version of the Idylla™ GeneFusion Assay, which also covered NTRK1/2/3 rearrangements, was launched before the CE-IVD Idylla™ GeneFusion Panel. In initial studies, this RUO Idylla™ GeneFusion Assay showed high sensitivity (82–100%) and specificity (98–100%) for the detection of ALK, ROS1, RET fusions and MET exon 14 skipping [22, 23]. In a recently published study, the Idylla™ GeneFusion Assay had an equal level of gene fusion detection compared with the Genexus NGS system, which has a turnaround time (not including RNA purification) of about 24 h; with each ultrafast gene fusion assay having its own specific technology-related limitations [24]. Due to sample-to-result automatization and only 2 min hands-on time required to load tissue slides into the single-use cartridge, the Idylla™ GeneFusion Assay can be easily established in on-site routine screening. Together with the absence of batching need and its short turnaround time of approximately 3 h, the technology may as such address the relatively long turnaround times associated with NGS [25,26,27,28,29].
The current multicenter study investigated the performance of the Idylla™ GeneFusion Assay (RUO) in a real-life clinical setting involving 12 clinical centers across Europe. These centers selected and tested a total of 326 archival histologically proven advanced NSCLC (stage IV) FFPE tissue samples with the Idylla™ GeneFusion Assay of which results were compared with the biomarker status determined earlier with routine reference methods (including FISH, IHC, RT-PCR, and NGS).
Materials and methods
Tissue sample collection and study design
Twelve clinical centers from nine different European countries participated in this multicenter observational non-interventional retrospective study that assessed the mutational status of 326 archival, histologically proven, advanced NSCLC (stage IV) tissue or cytological FFPE samples (Table 1). All samples had been tested previously with a routine reference method for ALK, ROS1, RET fusions and/or MET exon 14 skipping, and the current study retested them at the same clinical centers using the Idylla™ GeneFusion Assay.
Patients provided informed consent. The use of these patient samples was approved by the respective local Ethics Committees and was in accordance with the Declaration of Helsinki.
For inclusion, each sample had to originate from the same block that was used with the original reference method test and was required to have ≥10% neoplastic cells and a tissue area >20 mm2 when 1 slice/slide was used, or a tissue area ≤20 mm2 if three slices/slides were used. FFPE cell block requirements were ≥10% neoplastic cells and total cell number >2000 per slide when 1 slice/slide was used or neoplastic cell number ≤2000 per slide when three slices/slides were used. Stained samples, non-FFPE samples, decalcified samples, and samples older than 9 years (or 3 years if compared to FISH) were excluded. Macrodissection was allowed to increase neoplastic cell content to above the required 10%.
Sampling of FFPE tissue sections (mostly one or two, up to five slides) for the Idylla™ GeneFusion Assay was performed consecutively, if possible, to the sections used in the earlier routine reference method. For each sample, the tissue section(s) were placed in the lysis chamber of a new cartridge, which was next loaded onto the Idylla™ instrument. The whole procedure required about 2 min of hands-on time.
The results obtained with the Idylla™ GeneFusion Assay on archival material were not used for diagnostic purposes.
Idylla™ GeneFusion Assay
The real-time PCR-based Idylla™ GeneFusion Assay (RUO) was used in the current study. This is a fully automated in vitro diagnostic test qualitatively detecting specific ALK, ROS1, RET gene fusions as well as MET exon 14 skipping from RNA transcripts. The Idylla™ GeneFusion Assay gene panel is detailed in Supplementary Table 1. Apart from the detection of these specific fusions, the Idylla™ GeneFusion Assay (RUO) does also assess ALK, ROS1, RET, and NRTK1/2/3 expression imbalance. NRTK1/2/3 expression imbalance results are not reported here as NRTK1/2/3 fusion detection was not in the scope of the current study.
The Idylla™ GeneFusion Assay covers the entire sample-to-result process, including fully integrated RNA extraction (which is based on a combination of enzymatic degradation, heat, and high-frequency ultrasound), reverse transcription of RNA to cDNA, real-time PCR amplification and detection, as well as data analysis, and result reporting.
Routine methods used as reference method
Several commercial FISH assays were used as reference method following the manufacturer’s instructions: Abbot (Abbott, Abbott Park, IL) Vysis break apart probe (ALK fusion), Leica (Wetzlar, Germany) XL for BOND, ZytoVision (Bremerhaven, Germany) Zytolight SPEC Dual Color break apart probe (ALK, ROS1, RET fusions), and Empire Genomics (Buffalo, NY; ALK, ROS1, RET fusions).
The following commercial IHC methods were used according to the manufacturer’s instructions: Roche Diagnostics (Rotkreuz, Switzerland) Ventana D5F3 (ALK fusion), Ventana SP384 (ROS1 fusion), Abcam (Waltham, MA) 5A4 (ALK fusion), Cell Signaling (Danvers, MA) D4D6 (ROS1 fusion), Bond Leica D5F3 (ALK fusion), and Leica Bond D4D6 (ROS1 fusion).
Routine NGS methods that were used according to the manufacturer’s instructions were Archer (Boulder, CO) Fusionplex Lung v1.0 (ALK, ROS1, RET fusions, MET exon 14 skipping), Thermo Fisher Scientific (Waltham, MA) Oncomine Focus Assay (ALK, ROS1, RET fusions, MET exon 14 skipping), Thermo Fisher Scientific Oncomine Precision Assay (RET fusion, MET exon 14 skipping), and Illumina (San Diego, CA) TruSight Tumor 170 (ALK, ROS1, RET fusions, MET exon 14 skipping).
The Diatech (Jesi, Italy) Easy PGX RT025 (ALK, ROS1, RET fusions, MET exon 14 skipping) and AmoyDx (Xiamen, China) Pan Lung Cancer panel (ALK fusion, MET exon 14 skipping) RT-PCR assays were used following the manufacturer’s instructions.
A Sanger assay was used to determine MET exon 14 skipping.
Statistical analysis
Concordance was calculated using overall concordance, sensitivity, and specificity, excluding invalid and error test results. Failure rate was calculated as the sum of errors and invalid test results on the total sample set.
Results
Sample description
The current study evaluated the Idylla™ GeneFusion Assay in a real-life clinical setting. To this end, a set of 326 archival, unstained FFPE advanced NSCLC (stage IV) tissue or cytological samples was selected by 12 centers across nine European countries. For two of these samples, the Idylla™ GeneFusion Assay resulted in an error, and therefore the final analysis set contained 324 samples of which the characteristics are summarized in Table 2. All samples had been tested before at these clinical centers to detect ALK, ROS1, RET fusions and/or MET exon 14 skipping using the previously described range of validated reference methods, and the current study assesses the concordance between the Idylla™ GeneFusion Assay results and results of these former routine reference methods. Of the 324 samples tested, 179 (55%) had been reported by the routine reference methods as positive for ALK, ROS1, RET fusions and/or for MET exon 14 skipping, and 145 (45%) had been reported as negative for these gene alterations.
Considering the neoplastic cell content, six samples (2%) deviated from the protocol instructions to have at least 10% neoplastic cell content, and for four samples neoplastic cell content was not recorded. About two-thirds (69%) of the slides tested had a thickness of 5 μm, while one-third (30%) had a thickness of 10 μm; in five cases, slide thickness was not reported. Most often one (30%) or two (37%) slides were tested; for 14 samples, the number of slides tested was not reported. The tissue surface varied from below 20 mm2 (19%), to between 20 and 50 mm2 (36%), between 50 and 100 mm2 (14%), and above 100 mm2 (20%), while tissue surface was not reported in 38 cases. Macrodissection to increase neoplastic cell content was performed on 156 samples (48%).
Overall, the sample set offered a large sample size, included several clinical routine workflows, and was considered representative for real-life clinical circumstances.
Results of Idylla™ GeneFusion Assay compared with reference methods
Slides of 324 archival samples tested with the Idylla™ GeneFusion Assay obtained 323 valid overall test results, which were compared with the results of the routine reference methods that were previously performed on the same tissue block (Table 3). One invalid test result was obtained using the Idylla™ GeneFusion Assay, for which the reference reported an ALK fusion; the sample tested was however smaller (0–20 mm2) with low tumor cell content (10–20%) and confirmed to be a less-quality sample as commented to be indicative for RNA degradation.
A first comparison of the data showed that the Idylla™ GeneFusion Assay reported one ALK fusion, three ROS1 fusions, and one RET fusion not detected by the routine reference method. Inversely, the Idylla™ GeneFusion Assay did not confirm 18 ALK fusions, eight ROS1 fusions, five RET fusions, and four MET exon 14 skipping events previously reported by routine reference methods.
In addition to the different particular fusion events, the Idylla™ GeneFusion assay also measures ALK, ROS1, and RET expression imbalance, which could be indicative for the presence of a fusion. When including in the analysis of the expression imbalance results, the Idylla™ GeneFusion Assay detected eight additional ALK fusion events, three ROS1 fusions, and four RET fusions.
Including the expression imbalance results, there was an agreement between the Idylla™ GeneFusion Assay and the routine reference methods for 312 (96.3%) samples regarding ALK fusion, 315 (97.2%) samples for ROS1 fusion, 321 (99.1%) samples for RET fusion, and 319 (98.5%) samples for MET exon 14 skipping. The samples with discordant results were further analyzed.
Analysis of discordant results
The samples for which the results of the Idylla™ GeneFusion Assay and the reference method were discordant and for which enough sample material was still available, were re-analyzed using an additional test on material from the same FFPE tissue block. The additional test method is indicated in Table 4, together with its results. The “true value” is considered the value confirmed by at least two different technologies. If the re-analyzed result or third-method test result confirmed the Idylla™ GeneFusion Assay result, the outcome was reclassified as concordant.
For sample 3, a second NGS assay confirmed the absence of ALK fusion and therefore confirmed the Idylla™ GeneFusion Assay result. As part of the analysis of discordant samples, samples 4 and 9 were retested at the original study center with NGS and FISH, respectively, both resulting in a non-detected ALK fusion result. Both cases were therefore classified as “not detected” for “true value” and concluded to be concordant. For samples 5, 6, 7, and 8, a third-method analysis was performed at an independent testing site using NGS. In three cases (samples 5, 6, 7), NGS resulted in a non-detected ALK fusion result confirming false positive results with FISH, while in one case (sample 8), it confirmed the IHC result and hence a false negative result with the Idylla™ GeneFusion Assay. Sample 11 was repeated twice with the Idylla™ GeneFusion Assay at the original study center, resulting in two positive ALK and one negative ALK detection overall. It was decided to label this sample as discordant. For samples 18 and 19, two biomarkers were detected with the Idylla™ GeneFusion Assay, i.e., ROS1 fusion + MET exon 14 skipping for sample 18 and ROS1 fusion + ALK fusion for sample 19. MET exon 14 skipping (sample 18) and ALK fusion (sample 19) were detected by NGS as well. However in both cases, a ROS1 fusion had been detected at very low read count by the Archer NGS panel used at the original study center but had not been reported. For sample 19, this ROS1 rearrangement involved an intergenic region as well, and based on a profound re-analysis of the Archer NGS results, it was decided to label this sample as concordant (Supplementary Figure 1). In sample 21, the Idylla™ GeneFusion Assay detected a double fusion (i.e., ALK fusion + RET fusion), of which the RET fusion was not reported by the reference methods as they did not test for this rearrangement. However, further analysis revealed that this detection of a RET fusion by the Idylla™ GeneFusion Assay was due to a software error, which the manufacturer planned to correct with a software update. Third-method testing of sample 22 with a second NGS test (i.e., Oncomine Focus Assay), this time at an independent testing site, confirmed the presence of MET exon 14 skipping (read count of 397 reads). For sample 25, the site confirmed that the initial MET exon 14 skipping detection with Sanger was not a true MET exon 14 skipping variant, and therefore the “true value” was classified as “not detected.” Sample 16, 23, and 24 did not have sufficient sample available for a discordant analysis with an additional method. For sample 16, a CCDC6::ROS1 fusion was detected with NGS; CCDC6 is a partner gene not included in the Idylla™ GeneFusion Assay fusion-specific design.
Performance of Idylla™ GeneFusion Assay
Based on the discordant analysis results, eight additional samples were classified as ALK fusion concordant, as well as five additional samples for ROS1 fusion, one additional sample for RET fusion, and one additional sample for MET exon 14 skipping (Table 5). The resulting overall concordance in this dataset of the Idylla™ GeneFusion Assay (including expression imbalance) with reference method results was 98.8% for ALK fusion, 98.8% for ROS1 fusion, 99.4% for RET fusion, and 98.8% for MET exon 14 skipping. The inconclusive cases were considered to be discordant for the final calculation. Given the three failures reported above (i.e., two errors and one invalid result), the validity of the Idylla™ GeneFusion Assay was 99.1% (323/326).
Discussion
In patients with advanced NSCLC with rapid disease progression, timely therapeutic decision making is essential. In the past decade, treatment options for NSCLC have expanded, which led to an increased number of biomarkers to be tested, often on very sparse material with multiple testing technologies. This may result in an undesired prolonged time to treatment for this vulnerable group of patients.
The current multicenter study investigated the performance of the Idylla™ GeneFusion Assay in a real-life clinical setting involving 12 clinical centers across Europe. These centers selected and tested a total of 326 archival histologically proven advanced NSCLC (stage IV) FFPE tissue samples with the Idylla™ GeneFusion Assay of which results were compared with the molecular status determined earlier with routine reference methods including FISH, IHC, RT-PCR, and NGS.
Among the 326 samples analyzed, a very low failure rate of 0.9% (3/326) was observed for the Idylla™ GeneFusion Assay. This despite the analysis been carried out on archival material with up to 9 years between the initial analysis and the current study, the included material originating from various metastatic sites with different pre-analytic tissue preparation procedures, and the analysis being carried out at 12 different sites with different tissue processing and storage procedures. This observation supports the robustness of the assay as a fast and reliable test in a real-life diagnostic setting.
The 324 remaining samples tested comprised of 179 (55%) samples reported as positive for either ALK, ROS1, RET fusions and/or MET exon 14 skipping, and 145 (45%) samples reported as negative for these gene alterations by the routine reference method, providing a large sample size to assess both sensitivity and specificity of the Idylla™ GeneFusion Assay, with different tissues and different fusion compositions, but also offering a unique opportunity to test it against the routine reference methods currently used in routine NSCLC molecular testing.
It was found that the Idylla™ GeneFusion Assay has a high sensitivity/specificity, respectively, of 96.1%/99.6% for ALK, 96.7%/99.0% for ROS1, 100%/99.3% for RET fusions and 92.5%/99.6% for MET exon 14 skipping. It was clearly demonstrated that the expression imbalance technology has its value in increasing the sensitivity of the assay and thereby acts as the complement for detection of fusion transcripts with uncommon or novel fusion partners in NSCLC, especially in the case of ALK and RET fusions. In this study, only expression-imbalance-positive results that were confirmed by an alternative method were considered true positive, as per the manufacturer’s assay instructions. This makes the Idylla™ GeneFusion Assay expression imbalance technology highly relevant as a time-efficient upfront screening tool where detected imbalances can be verified quickly with either IHC or FISH, and the relatively slower NGS can be performed afterward to establish the exact fusion present, without delaying treatment initiation. In a recent investigation conducted by Gilson et al. in 2023, diverse potential applications of the Idylla™ system within laboratory workflows have been elucidated. These applications encompass integration in the form of a fast-track precursor to comprehensive testing, a sequential approach, or as a rescue test. It is imperative for each institution to conduct a thorough assessment of its unique clinical requirements and available resources, taking into consideration logistical and financial parameters, in order to determine the most suitable integration strategy [30]. The Idylla™ GeneFusion Assay can be used on demand without the need of batching, directly starting from limited FFPE material and with results available within 3 h.
These findings make the Idylla™ GeneFusion Assay an obvious choice for fast and reliable detection of treatment-relevant fusions in the initial NSCLC diagnostic workup. Fast detection of targetable fusions may however also be highly relevant for NSCLC patients progressing on current tyrosine kinase inhibitor treatment, as oncogenic fusions are one of many known tyrosine kinase inhibitor resistance mechanisms [31, 32].
As with all retrospective studies, one of the study limitations is the retrospective design and hence the sample selection bias. In an ideal setting, all the included samples would have been tested with several or all reference methods considered but given the limited sample availability and the diversity in methods applied at the different sites, like in a real-life setting, this was not an option.
The study design tried to ensure that only samples with enough material available to perform an additional in-depth discordant investigation were included, but this was unfortunately not the case for three of the 25 discordant samples. It can be quite difficult to assess the amount of available material left in a FFPE tissue block containing small lung biopsies or cell block material, and the availability can be further limited for fusion-positive samples if additional analysis has been performed as part of the initial diagnostic workup. One could of course exclude these samples from the study, but as they reflect the limitations of a retrospective study and the normal challenges faced in a thoracic oncology testing facility, they were included.
The same rationale was behind inclusion of the inconclusive cases in the final calculations of sensitivity and specificity for detection of ALK, ROS1, RET fusions and MET exon 14 skipping. They represent a real-life clinical cohort, and the result obtained with the reference method was therefore considered the “true value” if a third method could not be performed or its result was inconclusive. Retrospective studies can have difficulties to fully elucidate the real “true value” due to limited material and resources.
The true value for the discordant MET exon 14 skipping cases may be hard to establish if the reference or additional NGS method used is the Oncomine technology, which can create false positive calls, due to a homopolymeric error of the splice donor site [33]. These false positive calls can be distinguished by relatively low read counts compared to real MET exon 14 skipping events. The two inconclusive MET exon 14 skipping cases may reflect this pitfall.
To conclude, the Idylla™ GeneFusion Assay is a promising tool for rapid detection of ALK, ROS1, RET, or MET exon 14 alterations in NSCLC.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Change history
19 April 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00428-024-03800-0
References
Kazdal D, Hofman V, Christopoulos P, Ilié M, Stenzinger A, Hofman P (2022) Fusion-positive non-small cell lung carcinoma: biological principles, clinical practice, and diagnostic implications. Genes Chromosom Cancer 61(5):244–260. https://doi.org/10.1002/gcc.23022
Hallberg B, Palmer RH (2016) The role of the ALK receptor in cancer biology. Ann Oncol 27(Suppl 3):iii4–iii15. https://doi.org/10.1093/annonc/mdw301
Roskoski R Jr (2017) ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol Res 121:202–212. https://doi.org/10.1016/j.phrs.2017.04.022
Cascetta P, Sforza V, Manzo A, Carillio G, Palumbo G, Esposito G, Montanino A, Costanzo R, Sandomenico C, De Cecio R, Piccirillo MC, La Manna C, Totaro G, Muto P, Picone C, Bianco R, Normanno N, Morabito A (2021) RET inhibitors in non-small-cell lung cancer. Cancers (Basel) 13(17):4415. https://doi.org/10.3390/cancers13174415
Socinski MA, Pennell NA, Davies KD (2021) MET exon 14 skipping mutations in non-small-cell lung cancer: an overview of biology, clinical outcomes, and testing considerations. JCO Precis Oncol 5:PO.20.00516. https://doi.org/10.1200/PO.20.00516
Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, Chang MT, Ni A, Kundra R, Jonsson P, Jayakumaran G, Gao SP, Johnsen HC, Hanrahan AJ, Zehir A, Rekhtman N, Ginsberg MS, Li BT, Yu HA et al (2017) Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov 7(6):596–609. https://doi.org/10.1158/2159-8290.CD-16-1337
Levy MA, Lovly CM, Pao W (2012) Translating genomic information into clinical medicine: lung cancer as a paradigm. Genome Res 22(11):2101–2108. https://doi.org/10.1101/gr.131128.111
Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, Heng JC, Dahlberg SE, Jänne PA, Verma S, Christensen J, Hammerman PS, Sholl LM (2016) MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol 34(7):721–730. https://doi.org/10.1200/JCO.2015.63.4600
Jakobsen JN, Santoni-Rugiu E, Grauslund M, Melchior L, Sørensen JB (2018) Concomitant driver mutations in advanced EGFR-mutated non-small-cell lung cancer and their impact on erlotinib treatment. Oncotarget 9(40):26195–26208. https://doi.org/10.18632/oncotarget.25490
Cognigni V, Pecci F, Lupi A, Pinterpe G, De Filippis C, Felicetti C, Cantini L, Berardi R (2022) The landscape of ALK-rearranged non-small cell lung cancer: a comprehensive review of clinicopathologic, genomic characteristics, and therapeutic perspectives. Cancers (Basel) 14(19):4765. https://doi.org/10.3390/cancers14194765
Azelby CM, Sakamoto MR, Bowles DW (2021) ROS1 targeted therapies: current status. Curr Oncol Rep 23(8):94. https://doi.org/10.1007/s11912-021-01078-y
Servetto A, Esposito D, Ferrara R, Signorelli D, Belli S, Napolitano F, Santaniello A, Ciciola P, Formisano L (1877) Bianco R (2022) RET rearrangements in non-small cell lung cancer: evolving treatment landscape and future challenges. Biochim Biophys Acta Rev Cancer 6:188810. https://doi.org/10.1016/j.bbcan.2022.188810
Fujino T, Suda K, Mitsudomi T (2021) Lung cancer with MET exon 14 skipping mutation: genetic feature, current treatments, and future challenges. Lung Cancer (Auckl) 12:35–50. https://doi.org/10.2147/LCTT.S269307
Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, D’Amico TA, DeCamp M, Dilling TJ, Dowell J, Gettinger S, Grotz TE, Gubens MA, Hegde A, Lackner RP, Lanuti M, Lin J et al (2022) Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 20(5):497–530. https://doi.org/10.6004/jnccn.2022.0025
Hendriks LE, Kerr KM, Menis J, Mok TS, Nestle U, Passaro A, Peters S, Planchard D, Smit EF, Solomon BJ, Veronesi G, Reck M, Guidelines Committee ESMO (2023) ESMO Guidelines Committee. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol 34(4):339–357. https://doi.org/10.1016/j.annonc.2022.12.009
Hofman V, Rouquette I, Long-Mira E, Piton N, Chamorey E, Heeke S, Vignaud JM, Yguel C, Mazières J, Lepage AL, Bibeau F, Begueret H, Lassalle S, Lalvée S, Zahaf K, Benzaquen J, Poudenx M, Marquette CH, Sabourin JC et al (2019) Multicenter evaluation of a novel ROS1 immunohistochemistry assay (SP384) for detection of ROS1 rearrangements in a large cohort of lung adenocarcinoma patients. J Thorac Oncol 14(7):1204–1212. https://doi.org/10.1016/j.jtho.2019.03.024
Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, Colasacco C, Dacic S, Hirsch FR, Kerr K, Kwiatkowski DJ, Ladanyi M, Nowak JA, Sholl L, Temple-Smolkin R, Solomon B, Souter LH, Thunnissen E, Tsao MS et al (2018) Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 13(3):323–358. https://doi.org/10.1016/j.jtho.2017.12.001
Conde E, Rojo F, Gómez J, Enguita AB, Abdulkader I, González A, Lozano D, Mancheño N, Salas C, Salido M, Salido-Ruiz E, de Álava E (2022) Molecular diagnosis in non-small-cell lung cancer: expert opinion on ALK and ROS1 testing. J Clin Pathol 75(3):145–153. https://doi.org/10.1136/jclinpath-2021-207490
Baker JA, Sireci AN, Marella N, Cannon HK, Marquart TJ, Holzer TR, Reising LO, Cook JD, Wijayawardana SR, Bodo J, Hsi ED, Schade AE, Oakley GJ (2022) Analytical accuracy of RET fusion detection by break-apart fluorescence in situ hybridization. Arch Pathol Lab Med 146(3):351–359. https://doi.org/10.5858/arpa.2020-0376-OA
Brisudova A, Skarda J (2020) Gene rearrangement detection by next-generation sequencing in patients with non-small cell lung carcinoma. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 164(2):127–132. https://doi.org/10.5507/bp.2020.015
Wang R, Pan Y, Li C, Hu H, Zhang Y, Li H, Luo X, Zhang J, Fang Z, Li Y, Shen L, Ji H, Garfield D, Sun Y, Chen H (2012) The use of quantitative real-time reverse transcriptase PCR for 5′ and 3′ portions of ALK transcripts to detect ALK rearrangements in lung cancers. Clin Cancer Res 18(17):4725–4732 Erratum in: Clin Cancer Res 18(21):6079 (2012). https://doi.org/10.1158/1078-0432.CCR-12-0677
Chu YH, Barbee J, Yang SR, Chang JC, Liang P, Mullaney K, Chan R, Salazar P, Benayed R, Offin M, Drilon A, Ladanyi M, Nafa K, Arcila ME (2022) Clinical utility and performance of an ultrarapid multiplex RNA-based assay for detection of ALK, ROS1, RET, and NTRK1/2/3 rearrangements and MET exon 14 skipping alterations. J Mol Diagn 24(6):642–654. https://doi.org/10.1016/j.jmoldx.2022.03.006
Depoilly T, Garinet S, van Kempen LC, Schuuring E, Clavé S, Bellosillo B, Ercolani C, Buglioni S, Siemanowski J, Merkelbach-Bruse S, Tischler V, Demes MC, Paridaens H, Sibille C, de Montpreville VT, Rouleau E, Bartczak A, Pasieka-Lis M, Teo RYW et al (2022) Multicenter evaluation of the Idylla GeneFusion in non-small-cell lung cancer. J Mol Diagn 24(9):1021–1030. https://doi.org/10.1016/j.jmoldx.2022.05.004
Hofman V, Heeke S, Bontoux C, Chalabreysse L, Barritault M, Bringuier PP, Fenouil T, Benzerdjeb N, Begueret H, Merlio JP, Caumont C, Piton N, Sabourin JC, Evrard S, Syrykh C, Vigier A, Brousset P, Mazieres J, Long-Mira E et al (2022) Ultrafast gene fusion assessment for nonsquamous NSCLC. JTO Clin Res Rep 4(2):100457. https://doi.org/10.1016/j.jtocrr.2022.100457
Finall A, Davies G, Jones T, Emlyn G, Huey P, Mullard A (2022) Integration of rapid PCR testing as an adjunct to NGS in diagnostic pathology services within the UK: evidence from a case series of non-squamous, non-small cell lung cancer (NSCLC) patients with follow-up. J Clin Pathol 76(6):391–399. https://doi.org/10.1136/jclinpath-2021-207987
Behnke A, Cayre A, De Maglio G, Giannini G, Habran L, Tarsitano M, Chetta M, Cappellen D, Lespagnol A, Le Naoures C, Massazza G, Destro A, Bonzheim I, Rau A, Battmann A, Kah B, Watkin E, Hummel M (2023) FACILITATE: a real-world, multicenter, prospective study investigating the utility of a rapid, fully automated real-time PCR assay versus local reference methods for detecting epidermal growth factor receptor variants in NSCLC. Pathol Oncol Res 29:1610707. https://doi.org/10.3389/pore.2023.1610707
Normanno N, Apostolidis K, Wolf A, Al Dieri R, Deans Z, Fairley J, Maas J, Martinez A, Moch H, Nielsen S, Pilz T, Rouleau E, Patton S, Williams V (2022) Access and quality of biomarker testing for precision oncology in Europe. Eur J Cancer 176:70–77. https://doi.org/10.1016/j.ejca.2022.09.005
Adizie JB, Tweedie J, Khakwani A, Peach E, Hubbard R, Wood N, Gosney JR, Harden SV, Beckett P, Popat S, Navani N (2021) Biomarker testing for people with advanced lung cancer in England. JTO Clin Res Rep 2(6):100176. https://doi.org/10.1016/j.jtocrr.2021.100176
Petiteau C, Robinet-Zimmermann G, Riot A, Dorbeau M, Richard N, Blanc-Fournier C, Bibeau F, Deshayes S, Bergot E, Gervais R, Levallet G (2021) Contribution of the Idylla™ system to improving the therapeutic care of patients with NSCLC through early screening of EGFR mutations. Curr Oncol 28(6):4432–4445. https://doi.org/10.3390/curroncol28060376
Gilson P, Pouget C, Belmonte R, Fadil S, Demange J, Rouyer M, Lacour J, Betz M, Dardare J, Witz A, Merlin JL (2023) Harlé A (2023) Validation of the Idylla GeneFusion assay to detect fusions and MET exon-skipping in non-small cell lung cancers. Sci Rep 13(1):12909. https://doi.org/10.1038/s41598-023-39749-4
Urbanska EM, Sørensen JB, Melchior LC, Costa JC, Santoni-Rugiu E (2022) Durable response to combined osimertinib and pralsetinib treatment for osimertinib resistance due to novel intergenic ANK3-RET fusion in EGFR-mutated non-small-cell lung cancer. JCO Precis Oncol 6:e2200040. https://doi.org/10.1200/PO.22.00040
Santoni-Rugiu E, Melchior LC, Urbanska EM, Jakobsen JN, Stricker K, Grauslund M, Sørensen JB (2019) Intrinsic resistance to EGFR-tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: differences and similarities with acquired resistance. Cancers (Basel) 11(7):923. https://doi.org/10.3390/cancers11070923
Teishikata T, Shiraishi K, Shinno Y, Kobayashi Y, Kashima J, Ishiyama T, Yoshida T, Mori T, Yatabe Y (2021) An alert to possible false positives with a commercial assay for MET exon 14 skipping. J Thorac Oncol 16(12):2133–2138. https://doi.org/10.1016/j.jtho.2021.07.028
Acknowledgements
The authors thank the skillful lab technicians and data analysts for their valuable contribution to this paper, and Luc Geeraert (Bench to Pen-Scientific Writing) for his writing support.
Funding
Open access funding provided by Copenhagen University
Author information
Authors and Affiliations
Contributions
M.L., Hi.A., H.P., B.C., Co.A., M.-D.S., V.P., W.E., P.M., S.S., Ca.A., M.P., S.R., and Ha.A. took part in the development of the methodology, in the choice of the samples, in the technique and interpretation of the various assays carried out on these samples, in the interpretation of the data, and in proofreading and correction of the final paper. M.L and Ha.A. wrote the paper. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Patients provided informed consent. The use of these patient samples was approved by the respective local Ethics Committees and was in accordance with the Declaration of Helsinki.
Conflict of interest
Bontoux Christophe: nothing to declare
Cayre Anne: nothing to declare
Concha Angel: nothing to declare
Hartmann Arndt: honoraria for lectures or consulting/advisory boards for Abbvie, Agilent, AstraZeneca, Biocartis, BMS, Boehringer Ingelheim, Cepheid, Diaceutics, Gilead, Illumina, Ipsen, Janssen, Lilly, Merck, MSD, Nanostring, Novartis, Pfizer, Qiagen, QUIP GmbH, Roche, Sanofi, 3DHistotech and other research support from AstraZeneca, Biocartis, Cepheid, Gilead, Illumina, Janssen, Nanostring, Novartis, Owkin, Qiagen, QUIP GmbH, Roche, Sanofi
Hirschmann Astrid: nothing to declare
Hofman Paul: member of the Biocartis advisory board
Martin Paloma: nothing to declare
Melchior Linea: nothing to declare
Mrabet-Dahbi Salima: nothing to declare
Putzová Martina: nothing to declare
Scarpino Stefania: nothing to declare
Stoehr Robert: nothing to declare
Vannuffel Pascal: nothing to declare
Watkin Emmanuel: honoraria for lectures or consulting/advisory boards for AstraZeneca, Janssen, MSD, Incyte, Pierre Fabre
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The presentation of authors' given and family names has been corrected.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Melchior, L., Hirschmann, A., Hofman, P. et al. Multicenter evaluation of an automated, multiplex, RNA-based molecular assay for detection of ALK, ROS1, RET fusions and MET exon 14 skipping in NSCLC. Virchows Arch 484, 677–686 (2024). https://doi.org/10.1007/s00428-024-03778-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-024-03778-9