Abstract
Main conclusion
The entire process of embryo development is under the tight control of various transcription factors. Together with other proteins, they act in a combinatorial manner and control distinct events during embryo development.
Abstract
Seed development is a complex process that proceeds through sequences of events regulated by the interplay of various genes, prominent among them being the transcription factors (TFs). The members of WOX, HD-ZIP III, ARF, and CUC families have a preferential role in embryonic patterning. While WOX TFs are required for initiating body axis, HD-ZIP III TFs and CUCs establish bilateral symmetry and SAM. And ARF5 performs a major role during embryonic root, ground tissue, and vasculature development. TFs such as LEC1, ABI3, FUS3, and LEC2 (LAFL) are considered the master regulators of seed maturation. Furthermore, several new TFs involved in seed storage reserves and dormancy have been identified in the last few years. Their association with those master regulators has been established in the model plant Arabidopsis. Also, using chromatin immunoprecipitation (ChIP) assay coupled with transcriptomics, genome-wide target genes of these master regulators have recently been proposed. Many seed-specific genes, including those encoding oleosins and albumins, have appeared as the direct target of LAFL. Also, several other TFs act downstream of LAFL TFs and perform their function during maturation. In this review, the function of different TFs in different phases of early embryogenesis and maturation is discussed in detail, including information about their genetic and molecular interactors and target genes. Such knowledge can further be leveraged to understand and manipulate the regulatory mechanisms involved in seed development. In addition, the genomics approaches and their utilization to identify TFs aiming to study embryo development are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In most angiosperms, the seed is an outcome of a double fertilization process, in which one of the two sperm nuclei fuses with the egg cell to produce a diploid zygote, and the second sperm nucleus fuses with the binucleate central cell to generate the triploid endosperm (Baroux and Grossniklaus 2019). Subsequently, the single cellular zygote undergoes highly coordinated cell divisions and cellular differentiation to develop into a multicellular embryo in a process termed embryogenesis (Verma et al. 2021). In many dicots, including Arabidopsis, the endosperm develops as a coenocyte (series of mitosis without cytokinesis) followed by endosperm cellularization (Fig. 1; Brown et al. 1999; Li and Berger 2012; Batista et al. 2019). Later, in Arabidopsis, the endosperm is absorbed in part by the growing embryo. The seed coat is developed from the ovule’s integuments and consists of five cell layers. Two cell layers are derived from the outer integuments (OI): OI2 and OI1, and three cell layers are derived from the inner integuments (II): II2, II1′ and II1 (Fig. 1). During the development of the seed, cells of the outermost layer, i.e., OI2, produce mucilage which accumulates specifically at the outer corners of the cell. Besides, both outer integument cell layers (OI1 and OI2) produce starch granules, flavanols, and suberin (a lipophilic polymer) (Golz et al. 2018; Francoz et al. 2018). In contrast, cells of the innermost layer accumulate proanthocyanidins (PAs) which later oxidize and give the characteristic brown colour to the seed coat. In the end, the mature seed contains a filial embryo, a single-cell endosperm layer, and a protecting covering, i.e., a seed coat developed from the ovule's integuments.
Overall depiction of Arabidopsis seed development. The development of three components of the seed (seed coat, endosperm, and embryo) is illustrated. All five layers of the seed coat derived from ovules integuments are shown in different colors. Polymers accumulated in the seed coat layers are shown. Endosperm development is defined by the syncytial stage, followed by cellularization and absorption. Differentiation of the three mitotic domains of the endosperm (micropylar, peripheral, and chalazal) is depicted with nuclei in different colors in the globular stage. Embryo development stages are shown from one-cell to mature stages. At the top of the figure, bars represent the major events that occur during embryo and endosperm development. MEN, micropylar endosperm nuclei; PEN, peripheral endosperm nuclei; CEN, chalazal endosperm nuclei
Overall, embryo development can be classified into two distinct phases, (i) morphogenesis and (ii) maturation (Fig. 1; Goldberg et al. 1994; O'Neill et al. 2019; Jo et al. 2019). The morphogenesis phase begins immediately after fertilization and lasts until the late-heart stage. This phase is characterized by highly coordinated cell divisions and differentiation, during which the basic body organization of the embryo is established. By contrast, cell division and proliferation are ceased during embryo maturation. However, cell expansion occurs along with the accumulation of seed storage reserves such as carbohydrates, proteins, and fatty acids (Jo et al. 2019). Embryo maturation partially overlaps with the morphogenesis phase. It begins around the early-heart stage and lasts until the seed is filled with nutrients, becomes dry, and acquires a dormant state (e.g., seed maturation). It has been observed that the developmental stage, not the time elapsed since fertilization, determines the onset of maturation (O'Neill et al. 2019).
Seed development depends on the spatiotemporal expression of various genes involved in different processes occurring during this period, including cell division, differentiation, seed filling, desiccation, and dormancy (Le et al. 2010). Therefore, control of the spatiotemporal pattern and expression levels of such genes is crucial, which results from the regulation of their transcription by different transcription factors (TFs). TFs are critical regulatory proteins that act in a combinatorial manner together with other proteins to orchestrate this transcriptional regulation (Agarwal et al. 2011). They work through binding to specific DNA sequences (cis-regulatory elements) present over the promoters of their target genes (Spitz and Furlong 2012). These regulatory proteins are themselves regulated by other TFs, miRNAs, hormones, etc., thereby creating multiple layers and networks of regulation to control a particular developmental aspect. Over the last few years, several TFs regulating seed and/or embryo development have been identified using genetics and genomics approaches in different plant species (Wu et al. 2020; Ren et al. 2021; Hofmann et al. 2019). However, the mechanisms through which they perform their function, including their interacting partners, target promoters, etc., are still not clear for most of them.
This review aims to enhance our understanding of transcriptional regulation of the distinct events of embryo morphogenesis and maturation by different classes of TFs in Arabidopsis seed development. Additionally, the identification of novel TFs using genome-wide approaches will be discussed, and how this will improve future research in studying the transcriptional landscape of seed development.
Transcriptional control of early embryogenesis (morphogenesis)
Embryogenesis begins when the zygote undergoes an asymmetric division that generates a small apical cell and a large basal cell (Capron et al. 2009). The apical cell gives rise to the spherical embryo proper (proembryo) that will create most of the mature embryo. Two rounds of longitudinal and one round of transverse divisions convert the apical cell into an eight-celled embryo. In this stage, two domains, termed the upper tier and lower tier, are distinguished. The basal cell divides transversely and gives rise to a 7–9 celled filamentous suspensor. Later, the uppermost suspensor cell is specified as hypophysis, which ultimately protrudes into the embryo. The upper tier gives rise to the cotyledons and shoot apical meristem (SAM). The lower tier and hypophysis generate the cotyledons' abaxial part, hypocotyl, root apical meristem (RAM), and embryonic root. After a periclinal division, a 16-celled embryo results from the first visible cell differentiation when the outermost layer is specified as protoderm. Subsequent divisions give the embryo a globular appearance. At this stage, precursor cells of ground tissue and vascular tissue are specified. Later, with the development of cotyledon primordia, the embryo takes the shape of a heart. At this stage, SAM is established, cotyledons are formed, and they begin to elongate, marking the beginning of the maturation phase.
Transcription factors for initiating body axis
The first asymmetric division of the zygote lays the foundation for apical-basal patterning in the embryo. Members of the WUSCHEL-RELATED HOMEOBOX (WOX) TFs play key roles during embryo patterning. Different WOX genes exhibit distinct expression patterns throughout embryogenesis. For instance, WOX8/STIMPY is expressed in the zygote, where another TF, i.e., WRKY2, regulates its expression. This WRKY2-WOX8 interaction is required for the polar distribution of the cell organelles in the zygote, creating the zygotic polarity required to make the first division asymmetric (Fig. 2A; Ueda et al. 2011). WOX2, co-expressed with WOX8 in the zygote, expresses in the apical cell after the asymmetric zygotic division. WOX2 is required to regulate the apical patterning and development of SAM during embryogenesis. In contrast, WOX8 is expressed in the basal cell along with its closest homolog WOX9/STIMPY-LIKE after the zygotic division, redundantly regulating the basal cell lineage (Fig. 2B). They also regulate the embryo proper by activating WOX2 expression in the apical region (Breuninger et al. 2008). A shift of WOX9 expression from hypophysis to the embryo proper's basal region indicates its function in establishing the lower tier domain identity (Haecker et al. 2004). Additionally, other WOX TF family members, such as WOX1, WOX3, and WOX5, expressed in cotyledon primordia, vascular primordia, and hypophysis, respectively, contribute to embryo patterning (Haecker et al. 2004; Breuninger et al. 2008).
Transcription factors involved in embryo patterning. A In the zygote, WRKY2 activates the expression of WOX8 to promote the polarization of the nucleus (brown) and vacuoles (light yellow) and asymmetric division of the zygote. B In the resulting asymmetric embryo, WRKY2-WOX8/9 non-cell-autonomously regulates the development of the embryo proper by activating WOX2 expression (green) in the apical region. C At the early globular stage, the WOX2 module (WOX1/2/3/5) is required for the initiation of SAM. The WOX2 module activates the expression of HD-ZIP III TFs to protect the shoot meristem stem cells from differentiation. The HD-ZIP III TFs are targeted by miRNA165/166. These TFs act with HD-ZIP II TFs to regulate apical patterning during embryogenesis. The uppermost suspensor cell is specified as hypophysis via auxin-dependent activation of TMO5 and TMO7 by MP/ARF5. D Other targets of ARF5 such as NTT, WIP4/5, and PLTs are required for asymmetric division of the hypophysis and correct specification of the QC at the late globular stage. E CUC TFs regulate the boundary formation between the developing cotyledons and act synergistically with STM to control SAM formation
Transcription factors in regulating apical patterning
Members of the class III homeodomain-leucine zipper (HD-ZIP III) TF family are known regulators of apical patterning during embryogenesis (Fig. 2C; Emery et al. 2003; Prigge et al. 2005; Smith and Long 2010). This family consists of five genes that encode PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), ARABIDOPSIS THALIANA HOMEOBOX-8 (ATHB8), and INCURVATA4/CORONA/CNA/ATHB15. All five have been predicted as the targets of miRNA165/166 (Smith and Long 2010; Floyd and Bowman 2004; Mallory et al. 2004). They perform an overlapping function in establishing bilateral symmetry and shoot apical meristem, as demonstrated by the phenotypes of their mutant combinations (Emery et al. 2003; Prigge et al. 2005). For instance, rev phb double mutant embryos occasionally display patterning defects such as single or radially symmetric cotyledon. Such defects are more severe when combined with phv and/or cna. However, athb8 mutation does not affect these patterning defects (Prigge et al. 2005). Besides, members of HD-ZIP II TFs, i.e., HOMEOBOX ARABIDOPSIS THALIANA 3 (HAT3), ARABIDOPSIS THALIANA HOMEOBOX 2 (ATHB2), and ATHB4, respond to changes in light conditions and regulate cotyledon development, thus the establishment of bilateral symmetry during embryogenesis (Turchi et al. 2013). In contrast to single mutants where defects are not obvious, hat3 athb4 double mutants display aberrantly developed cotyledons such as fused or single, and/or vasculature-less cotyledons. These defects are attenuated by a gain-of-function mutation in ATHB2, suggesting that ATHB2 is redundant to HAT3 and ATHB4. Moreover, most hat3 athb2 athb4 mutant seedlings lack an active SAM. The defects in cotyledon development and SAM activity are enhanced when hat3 athb4 is combined with mutations in HD-ZIP III genes. This indicates that HD-ZIP II and HD-ZIP III proteins act in the same genetic pathway to establish bilateral symmetry in the embryo and control SAM activity in seedlings. The direct interaction of REV on the ATHB2 promoter indicates that HD-ZIP III proteins control HD-ZIP II proteins' expression. These findings suggest that HD-ZIP II and HD-ZIP III TFs act in a combinatorial manner to regulate apical patterning during embryogenesis (Turchi et al. 2013).
Besides, PHB, PHV, and REV are thought to be involved in defining the adaxial fate of the embryo. Their expression in the adaxial regions of the cotyledons and vasculature in heart-stage embryos supports this notion (Emery et al. 2003; McConnell et al. 2001). Moreover, phb phv rev triple mutant embryos appear to be fully abaxialized (Emery et al. 2003). In addition, KANADI (KAN) genes that encode the members of the GARP TFs family are found to be involved in embryo patterning along the abaxial side of the cotyledons and hypocotyl (Eshed et al. 2004; Kerstetter et al. 2001; Izhaki and Bowman 2007). Their mutant combination (kan1 kan2 kan4) displays abnormal phenotypes in the region that will generate the abaxial side of the respective tissues. Further genetic analysis indicates that HD-ZIP III and KAN act antagonistically to establish abaxial-adaxial identity in the embryo (Izhaki and Bowman 2007; Emery et al. 2003). Furthermore, the YABBY (YAB) gene family members (FILAMENTOUS FLOWER (FIL), YAB2, YAB3) are expressed in the abaxial domain of cotyledons at the mid-heart stage indicating its role in specifying abaxial cell fate (Siegfried et al. 1999). Despite all this, a detailed mechanism of adaxial-abaxial polarity during embryogenesis is still lacking and needs to be addressed. AP2/ERF-type TFs, DORNRÖSCHEN (DRN), and its paralogues DORNRÖSCHEN-LIKE (DRNL) interact with HD-ZIP III TFs. They act redundantly to regulate embryonic cell patterning and cotyledon development (Chandler et al. 2007; Cole et al. 2009). DRN and DRNL exhibit overlapping expression patterns till the early-heart stage (Chandler et al. 2007). DRN is expressed throughout the embryo proper from the 2 to 16-cell stage. Later, it becomes restricted to the apical domain at the site of cotyledon development (globular stage), then to the cotyledon tips at the heart stage (Chandler et al. 2007; Cole et al. 2009). The drn and drn drnl mutants are also defective in the hypophysis division, which gives rise to the embryonic root meristem. However, this function appears inconsistent with their expression, suggesting their non-cell-autonomous action in embryonic root formation (Cole et al. 2009).
The transition from radial to bilateral symmetry appears when the two distinct cotyledon primordia arise from the apical part of the globular embryo. Boundary formation between the developing cotyledons is essential for the establishment of SAM which is regulated by a gene regulatory network involving NAC (NAM/ATAF1/ATAF2/CUC) family TFs, i.e., CUP-SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3 (Fig. 2D, E; Aida et al. 1999; Takada et al. 2001; Vroemen et al. 2003). CUC TFs act synergistically with another TF of class 1 KNOTTED1-LIKE HOMEOBOX (KNOX) TF family, SHOOT MERISTEMLESS (STM), to control SAM formation. Consistent with their function, CUC and STM genes are expressed in the central strip by the late globular or heart stage. Furthermore, the expression of the three CUC genes is differentially regulated by WOX2 and WOX8 TFs during the establishment of the cotyledon boundary (Lie et al. 2012). It has been shown that WOX2 collectively with its redundant paralogs (i.e., WOX1, WOX3, and WOX5) is essential for the initiation of shoot meristem stem cells (Fig. 2C; Zhang et al. 2017). This module protects stem cells from differentiation by activating the expression of HD-ZIP III TFs in the presumptive SAM region. In addition, the WOX2 module reduces auxin activity by downregulating PIN1 expression in that region. This reduced auxin activity contributes to the initiation of stem cells therein.
Transcription factors in embryonic root formation
The establishment of RAM is initiated with the specification of the extraembryonic/uppermost suspensor cell as hypophysis (Fig. 2C, D). It divides asymmetrically and generates the quiescent centre (QC), which functions in maintaining stem cells (Capron et al. 2009). The monopteros (mp) mutants that lack the activity of the gene encoding the ARF5 TF, fail to initiate root meristem (Berleth and Jurgens 1993; Hamann et al. 1999, 2002). ARF5 is a downstream component of the auxin response pathway. It forms a complex with AUX/IAA12 (BODENLOS). In response to auxin, ARF5 releases from this complex and regulates the expression of its target genes by binding to the auxin response element (AuxRE) containing the core sequence TGTCTC. Two genes encoding bHLH TFs, i.e., TARGET OF MONOPTEROS5 (TMO5) and TMO7, have been identified as the direct targets of ARF5/MP that function in MP-mediated embryonic root formation (Schlereth et al. 2010). Other targets of ARF5/MP such as the gene encoding the zinc finger TF, NO TRANSMITTING TRACT (NTT), and its paralogs WIP domain protein 4 (WIP4) and WIP5 are required for root meristem initiation (Crawford et al. 2015). AP2 domain-containing TFs PLETHORA1 (PLT1) and PLT2 are redundantly required for the correct specification of QC, leading to the maintenance of the stem cells in the root meristem (Aida et al. 2004). Their basal embryonic expression requires ARF5 and ARF7/NPH4. Besides, the TFs that belong to the GRAS family, i.e., SHORT-ROOT (SHR) and SCARECROW (SCR), are required for QC identity and stem cell maintenance (Aida et al. 2004). Both SCR and SHR have different expression domains in the embryo. SCR is expressed in the hypophysis and ground tissue precursors at the globular stage. Later, after the asymmetric division of the hypophysis, SCR is expressed in the QC and nearby ground tissue (Helariutta et al. 2000; Wysocka-Diller et al. 2000). In comparison, SHR is expressed in the precursors of the vasculature (stele) but not in the hypophysis and QC (Helariutta et al. 2000). Nevertheless, in contrast to its expression region, SHR proteins were found in the QC in the postembryonic root, suggesting that SHR proteins move from the stele to adjacent cells, including the QC (Nakajima et al. 2001). Furthermore, it was observed that SHR protein is required for SCR activity in determining QC fate. It indicates a non-cell-autonomous function of SHR, also in the embryo, promoting the expression of SCR in the presumptive QC to determine its fate and maintain the stem cell niche (Aida et al. 2004). Notably, the expression of SHR and SCR does not depend on the PLTs (Aida et al. 2004). Further genetic analysis suggests that PLTs and SHR/SCR independently specify QC and stem cell niche.
Transcriptional regulation of radial patterning
Radial patterning begins with a periclinal division that generates the protoderm, the outermost layer visible at the 16-celled embryo. TFs of the HD-ZIP class IV family, i.e., ARABIDOPSIS THALIANA MERISTEM LAYER1 (AtML1) and PROTODERMAL FACTOR2 (PDF2), are thought to control this cell differentiation. The process is marked by their expression as early as in the two-celled embryo, then later confined to the outermost layer in the subsequent developmental stages (Abe et al. 2003; Lu et al. 1996; Takada and Jürgens 2007; Iida et al. 2019). Whereas the single mutants have no apparent phenotypes, the double mutant (atml1-1 pdf2-1) embryos display defects in cell layer differentiation at the embryonic apex and a lack of epidermis in the leaves of their germinated seedlings (Abe et al. 2003). Moreover, the embryo development arrests around the globular stage in mutant combination with a strong allele of AtML1 (atml1-3) (Ogawa et al. 2015). It indicates that AtML1 and PDF2 act redundantly for protodermal cell differentiation during embryogenesis.
Vascular identity is specified in the 16-cell embryo (Smit et al. 2020). A multigenic regulatory network, including TF-encoding genes, controls vascular tissue specification (Table 1). Among them, ARF5/MP and its interacting partner GBF2, a G-class bZIP TF, regulate the expression of many vascular-specific genes to specify the vascular tissue identity. GBF2 modulates and/or stabilizes the ARF5 binding to its target promoters (Smit et al. 2020). Subsequently, vascular initial cells undergo periclinal divisions to increase the number of cell files. It has been shown that ARF5 is also required for vascular tissue establishment and maintenance (De Rybel et al. 2013). Its mutant embryos contain fewer cell files due to abnormal periclinal divisions. ARF5 performs its function through its direct target, TMO5, and its closest homolog, TMO5-LIKE1 (T5L1). Also, the heterodimer composed of TMO5 and another bHLH TF, LONESOME HIGHWAY (LHW), promotes periclinal divisions and establishes the vascular tissue in the embryo. In addition, the TMO5/LHW dimer controls the growth and patterning in the vascular tissue by regulating the expression of LONELY GUY 4 (LOG4), required for the final step of cytokinin biosynthesis (De Rybel et al. 2014). This TMO5/LHW-LOG4 module creates distinct cytokinin and auxin response domains for growth and patterning (xylem and cambium) of the vascular tissue during embryogenesis.
There has been significant research on ground tissue patterning and maintenance in the postembryonic tissues around the SHR network. However, the molecular mechanisms for ground tissue initiation in the early embryo are still ambiguous. It was observed that ARF5/MP initiates the first ground tissue cells and controls their earliest asymmetric division in the early embryo (Möller et al. 2017). This activity of ARF5 does not depend on the SHR network genes. Also, it was observed that the SHR network is not required for the initiation of embryonic ground tissue cells. However, ARF5 transcriptionally regulates the expression of SHR and SCR in the early embryo, which indicates an unresolved role of these well-known regulators of ground tissue patterning in the embryonic ground tissue cells initiation (Möller et al. 2017). All TFs involved in embryo morphogenesis and mentioned in this review are compiled in Table 1.
Transcriptional regulation of seed maturation
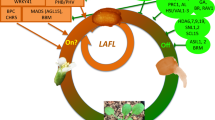
Seed maturation is a crucial phase of seed development as it ensures reproductive success by accumulating nourishment for the future seedling, helping to withstand desiccation, and promoting seed dispersal. In crop plants, including cereals and legumes, this phase is crucial for nutritional and economic purposes (Verma and Bhatia 2019; Gacek et al. 2018; Mathew et al. 2020). Maturation begins with the increase in the numbers of chloroplasts and chlorophyll accumulation (greening of the embryo), followed by the synthesis and accumulation of seed storage reserves (O'Neill et al. 2019; Baud et al. 2008). In oilseeds, like Arabidopsis, embryo greening appears necessary for photosynthesis and eventually accumulation of storage lipids (Mansfield and Briarty 1991). In Arabidopsis seeds, lipids as triacylglycerols (TAGs) and seed storage proteins (12S globulins and 2S albumins) are the major constituents of seed storage reserves (Baud et al. 2002, 2008). During late maturation, the embryo begins to lose water and chlorophyll (degreening) and simultaneously commences acquiring desiccation tolerance and dormancy (Leprince et al. 2017; Baud et al. 2008; Delmas et al. 2013). In Arabidopsis, the entire process of seed maturation has been shown under the tight control of several TFs that cooperate and act in a combinatorial manner (Fig. 3; Table 2; Boulard et al. 2017; Lepiniec et al. 2018; Jo et al. 2019).
Regulatory network involving TFs in the regulation of seed dormancy. The regulation of seed dormancy by different transcription factors is shown. Arrows and T-shaped lines represent activation and repression, respectively. Direct targets are shown by blue lines. Dotted arrows indicate indirect regulation by To et al. (2006). SPT targets are from Vaistij et al. (2013). Thick arrows denote the promotion of seed dormancy
LAFL transcription factors: the master regulators of maturation
To date, several TFs associated with the maturation phase have been identified and characterized in Arabidopsis. Among them, LAFL [LEAFY COTYLEDON1 (LEC1), ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2] TFs have been demonstrated to play central roles in most aspects of seed maturation, including accumulation of seed storage reserves, ability to withstand desiccation, and establishment of dormancy (Boulard et al. 2017; Lepiniec et al. 2018; Jo et al. 2019). They are required for the onset of embryo maturation as they, at least partially, regulate chloroplast development and subsequent embryo greening, which is the first visible sign of embryo maturation (O'Neill et al. 2019). ABI3, FUS3, and LEC2 (hereafter referred to as B3-AFL) are the members of the B3 domain-containing TFs, whereas LEC1 encodes the member of the NF-YB protein family (NF-YB9, HAP3 subunit of the CCAAT box-binding factors) (Braybrook and Harada 2008; Santos-Mendoza et al. 2008).
Mutations in LAFL genes revealed their partially overlapping functions during maturation (Meinke et al. 1994; West et al. 1994; Lotan et al. 1998; Nambara et al. 1995; Keith et al. 1994; Kagaya et al. 2005). However, they display some functional specificities. For instance, ABI3 controls chlorophyll degradation to mediate embryo degreening by regulating two functionally redundant genes, STAY-GREEN1 (SGR1) and SGR2 (Delmas et al. 2013). Similarly, LEC1 and LEC2 have a distinct role in initiating and maintaining embryonic fate. Their ectopic expression is sufficient to induce somatic embryogenesis in vegetative tissues. And their loss-of-function mutants exhibit trichomes and anthocyanin accumulation on cotyledon’s surface (Lotan et al. 1998; Stone et al. 2008, 2001). Recently, it has been observed that the expression of LEC1 in the embryo is not sufficient to initiate the maturation process. The endosperm-produced LEC1 protein, trafficked to the embryo through the suspensor, performs this function (Song et al. 2021). Moreover, detailed genetic analyses using complementation approach and mutant combinations of LAFL genes revealed the existence of a regulatory cascade with synergistic and/or redundant functions in parallel pathways (Bäumlein et al. 1994; Keith et al. 1994; Meinke et al. 1994; Parcy et al. 1997; To et al. 2006; Roscoe et al. 2015). Cross-regulation and feedback control of LAFL TFs have been observed. For instance, ABI3 and FUS3 regulate each other’s expression and can autoactivate themselves. Furthermore, their expression is positively regulated by LEC1 and LEC2 (To et al. 2006). One well-documented function of the LAFL network is to control protein and lipid accumulation during maturation (Roscoe et al. 2015; Baud et al. 2016). However, discrepancies in the level of control imposed by each LAFL on synthesis and accumulation of fatty acids and storage proteins have been observed (Baud et al. 2016; Roscoe et al. 2015; Kroj et al. 2003). For example, ABI3 exerts greater control over seed protein content, whereas FUS3 affects the lipid content to a greater extent (Roscoe et al. 2015). In another report, Baud et al. (2016) observed the strongest activation of an oleosin promoter fragment by ABI3 and the weakest by FUS3. Although LEC1 cannot activate the oleosin promoter alone, it enhances the synergistic activity of LEC2 and ABI3 on oleosin promoter expression. Oleosins are the structural proteins found on the surfaces of oil bodies of mature seeds (Frandsen et al. 2001).
Considerable efforts have been made to understand how LAFL genes control seed maturation. B3-AFL directly regulate maturation genes by binding to the RY element (CATGCA) and its variants found in many seed-specific promoters (Braybrook et al. 2006; Reidt et al. 2000; Kroj et al. 2003; Mönke et al. 2004; Baud et al. 2016; Sasnauskas et al. 2018). However, the binding specificity of the corresponding TFs depends on the flanking nucleotides of the core element (5′-CATG-3′) (Braybrook et al. 2006; Baud et al. 2016). Moreover, other motifs adjacent to the RY element are required to modulate this binding. In contrast, LEC1 does not exhibit specific DNA-binding activity. It interacts with NF-YA and NF-YC subunits to form the NF-Y complex, recognizing the CCAAT motif to regulate the transcription of its target genes (Dolfini et al. 2012; Baud et al. 2016). Nevertheless, an in vitro interaction of LEC1 with the OLEOSIN1 (OLE1) promoter has been observed (Baud et al. 2016).
Moreover, these TFs act in concert with other TFs in the regulation of maturation-specific genes. For instance, ABI3 cooperates with the members of bZIP TFs such as bZIP10, 25, and bZIP53 in regulating SSP (At2S) gene expression (Alonso et al. 2009; Lara et al. 2003). Although ABI3 does not interact with bZIP53, it forms a ternary complex with the heterodimers of bZIP53 and bZIP10 or 25 to enhance the maturation gene expression. Likewise, LEC1 and its paralogs LEC1-like (L1L) interact with an NF-YC subunit and bZIP67. This trimeric complex (LEC1/L1L-NFYC2-bZIP67) binds to ABRE/G-box elements through bZIP67 to activate the expression of genes involved in storage, such as CRUCIFERIN and FATTY ACID DESATURASE3 (FAD3), an enzyme involved in fatty acid biosynthesis (Mendes et al. 2013; Yamamoto et al. 2009).
Direct and indirect targets of LAFL TFs have been identified using genome-wide approaches, including microarray-based profiling and chromatin immunoprecipitation (ChIP) assay (Wang and Perry 2013; Mönke et al. 2012; Tian et al. 2020; Pelletier et al. 2017; Braybrook et al. 2006). Several maturation-specific genes were identified as the targets of LAFL. Among them, the genes encoding albumins and oleosins appear to be regulated by all four TFs (Braybrook et al. 2006; Tian et al. 2020; Wang and Perry 2013). As mentioned above, LAFL regulate each other in planta. For instance, genomic regions of ABI3, LEC1, and FUS3 are directly bound by FUS3 (Wang and Perry 2013). ABI3, LEC2, and FUS3 have been identified as the direct targets of the LEC1/NF-Y complex (Pelletier et al. 2017). These findings further corroborate the genetic interactions that exist between LAFL (To et al. 2006). Moreover, ChIP tilling array highlighted the abundant presence of RY elements and G-boxes (CACGTG) in the ABI3- and FUS3-bound regions. This suggests that G-box elements modulate the known combinatorial activity of ABI3 and bZIP TFs on RY elements. However, the interaction of FUS3 with bZIP TFs in the regulation of seed storage proteins (SSPs) is not yet identified. Likewise, apart from the LEC1-binding CCAAT motif, other elements such as RY, ABRE-like, and G-box-like are overrepresented in LEC1-bound regions (Pelletier et al. 2017). Together, these findings imply that the combinatorial function of LAFL with other TFs is mediated by cis-regulatory modules containing the clustered binding sites of different TFs and by physical interactions that are known to occur between them.
The strict control of LAFL expression is crucial during the transition from seed maturation to seed germination and seedling growth. HIGH-LEVEL EXPRESSION OF SUGAR INDUCIBLE GENE2 (HSI2)/VIVIPAROUS1 ABI3-LIKE1 (VAL1), HSI2-LIKE1 (HSL1)/VAL2, and VAL3 mediate repression of the LAFL network, thereby restraining seed maturation program after germination (Tsukagoshi et al. 2007; Suzuki et al. 2007; Veerappan et al. 2012). Recently, Chen et al. (2018) reported that HSI2/VAL1 indirectly controls LAFL expression through down-regulation of AGAMOUS-Like 15 (AGL15) during germination and seedling growth. Moreover, AGL15 is also regulated by LAFL TFs. For example, AGL15 is the direct target of LEC1 and FUS3, and its expression is induced by LEC2 (Braybrook et al. 2006).
Other transcription factors regulating synthesis and accumulation of seed storage reserves
The AP2 domain-containing TF, WRINKELED1 (WRI1), is a crucial regulator of triacylglyceride (TAG) biosynthesis (Kong et al. 2019). It directly regulates genes involved in fatty acid (FAs) biosynthesis by binding to an AW-box present in their 5` upstream region (UTR) (Maeo et al. 2009). Genetic and molecular analysis has revealed that WRI1 acts downstream to LEC1 and LEC2 (Baud et al. 2007; Mu et al. 2008). Previously, FUS3 has been shown to affect the expression of fatty acid biosynthetic genes, probably by activating WRI1 expression (Yamamoto et al. 2010). In recent reports, WRI1 has appeared as the direct target of LEC1 and FUS3 (Wang and Perry 2013; Pelletier et al. 2017). Also, ABI3 directly induces WRI1 and other fatty acid biosynthesis-related genes such as FAD3 and SSI2 (Tian et al. 2020). Another TF, MYB89, a member of the R2R3 MYB TF family, negatively regulates oil accumulation by directly repressing WRI1 and other essential genes involved in oil synthesis and accumulation during maturation (Li et al. 2017). Three members of the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) TF family, i.e., TCP4, TCP10, and TCP24, have been identified as the interacting proteins of WRI1 (Kong et al. 2020). However, only interaction between TCP4 and WRI1 negatively affects WRl1-mediated oil biosynthesis in the seed.
TRANSPARENT TESTA GLABRA1 (TTG1), a WD40 repeat protein, suppresses the accumulation of seed storage reserves by negatively affecting the expression of genes encoding SSPs and enzymes involved in FA biosynthesis (Chen et al. 2015). It also indirectly suppresses the expression of genes that encode the major regulators of maturation and enzymes for the synthesis and modification of FAs. Moreover, TTG1 acts downstream to FUS3, which negatively regulates TTG1 expression via direct binding. Similarly, TRANSPARENT TESTA8 (TT8), a bHLH TF, inhibits FA content by down-regulating the expression of genes involved in FA biosynthesis and directly affecting the expression of LEC1, LEC2, and FUS3 (Chen et al. 2014). Likewise, three bHLH TFs, MYC2, MYC3, and MYC4, were found to redundantly control SSPs’ accumulation and their relative proportion in the seed. The loss-of-function triple mutants (myc2 myc3 myc4) accumulate fewer 2S albumins but more cruciferins (Gao et al. 2016). Recently, the E2F TFs, E2FA and E2FB, have been identified to act as repressors on the cell cycle genes and the maturation genes such as LEC2 and WRI1 during the transition from proliferation to maturation (Leviczky et al. 2019). They also restrict the premature accumulation of storage proteins during early embryo development.
Transcriptional regulation of acquisition of desiccation tolerance and onset of dormancy
To survive extreme water loss, seeds need to develop desiccation tolerance (DT) during the mid-maturation phase. Like other seed maturation events, LAFL TFs are essential for seed DT. However, genetic evidence suggests that several other TFs act downstream of the LAFL network to control this process. González-Morales et al. (2016) identified genes that act downstream to the LAFL network to acquire DT using an integrated approach combining genetics, genomics, and metabolomics. The genes encoding three TFs, i.e., the plant AT-rich sequence and zinc-binding protein 1 (PLATZ1), PLATZ2, and AGL67, were shown to play a critical role in seed desiccation tolerance. Moreover, these TFs have been identified as the direct targets of ABI3 (Tian et al. 2020). In addition, many genes encoding Late Embryogenesis-Abundant (LEA) proteins that play a protective role during the acquisition of desiccation tolerance are directly induced by ABI3 (Tian et al. 2020; Kijak and Ratajczak 2020).
Primary seed dormancy (PD) is defined as the inability of a freshly matured seed to complete germination even under optimal conditions (Carrillo-Barral et al. 2020; Née et al. 2017). Seeds will germinate only after the release of dormancy by a period of dry storage at room temperature (after-ripening) or moist cold treatment (stratification) (Née et al. 2017). Some non-dormant seeds may experience a state of secondary dormancy upon exposure to unfavourable conditions such as very high or low temperatures or osmotic stress (Buijs 2020; Finkelstein et al. 2008). The network involving TFs in the establishment and control of seed dormancy is shown in Fig. 3.
The plant hormone abscisic acid (ABA) plays a central role in inducing and maintaining seed dormancy (Ali et al. 2021). Several mutants in which ABA signalling is attenuated display a reduced seed dormancy. Three TFs, including ABI3, ABI4, and ABI5, are the key downstream components of the seed-specific ABA response pathway. However, a loss-of-function mutation in ABI3 and ABI4, but not in ABI5, reduces seed dormancy (Finkelstein 1994; Shu et al. 2013). A WRKY domain-containing TF, WRKY41, has been found to regulate PD by directly regulating ABI3 expression in a maturing seed. However, its activity on ABI3 appears to be independent of ABA signalling (Ding et al. 2014). ABI4, a member of the AP2 domain family, positively regulates PD by maintaining the balance between ABA and gibberellic acid (GA) biogenesis (Shu et al. 2013). Moreover, auxin also controls seed dormancy via activation of ABI3 expression by two auxin-responsive TFs, ARF10 and ARF16 (Liu et al. 2013). MicroRNA160 post-transcriptionally regulates these two TFs. A feedback loop has been observed where ABI3 represses the expression of miR160, leading to increased transcripts of ARF10 and ARF16 that upregulate ABI3, resulting in seed dormancy (Liu et al. 2013; Tian et al. 2020). SPATULA (SPT), a bHLH TF, differentially regulates the establishment of seed dormancy in Arabidopsis ecotypes as Landsberg erecta (Ler) seeds become highly dormant due to lack of SPT, whereas Columbia (Col-0) seeds are less dormant. SPT represses the expression of a gene encoding MOTHER-OF-FT-AND-TFL1 (MFT) that promotes seed dormancy. It interferes with ABA and GA signalling by repressing ABI4 and REPRESSOR-OF-GA (RGA) expression, respectively. In contrast, it induces the expression of ABI5 and RGA-Like3 (RGL3) to control primary seed dormancy (Vaistij et al. 2013). Two bHLH TFs, ZHOUPI (ZOU) and INDUCER OF CBF EXPRESSION1 (ICE1) are preferentially expressed in the endosperm and play a role in determining the depth of PD (MacGregor and Zhang 2019). Lack of ICE1 and ZOU activity increases seed dormancy that is accompanied by increased ABA levels. Moreover, ICE1 directly represses the expression of ABI3, thereby modulating the LAFL network to regulate seed dormancy.
The gene that encodes DELAY OF GERMINATION (DOG1) is essential for seed dormancy in Arabidopsis (Nonogaki 2019; Carrillo-Barral et al. 2020). It acts in parallel but independently to the ABA signalling pathway. The amount of active DOG1 protein determines the after-ripening period of freshly harvested seeds. Thus, it may serve as a timer for seed dormancy release. It was found that the dog1-1 mutation can enhance the weak effects of the abi3-1 allele, thus revealing the genetic interaction between ABI3 and DOG1 (Dekkers et al. 2016). This raises the possibility of ABI3 being at the point of convergence for both pathways. Another TF, bZIP67, acts downstream to LEC1 and regulates the expression of DOG1 to establish primary seed dormancy (Bryant and Hughes 2019). ETHYLENE RESPONSE FACTOR12 (ERF12), a member of the AP2/ERF TF family, acts downstream of ETHYLENE RESPONSE1 (ETR1)/ REDUCED DORMANCY3 (RDO3) in the ethylene response pathway and negatively regulates seed dormancy by inhibiting the expression of DOG1 (Li et al. 2019). It physically interacts with TOPLESS (TPL), and this complex binds to the DEHYDRATION-RESPONSIVE ELEMENT (DRE)/C-repeat (CRT) motif (5′-RCCGAC-3′) present on the promoter of DOG1.
Methodologies used to identify TFs involved in embryo development
Known and new TFs from different (plant and animal) tissues and developmental stages are often identified by methods such as yeast one-hybrid library screening (Reece-Hoyes and Marian Walhout 2012), transcriptome analysis (Andrilenas et al. 2015; Kodama et al. 2018), DNA affinity purification followed by Mass Spectrometry (Tacheny et al. 2013) and protein arrays (Hu et al. 2009; Fig. 4). The newly identified DNA-binding proteins are scanned against known plant TF databases such as the “Plant TF database” to annotate and assign them into a TF family (Jin et al. 2017). The classification of TFs under a specific family depends on the presence of DNA binding domain (DBD) in their protein sequence that shows sequence homology to the previously characterized DBDs such as B3, NAC, C2H2, AP2, etc. These DBDs are catalogued in different databases such as InterPro (Blum et al. 2021), Pfam (Mistry et al. 2021), SMART (Letunic et al. 2021) as multiple sequence alignments, and hidden Markov models (HMM). Alternatively, these HMM profiles can be used to scan the protein dataset for the presence of a particular DBD, thereby classifying the proteins into a corresponding TF family. Indeed, transcriptome data provide valuable information on the gene expression pattern. Potential candidate TFs having differential expression can be identified by comparing transcriptomes of different tissues and/or different developmental stages. For Arabidopsis, such expression profiles can be found in the gene expression tool, ePlant (http://bar.utoronto.ca/eplant/). In addition, putative TFs regulating different sets of genes can be identified by performing co-expression analysis (Zogopoulos et al. 2021). These methods have successfully been utilized in the past decade to identify TFs involved in seed and/or embryo development in different plant species (Wu et al. 2020; Gu et al. 2020; Pradhan et al. 2014; Yi and Gu 2019). The identified TFs can be molecularly characterised using genomic approaches to identify binding promoters (e.g., ChIP assay) and to identify interacting proteins (e.g., yeast two-hybrid library screening, Mass spectrometry).
Among the methods mentioned above, transcriptomics has gained widespread interest in identifying gene regulatory networks, including TFs. However, most transcriptomics were performed utilizing whole or parts of embryos. It limits the understanding of transcriptome changes at the cellular level during early embryogenesis. To identify TFs for cell fate specification, the utilization of cell-type-specific transcriptome would be a better approach. Various methods such as laser capture microdissection (LCM), fluorescence-activated cell sorting (FACS), translating ribosome affinity purification (TRAP), and isolation of nuclei tagged in specific cell types (INTACT) are available to isolate specific cell types for transcriptome profiling (reviewed in Palovaara et al. 2013).
Although significant transcriptome profiling has been carried out to study seed/embryo development, only a few have focused exclusively on TFs required for the development of different stages and/or tissues of seeds. For example, Le et al. (2010) identified 48 seed-specific TFs using Affymetrix Gene Chips. These TF genes exhibit stage-specific expression, suggesting their active involvement in that particular stage. Similarly, 57 candidate TFs have been identified using RNA-seq data of developing embryos (Hofmann et al. 2019). In addition to known TF genes, some novel TFs involved in embryo morphogenesis were identified, such as storekeeper protein-related transcripts, MYB62, and REGULATOR OF AXILLARY MERISTEMS2 (RAX2) (Hofmann et al. 2019). Likewise, distinct TFs involved in the apical and basal cell lineages specification have been identified using the RNA-seq data of the apical and basal domains of the proembryo (Zhou et al. 2020). Differential expression profiling of the identified TFs resulted in 73 and 39 apical and basal cell lineage-maintained TFs, respectively. Moreover, putative TFs that may regulate the other lineage-specific genes were identified by co-expression analysis and by analysing the promoter sequences of lineage-specific genes for the presence of TF binding motifs. Recently, using single-nucleus mRNA-sequencing, Kao et al. (2021) identified many candidate TFs involved in the development of different cell types of early embryos.
Besides, a yeast-one hybrid library screening identifies a repertoire of TFs that can bind to the DNA region of interest. However, utilization of this method for genome-wide identification of TFs associated with seed development is very limited in Arabidopsis. Recently, Smit et al. (2020) identified potential TFs that may contribute to vascular identity during embryogenesis using a large-scale enhanced yeast one-hybrid assay. Among the most robust candidate TFs, the G-class bZIP TF G-box-binding factor 2 (GBF2) was identified as an interacting protein of MP/ARF5 in modulating vascular gene expression.
The identified TFs can be molecularly characterised using genome-wide approaches to identify binding promoters (e.g., ChIP assay) and interacting proteins (e.g., yeast two-hybrid library screening, Mass spectrometry). Furthermore, reverse genetics methods are widely used to establish the function of identified TFs, such as generating mutants and over-expression lines of newly identified genes, followed by phenotype analysis. Non-targeted gene mutations are achieved by chemical mutagenesis, transgene and transposon insertions. These methods are generally used to knock out a gene where the gene is completely deactivated. For Arabidopsis, such mutants may be available to researchers through seed stock centers. In the last few years, type II CRISPR/Cas (CRISPR/Cas9) has emerged as a powerful genome-editing tool for targeted gene mutations and knocking out the function of a gene (Liu et al. 2017). A modified CRISPR/Cas9 system can also be used for transcriptional activation and repression in plants (Piatek et al. 2015). Furthermore, RNA interference (RNAi) is a potential method for gene knockdown where an exogenous or endogenous small double-stranded RNA (dsRNA) interferes with target gene expression (Hung and Slotkin 2021). In addition, over-expression lines are utilized to analyze the effects of gain-of-function of studied genes. However, over-expression of a gene using constitutive promoters may harm the plant, resulting in seed/embryo abortion, lack of germination, or reduced seed set. To overcome such issues, tissue-specific (restricted) promoters and/or inducible gene expression systems can be used (Borghi 2010). For example, to achieve early-embryo-specific expression, WOX2 promoter has been used in Arabidopsis, whereas the promoters of seed storage proteins have been a major choice to study the maturation stage (Liao and Weijers 2018; Jeong et al. 2014).
Conclusions and perspectives
It is evident that TFs are crucial members of regulatory networks involved in many biological processes, including seed development. Therefore, the mechanisms by which these TFs exert their function, including interactions with other proteins and target promoters, will provide strategic insights toward understanding this complex developmental process. To date, several TFs involved in seed development have been identified using forward and reverse genetic analysis. Nevertheless, their molecular and genetic interactors remain to be explored. To achieve this goal, it is essential to utilize global approaches such as affinity purification followed by mass spectrometry and ChIP assays, especially using seed tissues. For example, in recent years, using ChIP coupled with transcriptomics, the target genes of many TFs began to emerge to a greater extent. Such techniques would be helpful to uncover several new genes that would aid in understanding and manipulating the regulatory mechanisms involved in seed development.
Although many TFs involved in maturation have been identified and characterized at the molecular and genetic levels, knowledge on the transcriptional regulation of early embryogenesis is still minimal. For instance, most studies are biased towards the ARF5 TF for lower tier domain fate. Some other TFs most probably exist in this process that need to be detected. Also, hormones such as auxin and ABA are required for early embryogenesis and maturation. Identifying TFs involved in their biosynthesis and transport would open the door to devising new strategies to control these processes of seed development. Moreover, the activity of TFs is tightly controlled by several genetic and epigenetic factors, the knowledge of which is still fragmented. Systemic analysis of such factors will aid in expanding regulatory networks and filling the knowledge gaps for better comprehension of seed development. Furthermore, to deepen our knowledge, cutting-edge approaches such as cell-type-specific transcriptomics are available to identify cell and/or tissue-specific TFs in developing embryos. Overall, this review provides a repository of TFs as potential candidates that can be functionally studied in crop plants to improve seed quality and agronomic practices.
Author contribution statement
SV reviewed the literature and wrote the manuscript. VPSA prepared the illustrations. HSR reviewed and edited the manuscript. All authors read and approved the manuscript.
Data availability
Data sharing does not apply to this article as no datasets were generated or analyzed during the current study.
References
Abe M, Katsumata H, Komeda Y, Takahashi T (2003) Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development (Cambridge, England) 130(4):635–643. https://doi.org/10.1242/dev.00292
Agarwal P, Kapoor S, Tyagi AK (2011) Transcription factors regulating the progression of monocot and dicot seed development. BioEssays 33(3):189–202. https://doi.org/10.1002/bies.201000107
Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh Y-S, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119(1):109–120. https://doi.org/10.1016/j.cell.2004.09.018
Aida M, Ishida T, Tasaka M (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development (Cambridge, England) 126(8):1563–1570
Ali F, Qanmber G, Li F, Wang Z (2021) Updated role of ABA in seed maturation, dormancy, and germination. J Adv Res. https://doi.org/10.1016/j.jare.2021.03.011
Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21(6):1747–1761. https://doi.org/10.1105/tpc.108.062968
Andrilenas KK, Penvose A, Siggers T (2015) Using protein-binding microarrays to study transcription factor specificity: homologs, isoforms and complexes. Brief Funct Genomics 14(1):17–29. https://doi.org/10.1093/bfgp/elu046
Baroux C, Grossniklaus U (2019) Seeds-An evolutionary innovation underlying reproductive success in flowering plants. Curr Top Dev Biol 131:605–642. https://doi.org/10.1016/bs.ctdb.2018.11.017
Batista RA, Moreno-Romero J, Qiu Y, van Boven J, Santos-González J, Figueiredo DD, Köhler C (2019) The MADS-box transcription factor PHERES1 controls imprinting in the endosperm by binding to domesticated transposons. eLife 8:e50541. doi:https://doi.org/10.7554/eLife.50541
Baud S, Boutin J-P, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40(2):151–160. https://doi.org/10.1016/S0981-9428(01)01350-X
Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. Arabidopsis Book 6:e0113–e0113. https://doi.org/10.1199/tab.0113
Baud S, Kelemen Z, Thévenin J (2016) Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiol 171(2):1099–1112. https://doi.org/10.1104/pp.16.00034
Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50(5):825–838. https://doi.org/10.1111/j.1365-313X.2007.03092.x
Bäumlein H, Miséra S, Luerssen H, Kölle K, Horstmann C, Wobus U, Müller A (1994) The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J 6:379–387
Berleth T, Jurgens G (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development (Cambridge, England) 118(2):575–587. https://doi.org/10.1242/dev.118.2.575
Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49(D1):D344–D354. https://doi.org/10.1093/nar/gkaa977
Borghi L (2010) Inducible gene expression systems for plants. Methods Mol Biol (Clifton, NJ) 655:65–75. https://doi.org/10.1007/978-1-60761-765-5_5
Boulard C, Fatihi A, Lepiniec L, Dubreucq B (2017) Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits. Biochim Biophys Acta 1860(10):1069–1078. https://doi.org/10.1016/j.bbagrm.2017.08.008
Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13(12):624–630. https://doi.org/10.1016/j.tplants.2008.09.008
Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103(9):3468–3473. https://doi.org/10.1073/pnas.0511331103
Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14(6):867–876. https://doi.org/10.1016/j.devcel.2008.03.008
Brown RC, Lemmon BE, Nguyen H, Olsen O-A (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12(1):32–42. https://doi.org/10.1007/s004970050169
Bryant FM, Hughes D (2019) Basic LEUCINE ZIPPER TRANSCRIPTION FACTOR67 transactivates DELAY OF GERMINATION1 to establish primary seed dormancy in Arabidopsis. Plant Cell 31(6):1276–1288. https://doi.org/10.1105/tpc.18.00892
Buijs G (2020) A perspective on secondary seed dormancy in Arabidopsis thaliana. Plants (Basel, Switzerland) 9(6):749. https://doi.org/10.3390/plants9060749
Capron A, Chatfield S, Provart N, Berleth T (2009) Embryogenesis: pattern formation from a single cell. Arabidopsis Book 7:e0126–e0126. https://doi.org/10.1199/tab.0126
Carrillo-Barral N, Rodríguez-Gacio MDC, Matilla AJ (2020) Delay of Germination-1 (DOG1): a key to understanding seed dormancy. Plants (Basel, Switzerland) 9(4):480. https://doi.org/10.3390/plants9040480
Chandler JW, Cole M, Flier A, Grewe B, Werr W (2007) The AP2 transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development (Cambridge, England) 134(9):1653–1662. https://doi.org/10.1242/dev.001016
Chen M, Xuan L, Wang Z, Zhou L, Li Z, Du X, Ali E, Zhang G, Jiang L (2014) TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol 165(2):905–916. https://doi.org/10.1104/pp.114.235507
Chen M, Zhang B, Li C, Kulaveerasingam H, Chew FT, Yu H (2015) TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol 169(1):391–402. https://doi.org/10.1104/pp.15.00943
Chen N, Veerappan V, Abdelmageed H, Kang M, Allen RD (2018) HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell 30(3):600–619. https://doi.org/10.1105/tpc.17.00655
Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W (2009) DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development (Cambridge, England) 136(10):1643–1651. https://doi.org/10.1242/dev.032177
Crawford BCW, Sewell J, Golembeski G, Roshan C, Long Jeff A, Yanofsky Martin F (2015) Genetic control of distal stem cell fate within root and embryonic meristems. Science 347(6222):655–659. https://doi.org/10.1126/science.aaa0196
De Rybel B, Adibi M, Breda Alice S, Wendrich Jos R, Smit Margot E, Novák O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J, Nijsse B, Boekschoten Mark V, Hooiveld G, Beeckman T, Wagner D, Ljung K, Fleck C, Weijers D (2014) Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345(6197):1255215. https://doi.org/10.1126/science.1255215
De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith Richard S, Borst Jan W, Weijers D (2013) A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 24(4):426–437. https://doi.org/10.1016/j.devcel.2012.12.013
Dekkers BJ, He H, Hanson J, Willems LA, Jamar DC, Cueff G, Rajjou L, Hilhorst HW, Bentsink L (2016) The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J 85(4):451–465. https://doi.org/10.1111/tpj.13118
Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, Northey JGB, McCourt P, Samuel MA (2013) ABI3 controls embryo degreening through Mendel’s locus. Proc Natl Acad Sci USA 110(40):E3888. https://doi.org/10.1073/pnas.1308114110
Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ (2014) WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J 79(5):810–823. https://doi.org/10.1111/tpj.12597
Dolfini D, Gatta R, Mantovani R (2012) NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol 47(1):29–49. https://doi.org/10.3109/10409238.2011.628970
Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by Class III HD-ZIP and KANADI genes. Curr Biol 13(20):1768–1774. https://doi.org/10.1016/j.cub.2003.09.035
Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsisare mediated by KANADI and YABBY activities. Development (Cambridge, England) 131(12):2997–3006. https://doi.org/10.1242/dev.01186
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415. https://doi.org/10.1146/annurev.arplant.59.032607.092740
Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5(6):765–771. https://doi.org/10.1046/j.1365-313X.1994.5060765.x
Floyd SK, Bowman JL (2004) Ancient microRNA target sequences in plants. Nature 428(6982):485–486. https://doi.org/10.1038/428485a
Francoz E, Lepiniec L, North HM (2018) Seed coats as an alternative molecular factory: thinking outside the box. Plant Reprod 31(3):327–342. https://doi.org/10.1007/s00497-018-0345-2
Frandsen GI, Mundy J, Tzen JTC (2001) Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 112(3):301–307. https://doi.org/10.1034/j.1399-3054.2001.1120301.x
Gacek K, Bartkowiak-Broda I, Batley J (2018) Genetic and molecular regulation of seed storage proteins (SSPs) to improve protein nutritional value of oilseed rape (Brassica napus L.) Seeds. Front Plant Sci 9:890. https://doi.org/10.3389/fpls.2018.00890
Gao C, Qi S, Liu K, Li D, Jin C, Li Z, Huang G, Hai J, Zhang M, Chen M (2016) MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiology and Biochemistry : PPB 108:63–70. https://doi.org/10.1016/j.plaphy.2016.07.004
Goldberg RB, de Paiva G, Yadegari R (1994) Plant Embryogenesis: Zygote to Seed. Science 266(5185):605–614. https://doi.org/10.1126/science.266.5185.605
Golz JF, Allen PJ, Li SF, Parish RW, Jayawardana NU, Bacic A, Doblin MS (2018) Layers of regulation – Insights into the role of transcription factors controlling mucilage production in the Arabidopsis seed coat. Plant Sci 272:179–192. https://doi.org/10.1016/j.plantsci.2018.04.021
González-Morales SI, Chávez-Montes RA, Hayano-Kanashiro C, Alejo-Jacuinde G, Rico-Cambron TY, de Folter S, Herrera-Estrella L (2016) Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 113(35):E5232-5241. https://doi.org/10.1073/pnas.1610985113
Gu W, Yu D, Guan Y, Wang H, Qin T, Sun P, Hu Y, Wei J, Zheng H (2020) The dynamic transcriptome of waxy maize (Zea mays L. sinensis Kulesh) during seed development. Genes Genomics 42(9):997–1010. https://doi.org/10.1007/s13258-020-00967-z
Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development (Cambridge, England) 131(3):657–668. https://doi.org/10.1242/dev.00963
Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16(13):1610–1615. https://doi.org/10.1101/gad.229402
Hamann T, Mayer U, Jürgens G (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development (Cambridge, England) 126(7):1387–1395
Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101(5):555–567. https://doi.org/10.1016/s0092-8674(00)80865-x
Hofmann F, Schon MA, Nodine MD (2019) The embryonic transcriptome of Arabidopsis thaliana. Plant Reprod 32(1):77–91. https://doi.org/10.1007/s00497-018-00357-2
Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho H-s, Woodard C, Wang H, Jeong J-S, Long S, He X, Wade H, Blackshaw S, Qian J, Zhu H (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139(3):610–622. https://doi.org/10.1016/j.cell.2009.08.037
Hung Y-H, Slotkin RK (2021) The initiation of RNA interference (RNAi) in plants. Curr Opin Plant Biol 61:102014. https://doi.org/10.1016/j.pbi.2021.102014
Iida H, Yoshida A, Takada S (2019) ATML1 activity is restricted to the outermost cells of the embryo through post-transcriptional repressions. Development (Cambridge, England) 146(4):dev169300. https://doi.org/10.1242/dev.169300
Izhaki A, Bowman JL (2007) KANADI and Class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19(2):495–508. https://doi.org/10.1105/tpc.106.047472
Jeong HJ, Choi JY, Shin HY, Bae JM, Shin JS (2014) Seed-specific expression of seven Arabidopsis promoters. Gene 553(1):17–23. https://doi.org/10.1016/j.gene.2014.09.051
Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45(D1):D1040–D1045. https://doi.org/10.1093/nar/gkw982
Jo L, Pelletier JM, Harada JJ (2019) Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J Integr Plant Biol 61(5):564–580. https://doi.org/10.1111/jipb.12806
Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T (2005) Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol 46(2):300–311. https://doi.org/10.1093/pcp/pci031
Kao P, Schon MA, Mosiolek M, Enugutti B, Nodine MD (2021) Gene expression variation in Arabidopsis embryos at single-nucleus resolution. Development (Cambridge, England) 148(13):dev199589. https://doi.org/10.1242/dev.199589
Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6(5):589–600. https://doi.org/10.1105/tpc.6.5.589
Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411(6838):706–709. https://doi.org/10.1038/35079629
Kijak H, Ratajczak E (2020) What Do We Know About the Genetic Basis of Seed Desiccation Tolerance and Longevity? International Journal of Molecular Sciences 21(10):3612. https://doi.org/10.3390/ijms21103612
Kodama M, Brinch-Pedersen H, Sharma S, Holme IB, Joernsgaard B, Dzhanfezova T, Amby DB, Vieira FG, Liu S, Gilbert MTP (2018) Identification of transcription factor genes involved in anthocyanin biosynthesis in carrot (Daucus carota L.) using RNA-Seq. BMC Genomics 19(1):811. https://doi.org/10.1186/s12864-018-5135-6
Kong Q, Singh SK, Mantyla JJ (2020) TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 Interacts with WRINKLED1 to mediate seed oil biosynthesis. Plant Physiol 184(2):658–665. https://doi.org/10.1104/pp.20.00547
Kong Q, Yuan L, Ma W (2019) WRINKLED1, a “Master Regulator” in transcriptional control of plant oil biosynthesis. Plant (Basel) 8(7):238. https://doi.org/10.3390/plants8070238
Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development (Cambridge, England) 130(24):6065–6073. https://doi.org/10.1242/dev.00814
Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278(23):21003–21011. https://doi.org/10.1074/jbc.M210538200
Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, Drews GN, Fischer RL, Okamuro JK, Harada JJ, Goldberg RB (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107(18):8063. https://doi.org/10.1073/pnas.1003530107
Lepiniec L, Devic M, Roscoe TJ, Bouyer D, Zhou DX, Boulard C, Baud S, Dubreucq B (2018) Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod 31(3):291–307. https://doi.org/10.1007/s00497-018-0337-2
Leprince O, Pellizzaro A, Berriri S, Buitink J (2017) Late seed maturation: drying without dying. J Exp Bot 68(4):827–841. https://doi.org/10.1093/jxb/erw363
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49(D1):D458–D460. https://doi.org/10.1093/nar/gkaa937
Leviczky T, Molnár E, Papdi C, Őszi E, Horváth GV, Vizler C, Nagy V, Pauk J, Bögre L, Magyar Z (2019) E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development 146(22):dev179333. https://doi.org/10.1242/dev.179333
Li D, Jin C, Duan S, Zhu Y, Qi S, Liu K, Gao C, Ma H, Zhang M, Liao Y, Chen M (2017) MYB89 transcription factor represses seed oil accumulation. Plant Physiol 173(2):1211–1225. https://doi.org/10.1104/pp.16.01634
Li J, Berger F (2012) Endosperm: food for humankind and fodder for scientific discoveries. New Phytol 195(2):290–305. https://doi.org/10.1111/j.1469-8137.2012.04182.x
Li X, Chen T, Li Y, Wang Z, Cao H, Chen F, Li Y, Soppe WJJ, Li W, Liu Y (2019) ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 31(4):832–847. https://doi.org/10.1105/tpc.18.00449
Liao C-Y, Weijers D (2018) A toolkit for studying cellular reorganization during early embryogenesis in Arabidopsis thaliana. Plant J 93(6):963–976. https://doi.org/10.1111/tpj.13841
Lie C, Kelsom C, Wu X (2012) WOX2 and STIMPY-LIKE/WOX8 promote cotyledon boundary formation in Arabidopsis. Plant J 72(4):674–682. https://doi.org/10.1111/j.1365-313X.2012.05113.x
Liu X, Wu S, Xu J, Sui C, Wei J (2017) Application of CRISPR/Cas9 in plant biology. Acta Pharm Sin B 7(3):292–302. https://doi.org/10.1016/j.apsb.2017.01.002
Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang H-Q, Luan S, Li J, He Z-H (2013) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110(38):15485. https://doi.org/10.1073/pnas.1304651110
Lotan T, Ohto M-a, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93(7):1195–1205. https://doi.org/10.1016/S0092-8674(00)81463-4
Lu P, Porat R, Nadeau JA, O’Neill SD (1996) Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8(12):2155–2168. https://doi.org/10.1105/tpc.8.12.2155
MacGregor DR, Zhang N (2019) ICE1 and ZOU determine the depth of primary seed dormancy in Arabidopsis independently of their role in endosperm development. Plant J 98(2):277–290. https://doi.org/10.1111/tpj.14211
Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60(3):476–487. https://doi.org/10.1111/j.1365-313X.2009.03967.x
Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5’ region. EMBO J 23(16):3356–3364. https://doi.org/10.1038/sj.emboj.7600340
Mansfield SG, Briarty LG (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69(3):461–476. https://doi.org/10.1139/b91-063
Mathew IE, Priyadarshini R, Mahto A, Jaiswal P, Parida SK, Agarwal P (2020) SUPER STARCHY1/ONAC025 participates in rice grain filling. Plant Direct 4(9):e00249. https://doi.org/10.1002/pld3.249
McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411(6838):709–713. https://doi.org/10.1038/35079635
Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6(8):1049–1064. https://doi.org/10.1105/tpc.6.8.1049
Mendes A, Kelly AA, van Erp H, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013) bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating FATTY ACID DESATURASE3. Plant Cell 25(8):3104–3116. https://doi.org/10.1105/tpc.113.116343
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar Gustavo A, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A (2021) Pfam: The protein families database in 2021. Nucleic Acids Res 49(D1):D412–D419. https://doi.org/10.1093/nar/gkaa913
Möller BK, ten Hove CA, Xiang D, Williams N, López LG, Yoshida S, Smit M, Datla R, Weijers D (2017) Auxin response cell-autonomously controls ground tissue initiation in the early <em>Arabidopsis</em> embryo. Proc Natl Acad Sci USA 114(12):E2533. https://doi.org/10.1073/pnas.1616493114
Mönke G, Altschmied L, Tewes A, Reidt W, Mock H-P, Bäumlein H, Conrad U (2004) Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219(1):158–166. https://doi.org/10.1007/s00425-004-1206-9
Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, Altschmied L (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40(17):8240–8254. https://doi.org/10.1093/nar/gks594
Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang X-J, Zuo J (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148(2):1042–1054. https://doi.org/10.1104/pp.108.126342
Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413(6853):307–311. https://doi.org/10.1038/35095061
Nambara E, Nambara E, McCourt P, Naito S (1995) A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development (Cambridge, England) 121(3):629–636. https://doi.org/10.1242/dev.121.3.629
Née G, Xiang Y, Soppe WJJ (2017) The release of dormancy, a wake-up call for seeds to germinate. Curr Opin Plant Biol 35:8–14. https://doi.org/10.1016/j.pbi.2016.09.002
Nonogaki H (2019) Seed germination and dormancy: The classic story, new puzzles, and evolution. J Integr Plant Biol 61(5):541–563. https://doi.org/10.1111/jipb.12762
O’Neill JP, Colon KT, Jenik PD (2019) The onset of embryo maturation in Arabidopsis is determined by its developmental stage and does not depend on endosperm cellularization. Plant J 99(2):286–301. https://doi.org/10.1111/tpj.14324
Ogawa E, Yamada Y, Sezaki N, Kosaka S, Kondo H, Kamata N, Abe M, Komeda Y, Takahashi T (2015) ATML1 and PDF2 play a redundant and essential role in Arabidopsis embryo development. Plant Cell Physiol 56(6):1183–1192. https://doi.org/10.1093/pcp/pcv045
Palovaara J, Saiga S, Weijers D (2013) Transcriptomics approaches in the early Arabidopsis embryo. Trends Plant Sci 18(9):514–521. https://doi.org/10.1016/j.tplants.2013.04.011
Parcy F, Valon C, Kohara A, Miséra S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9(8):1265–1277. https://doi.org/10.1105/tpc.9.8.1265
Pelletier JM, Kwong RW, Park S, Le BH, Baden R, Cagliari A, Hashimoto M, Munoz MD, Fischer RL, Goldberg RB, Harada JJ (2017) LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc Natl Acad Sci USA 114(32):E6710. https://doi.org/10.1073/pnas.1707957114
Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2015) RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13(4):578–589. https://doi.org/10.1111/pbi.12284
Pradhan S, Bandhiwal N, Shah N, Kant C, Gaur R, Bhatia S (2014) Global transcriptome analysis of developing chickpea (Cicer arietinum L.) seeds. Front Plant Sci 5:698–698. https://doi.org/10.3389/fpls.2014.00698
Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17(1):61–76. https://doi.org/10.1105/tpc.104.026161
Reece-Hoyes JS, Marian Walhout AJ (2012) Yeast one-hybrid assays: a historical and technical perspective. Methods (San Diego, Calif) 57(4):441–447. https://doi.org/10.1016/j.ymeth.2012.07.027
Reidt W, Wohlfarth T, Ellerström M, Czihal A, Tewes A, Ezcurra I, Rask L, Bäumlein H (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21(5):401–408. https://doi.org/10.1046/j.1365-313x.2000.00686.x
Ren Y, Huang Z, Jiang H, Wang Z, Wu F, Xiong Y, Yao J (2021) A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J Exp Bot 72(8):2947–2964. https://doi.org/10.1093/jxb/erab027
Roscoe TT, Guilleminot J, Bessoule J-J, Berger F, Devic M (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol 56(6):1215–1228. https://doi.org/10.1093/pcp/pcv049
Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54(4):608–620. https://doi.org/10.1111/j.1365-313X.2008.03461.x
Sasnauskas G, Manakova E, Lapėnas K, Kauneckaitė K, Siksnys V (2018) DNA recognition by Arabidopsis transcription factors ABI3 and NGA1. FEBS J 285(21):4041–4059. https://doi.org/10.1111/febs.14649
Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464(7290):913–916. https://doi.org/10.1038/nature08836
Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q (2013) ABI4 Regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9(6):e1003577. https://doi.org/10.1371/journal.pgen.1003577
Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development (Cambridge, England) 126(18):4117–4128
Smit ME, Llavata-Peris CI, Roosjen M, van Beijnum H, Novikova D, Levitsky V, Sevilem I, Roszak P, Slane D, Jürgens G, Mironova V, Brady SM, Weijers D (2020) Specification and regulation of vascular tissue identity in the Arabidopsis embryo. Development (Cambridge, England) 147(8):dev186130. https://doi.org/10.1242/dev.186130
Smith ZR, Long JA (2010) Control of Arabidopsis apical–basal embryo polarity by antagonistic transcription factors. Nature 464(7287):423–426. https://doi.org/10.1038/nature08843
Song J, Xie X, Chen C, Shu J, Thapa RK, Nguyen V, Bian S, Kohalmi SE, Marsolais F, Zou J, Cui Y (2021) LEAFY COTYLEDON1 expression in the endosperm enables embryo maturation in Arabidopsis. Nat Commun 12(1):3963. https://doi.org/10.1038/s41467-021-24234-1
Spitz F, Furlong EEM (2012) Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13(9):613–626. https://doi.org/10.1038/nrg3207
Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc Natl Acad Sci USA 105(8):3151–3156. https://doi.org/10.1073/pnas.0712364105
Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98(20):11806–11811. https://doi.org/10.1073/pnas.201413498
Suzuki M, Wang HHY, McCarty DR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143(2):902–911. https://doi.org/10.1104/pp.106.092320
Tacheny A, Dieu M, Arnould T, Renard P (2013) Mass spectrometry-based identification of proteins interacting with nucleic acids. J Proteomics 94:89–109. https://doi.org/10.1016/j.jprot.2013.09.011
Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development (Cambridge, England) 128(7):1127–1135. https://doi.org/10.1242/dev.128.7.1127
Takada S, Jürgens G (2007) Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development (Cambridge, England) 134(6):1141–1150. https://doi.org/10.1242/dev.02803
Tian R, Wang F, Zheng Q, Niza V, Downie AB (2020) Direct and indirect targets of the arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J 103(5):1679–1694. https://doi.org/10.1111/tpj.14854
To A, Valon C, Savino G, Guilleminot J, Devic M, Jrm G, Fo P (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18(7):1642–1651. https://doi.org/10.1105/tpc.105.039925
Tsukagoshi H, Morikami A, Nakamura K (2007) Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA 104(7):2543–2547. https://doi.org/10.1073/pnas.0607940104
Turchi L, Carabelli M, Ruzza V, Possenti M, Sassi M, Peñalosa A, Sessa G, Salvi S, Forte V, Morelli G, Ruberti I (2013) Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development (Cambridge, England) 140(10):2118–2129. https://doi.org/10.1242/dev.092833
Ueda M, Zhang Z, Laux T (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20(2):264–270. https://doi.org/10.1016/j.devcel.2011.01.009
Vaistij FE, Gan Y, Penfield S, Gilday AD, Dave A, He Z, Josse EM, Choi G, Halliday KJ, Graham IA (2013) Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci USA 110(26):10866–10871. https://doi.org/10.1073/pnas.1301647110
Veerappan V, Wang J, Kang M, Lee J, Tang Y, Jha AK, Shi H, Palanivelu R, Allen RD (2012) A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to derepression of seed-specific gene expression. Planta 236(1):1–17. https://doi.org/10.1007/s00425-012-1630-1
Verma S, Attuluri VPS, Robert HS (2021) An essential function for auxin in embryo development. Cold Spring Harbor Perspect Biology. https://doi.org/10.1101/cshperspect.a039966
Verma S, Bhatia S (2019) Analysis of genes encoding seed storage proteins (SSPs) in chickpea (Cicer arietinum L.) reveals co-expressing transcription factors and a seed-specific promoter. Funct Integr Genomics 19(3):373–390. https://doi.org/10.1007/s10142-018-0650-8
Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15(7):1563–1577. https://doi.org/10.1105/tpc.012203
Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161(3):1251–1264. https://doi.org/10.1104/pp.112.212282
West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6(12):1731–1745. https://doi.org/10.1105/tpc.6.12.1731
Wu Q, Bai X, Wu X, Xiang D, Wan Y, Luo Y, Shi X, Li Q, Zhao J, Qin P, Yang X, Zhao G (2020) Transcriptome profiling identifies transcription factors and key homologs involved in seed dormancy and germination regulation of Chenopodium quinoa. Plant Physiol Biochem 151:443–456. https://doi.org/10.1016/j.plaphy.2020.03.050
Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN (2000) Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development (Cambridge, England) 127(3):595–603
Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58(5):843–856. https://doi.org/10.1111/j.1365-313X.2009.03817.x
Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol 51(12):2031–2046. https://doi.org/10.1093/pcp/pcq162
Yi F, Gu W (2019) High Temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31(5):974–992. https://doi.org/10.1105/tpc.18.00961
Zhang Z, Tucker E, Hermann M, Laux T (2017) A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev Cell 40(3):264-277.e264. https://doi.org/10.1016/j.devcel.2017.01.002
Zhou X, Liu Z, Shen K, Zhao P, Sun M-X (2020) Cell lineage-specific transcriptome analysis for interpreting cell fate specification of proembryos. Nat Commun 11(1):1366. https://doi.org/10.1038/s41467-020-15189-w
Zogopoulos VL, Saxami G, Malatras A, Angelopoulou A, Jen C-H, Duddy WJ, Daras G, Hatzopoulos P, Westhead DR, Michalopoulos I (2021) Arabidopsis Coexpression Tool: a tool for gene coexpression analysis in Arabidopsis thaliana. iScience 24(8):102848. doi:https://doi.org/10.1016/j.isci.2021.102848
Funding
SV is supported by the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 895295 (ITRABAE). This work was supported by the European Regional Development Fund-Project “Centre for Experimental Plant Biology” (No. CZ.02.1.01/0.0/0.0/16_019/0000738).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare relevant to the content of this article.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Verma, S., Attuluri, V.P.S. & Robert, H.S. Transcriptional control of Arabidopsis seed development. Planta 255, 90 (2022). https://doi.org/10.1007/s00425-022-03870-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03870-x