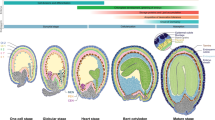
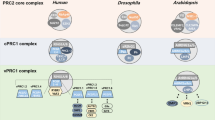
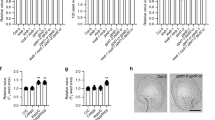
Abstract
The LAFL (i.e. LEC1, ABI3, FUS3, and LEC2) master transcriptional regulators interact to form different complexes that induce embryo development and maturation, and inhibit seed germination and vegetative growth in Arabidopsis. Orthologous genes involved in similar regulatory processes have been described in various angiosperms including important crop species. Consistent with a prominent role of the LAFL regulators in triggering and maintaining embryonic cell fate, their expression appears finely tuned in different tissues during seed development and tightly repressed in vegetative tissues by a surprisingly high number of genetic and epigenetic factors. Partial functional redundancies and intricate feedback regulations of the LAFL have hampered the elucidation of the underpinning molecular mechanisms. Nevertheless, genetic, genomic, cellular, molecular, and biochemical analyses implemented during the last years have greatly improved our knowledge of the LALF network. Here we summarize and discuss recent progress, together with current issues required to gain a comprehensive insight into the network, including the emerging function of LEC1 and possibly LEC2 as pioneer transcription factors.


Similar content being viewed by others
References
Abraham Z, Iglesias Fernandez R, Martinez M, Rubio-Somoza I, Diaz I, Carbonero P, Vicente-Carbajosa J (2016) A developmental switch of gene expression in the barley seed mediated by HvVP1 (Viviparous1) and HvGAMYB interactions. Plant Physiol 170:2146–2158
Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Kohler C (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet 5:e1000605
Aichinger E, Villar CB, Di Mambro R, Sabatini S, Kohler C (2011) The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23:1047–1060
Alonso R, Onate-Sanchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Droge-Laser W (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21:1747–1761
Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108:9709–9714
Baud S, Wuilleme S, To A, Rochat C, Lepiniec L (2009) Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J 60:933–947
Baud S, Kelemen Z, Thevenin J, Boulard C, Blanchet S, To A, Payre M, Berger N, Effroy-Cuzzi D, Franco-Zorrilla JM, Godoy M, Solano R, Thevenon E, Parcy F, Lepiniec L, Dubreucq B (2016) Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiol 171:1099–1112
Baumlein H, Misera S, Luerssen H, Kolle K, Horstmann C, Wobus U, Muller AJ (1994) The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J 6:379–387
Belluti S, Semeghini V, Basile V, Rigillo G, Salsi V, Genovese F, Dolfini D, Imbriano C (2018) An autoregulatory loop controls the expression of the transcription factor NF-Y. Biochim Biophys Acta 186:509–518
Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110:435–444
Berger N, Dubreucq B, Roudier F, Dubos C, Lepiniec L (2011) Transcriptional regulation of arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at Lysine-27. Plant Cell 23:4065–4078
Boulard C, Fatihi A, Lepiniec L, Dubreucq B (2017) Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits. Biochim Biophys Acta 1860:1069–1078
Boulard C, Thévenin J, Tranquet O, Laporte V, Lepiniec L, Dubreucq B (2018) LEC1 (NF-YB9) directly interacts with LEC2 to control gene expression in seed. Biochim Biophys Acta 1861:443–450
Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V, Schnittger A (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7:e1002014
Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, Colot V (2017) DNA methylation dynamics during early plant life. Genome Biol 18:179
Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, Ahnert S, Ohler U, Ware D, Walhout AJ, Benfey PN (2011) A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol 7:459
Bratzel F, Lopez-Torrejon G, Koch M, Del Pozo JC, Calonje M (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20:1853–1859
Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13:624–630
Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103:3468–3473
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ (2015) ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109:21–29
Cagliari A, Turchetto-Zolet AC, Korbes AP, Maraschin Fdos S, Margis R, Margis-Pinheiro M (2014) New insights on the evolution of Leafy cotyledon1 (LEC1) type genes in vascular plants. Genomics 103:380–387
Calonje M (2014) PRC1 marks the difference in plant PcG repression. Mol Plant 7:459–471
Carbonero P, Iglesias-Fernandez R, Vicente-Carbajosa J (2016) The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds. J Exp Bot 68:871–880
Carter B, Henderson JT, Svedin E, Fiers M, McCarthy K, Smith A, Guo C, Bishop B, Zhang H, Riksen T, Shockley A, Dilkes BP, Boutilier K, Ogas J (2016) Crosstalk between sporophyte and gametophyte generations is promoted by chd3 chromatin remodelers in Arabidopsis thaliana. Genetics 203:817–829
Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, Paz-Ares J, Onate-Sanchez L (2011) Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS ONE 6:e21524
Chan AC, Khan D, Girard IJ, Becker MG, Millar JL, Sytnik D, Belmonte MF (2016) Tissue-specific laser microdissection of the Brassica napus funiculus improves gene discovery and spatial identification of biological processes. J Exp Bot 67:3561–3571
Chan A, Carianopol C, Tsai AY, Varathanajah K, Chiu RS, Gazzarrini S (2017) SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J Exp Bot 68:4219–4231
Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon Y-H, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131:5263–5276
Chen D, Molitor A, Liu C, Shen WH (2010) The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res 20:1332–1344
Chen N, Veerappan V, Abdelmageed H, Kang M, Allen RD (2018) HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell 30:600–619
Cheng ZJ, Zhao XY, Shao XX, Wang F, Zhou C, Liu YG, Zhang Y, Zhang XS (2014) Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26:1053–1068
Chhun T, Chong SY, Park BS, Wong EC, Yin JL, Kim M, Chua NH (2016) HSI2 repressor recruits MED13 and HDA6 to down-regulate seed maturation gene expression directly during arabidopsis early seedling growth. Plant Cell Physiol 57:1689–1706
Chiu RS, Nahal H, Provart NJ, Gazzarrini S (2012) The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol 12:15
Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136:3660–3669
D’Ario M, Griffiths-Jones S, Kim M (2017) Small RNAs: big impact on plant development. Trends Plant Sci 22:1056–1068
Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, Northey JG, McCourt P, Samuel MA (2013) ABI3 controls embryo degreening through Mendel’s I locus. Proc Natl Acad Sci USA 110:3888–3894
Deng W, Buzas DM, Ying H, Robertson M, Taylor J, Peacock WJ, Dennis ES, Helliwell C (2013) Arabidopsis polycomb repressive complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genom 14:593
Derkacheva M, Hennig L (2014) Variations on a theme: polycomb group proteins in plants. J Exp Bot 65:2769–2784
Derkacheva M, Steinbach Y, Wildhaber T, Mozgova I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32:2073–2085
Devic M, Roscoe T (2016) Seed maturation: simplification of control networks in plants. Plant Sci 252:335–346
Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ (2014) WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J 79:810–823
Duong S, Vonapartis E, Li CY, Patel S, Gazzarrini S (2017) The E3 ligase ABI3-INTERACTING PROTEIN2 negatively regulates FUSCA3 and plays a role in cotyledon development in Arabidopsis thaliana. J Exp Bot 68:1555–1567
Ettaki H, Troncoso-Ponce MA, To A, Barthole G, Lepiniec L, Baud S (2018) Overexpression of MYB115, AAD2, or AAD3 in Arabidopsis thaliana seeds yields contrasting omega-7 contents. PLoS ONE 13:e0192156
Fatihi A, Boulard C, Bouyer D, Baud S, Dubreucq B, Lepiniec L (2016) Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Sci 250:198–204
Feeney M, Frigerio L, Cui Y, Menassa R (2013) Following vegetative to embryonic cellular changes in leaves of Arabidopsis overexpressing LEAFY COTYLEDON2. Plant Physiol 162:1881–1896
Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF (2014) Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J 80:654–668
Feng J, Chen D, Berr A, Shen WH (2016) ZRF1 chromatin regulators have polycomb silencing and independent roles in development. Plant Physiol 172:1746–1759
Gagete AP, Riera M, Franco L, Rodrigo MI (2009) Functional analysis of the isoforms of an ABI3-like factor of Pisum sativum generated by alternative splicing. J Exp Bot 60:1703–1714
Gaj M, Zhang S, Harada J, Lemaux P (2005) Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222:977–988
Gao MJ, Lydiate DJ, Li X, Lui H, Gjetvaj B, Hegedus DD, Rozwadowski K (2009) Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21:54–71
Gao Y, Liu J, Zhang Z, Sun X, Zhang N, Fan J, Niu X, Xiao F, Liu Y (2013) Functional characterization of two alternatively spliced transcripts of tomato ABSCISIC ACID INSENSITIVE3 (ABI3) gene. Plant Mol Biol 82:131–145
Gao MJ, Li X, Huang J, Gropp GM, Gjetvaj B, Lindsay DL, Wei S, Coutu C, Chen Z, Wan XC, Hannoufa A, Lydiate DJ, Gruber MY, Chen ZJ, Hegedus DD (2015) SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat Commun 6:7243
Gaudinier A, Zhang L, Reece-Hoyes JS, Taylor-Teeples M, Pu L, Liu Z, Breton G, Pruneda-Paz JL, Kim D, Kay SA, Walhout AJ, Ware D, Brady SM (2011) Enhanced Y1H assays for Arabidopsis. Nat Methods 8:1053–1055
Gaudinier A, Tang M, Bagman AM, Brady SM (2017) Identification of Protein-DNA interactions using enhanced yeast one-hybrid assays and a semiautomated approach. Methods Mol Biol 1610:187–215
Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7:373–385
Gil J, O’Loghlen A (2014) PRC1 complex diversity: where is it taking us? Trends Cell Biol 24:632–641
Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4:1251–1261
Gnesutta N, Saad D, Chaves-Sanjuan A, Mantovani R, Nardini M (2017) Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 family of NF-Y subunits in plants. Mol Plant 10:645–648
Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A (2002) Short Integuments1/suspensor1/carpel factory, a dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130:808–822
Gonzalez-Morales SI, Chavez-Montes RA, Hayano-Kanashiro C, Alejo-Jacuinde G, Rico-Cambron TY, de Folter S, Herrera-Estrella L (2016) Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 113:E5232–E5241
Grimault A, Gendrot G, Chaignon S, Gilard F, Tcherkez G, Thevenin J, Dubreucq B, Depege-Fargeix N, Rogowsky PM (2015) Role of B3 domain transcription factors of the AFL family in maize kernel filling. Plant Sci 236:116–125
Guo F, Liu C, Xia H, Bi Y, Zhao C, Zhao S, Hou L, Li F, Wang X (2013) Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS ONE 8:e71714
Gutzat R, Borghi L, Futterer J, Bischof S, Laizet Y, Hennig L, Feil R, Lunn J, Gruissem W (2011) Retinoblastoma-related protein controls the transition to autotrophic plant development. Development 138:2977–2986
Han SK, Wu MF, Cui S, Wagner D (2015) Roles and activities of chromatin remodeling ATPases in plants. Plant J 83:62–77
Harada JJ, Pelletier J (2012) Genome-wide analyses of gene activity during seed development. Seed Sci Res 22:S15–S22
Hecker A, Brand LH, Peter S, Simoncello N, Kilian J, Harter K, Gaudin V, Wanke D (2015) The Arabidopsis GAGA-binding factor basic pentacysteine6 recruits the polycomb-repressive complex1 component like heterochromatin protein1 to GAGA DNA Motifs. Plant Physiol 168:1013–1024
Henderson JT, Li H-C, Rider SD, Mordhorst AP, Romero-Severson J, Cheng J-C, Robey J, Sung ZR, de Vries SC, Ogas J (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134:995–1005
Ho KK, Zhang H, Golden BL, Ogas J (2013) PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim Biophys Acta 1829:199–210
Horstman A, Li M, Heidmann I, Weemen M, Chen B, Muino JM, Angenent GC, Boutilier K (2017) The baby boom transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol 175:848–857
Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H (2014) Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 5:4601
Huang M, Hu Y, Liu X, Li Y, Hou X (2015a) Arabidopsis leafy cotyledon1 mediates postembryonic development via interacting with phytochrome-interacting factor4. Plant Cell 27:3099–3111
Huang M, Hu Y, Liu X, Li Y, Hou X (2015b) Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front Plant Sci 6:955
Ikeuchi M, Iwase A, Rymen B, Harashima H, Shibata M, Ohnuma M, Breuer C, Morao AK, de Lucas M, De Veylder L, Goodrich J, Brady SM, Roudier F, Sugimoto K (2015) PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat Plants 1:15089
Iwase A, Mita K, Nonaka S, Ikeuchi M, Koizuka C, Ohnuma M, Ezura H, Imamura J, Sugimoto K (2015) WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J Plant Res 128:389–397
Jia H, McCarty DR, Suzuki M (2013) Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol 163:1293–1305
Jia H, Suzuki M, McCarty DR (2014) Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Interdiscip Rev Dev Biol 3:135–145
Jing Y, Zhang D, Wang X, Tang W, Wang W, Huai J, Xu G, Chen D, Li Y, Lin R (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25:242–256
Jr Dean Rider S, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35:33–43
Junker A, Baumlein H (2012) Multifunctionality of the LEC1 transcription factor during plant development. Plant Signal Behav 7:1718–1720
Junker A, Monke G, Rutten T, Keilwagen J, Seifert M, Thi TM, Renou JP, Balzergue S, Viehover P, Hahnel U, Ludwig-Muller J, Altschmied L, Conrad U, Weisshaar B, Baumlein H (2012) Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J 71:427–442
Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, Seo M (2010) Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol 51:1988–2001
Kawakatsu T, Nery JR, Castanon R, Ecker JR (2017) Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol 18:171
Kawashima T, Berger F (2014) Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet 15:613–624
Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: a heterochronic mutation affecting late embryo development in arabidopsis. Plant Cell 6:589–600
Kelsey G, Stegle O, Reik W (2017) Single-cell epigenomics: recording the past and predicting the future. Science 358:69–75
Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR (2012) EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet 8:e1002512
Kim D, Cho YH, Ryu H, Kim Y, Kim TH, Hwang I (2013) BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J 75:755–766
Kuwabara A, Gruissem W (2014) Arabidopsis retinoblastoma-related and Polycomb group proteins: cooperation during plant cell differentiation and development. J Exp Bot 65:2667–2676
Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-like defines a class of regulators essential for embryo development. Plant Cell 15:5–18
Lara P, Oñate-Sànchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278:21003–21011
Lee K, Seo PJ (2018) Dynamic epigenetic changes during plant regeneration. Trends Plant Sci 23:235–247
Lehti-Shiu MD, Panchy N, Wang P, Uygun S, Shiu SH (2017) Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochim Biophys Acta 1860:3–20
Li Y, Jin K, Zhu Z, Yang J (2010) Stepwise origin and functional diversification of the AFL subfamily B3 genes during land plant evolution. J Bioinform Comput Biol 8(Suppl 1):33–45
Libault M, Pingault L, Zogli P, Schiefelbein J (2017) Plant systems biology at the single-cell level. Trends Plant Sci 22:949–960
Lin JY, Le BH, Chen M, Henry KF, Hur J, Hsieh TF, Chen PY, Pelletier JM, Pellegrini M, Fischer RL, Harada JJ, Goldberg RB (2017) Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc Natl Acad Sci USA 114:E9730–E9739
Liu X, Hou X (2018) Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front Plant Sci 9:251
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007a) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52:133–146
Liu Y, Koornneef M, Soppe WJ (2007b) The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19:433–444
Liu J, Deng S, Wang H, Ye J, Wu HW, Sun HX, Chua NH (2016) CURLY LEAF regulates gene sets coordinating seed size and lipid biosynthesis in arabidopsis. Plant Physiol 171:424–436
Lopez-Molina L, Mongrand B, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32:317–328
Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205
Lu QS, Paz JD, Pathmanathan A, Chiu RS, Tsai AY, Gazzarrini S (2010) The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J 64:100–113
Luerssen H, Kirik V, Herrmann P, Misera S (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15:755–764
Magnani E, Jimenez-Gomez JM, Soubigou-Taconnat L, Lepiniec L, Fiume E (2017) Profiling the onset of somatic embryogenesis in Arabidopsis. BMC Genom 18:998
Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Kohler C (2006) Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7:947–952
Manan S, Ahmad MZ, Zhang G, Chen B, Haq BU, Yang J, Zhao J (2017) Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front Plant Sci 8:1604
Mayran A, Drouin J (2018) Pioneer transcription factors shape the epigenetic landscape. J Biol Chem. https://doi.org/10.1074/jbc.R117.001232
McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM, Lazzeri PA, Gale MD, Lenton JR, Holdsworth MJ (2002) Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc Natl Acad Sci USA 99:10203–10208
Mehdi S, Derkacheva M, Ramstrom M, Kralemann L, Bergquist J, Hennig L (2016) The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 28:42–54
Meinke DW (1992) A homeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258:1647–1650
Mendes A, Kelly AA, van Erp H, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013) bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25:3104–3116
Merini W, Calonje M (2015) PRC1 is taking the lead in PcG repression. Plant J 83:110–120
Merini W, Romero-Campero FJ, Gomez-Zambrano A, Zhou Y, Turck F, Calonje M (2017) The Arabidopsis polycomb repressive complex 1 (PRC1) components AtBMI1A, B, and C impact gene networks throughout all stages of plant development. Plant Physiol 173:627–641
Molitor AM, Bu Z, Yu Y, Shen WH (2014) Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet 10:e1004091
Monke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hahnel U, Junker A, Weisshaar B, Conrad U, Baumlein H, Altschmied L (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40:8240–8254
Mozgova I, Hennig L (2015) The polycomb group protein regulatory network. Annu Rev Plant Biol 66:269–296
Mozgova I, Kohler C, Hennig L (2015) Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. Plant J 83:121–132
Mozgova I, Munoz-Viana R, Hennig L (2017) PRC2 represses hormone-induced somatic embryogenesis in vegetative tissue of Arabidopsis thaliana. PLoS Genet 13:e1006562
Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, Zuo J (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148:1042–1054
Mu Y, Zou M, Sun X, He B, Xu X, Liu Y, Zhang L, Chi W (2017a) Basic pentacysteine proteins repress abscisic acid insensitive4 expression via direct recruitment of the polycomb-repressive complex 2 in Arabidopsis root development. Plant Cell Physiol 58:607–621
Mu Y, Liu Y, Bai L, Li S, He C, Yan Y, Yu X, Li Y (2017b) Cucumber CsBPCs regulate the expression of CsABI3 during seed germination. Front Plant Sci 8:459
Muller K, Bouyer D, Schnittger A, Kermode AR (2012) Evolutionarily conserved histone methylation dynamics during seed life-cycle transitions. PLoS ONE 7:e51532
Narro-Diego L, Lopez-Gonzalez L, Jarillo JA, Pineiro M (2017) The PHD-containing protein early bolting in short days regulates seed dormancy in Arabidopsis. Plant, Cell Environ 40:2393–2405
Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J (2017) Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol 18:172
Ng DW, Chandrasekharan MB, Hall TC (2004) The 5′ UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Mol Biol 54:25–38
Nodine MD, Bartel DP (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev 24:2678–2692
Nonogaki H (2017) Seed biology updates—highlights and new discoveries in seed dormancy and germination research. Front Plant Sci 8:524
Nowak K, Gaj MD (2016) Transcription factors in the regulation of somatic embryogenesis. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Somatic embryogenesis: fundamental aspects and applications. Springer, Cham, pp 53–79
Parcy F, Valon C, Kohara A, Misera S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9:1265–1277
Park W, Li J, Song R, Messing J, Chen X (2002) Carpel factory, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495
Pelletier JM, Kwong RW, Park S, Le BH, Baden R, Cagliari A, Hashimoto M, Munoz MD, Fischer RL, Goldberg RB, Harada JJ (2017) LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc Natl Acad Sci USA 114:E6710–E6719
Peng FY, Weselake RJ (2013) Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor Appl Genet 126:1305–1319
Questa JI, Song J, Geraldo N, An H, Dean C (2016) Arabidopsis transcriptional repressor VAL1 triggers polycomb silencing at FLC during vernalization. Science 353:485–488
Ramirez-Parra E, Gutierrez C (2007) The many faces of chromatin assembly factor 1. Trends Plant Sci 12:570–576
Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128:243–252
Ringrose L, Paro R (2007) Polycomb/trithorax response elements and epigenetic memory of cell identity. Development 134:223–232
Rodrigues AS, Miguel CM (2017) The pivotal role of small non-coding RNAs in the regulation of seed development. Plant Cell Rep 36:653–667
Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol 56:1215–1228
Ryu H, Cho H, Bae W, Hwang I (2014) Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun 5:4138
Sakai K, Taconnat L, Borrega N, Yansouni J, Brunaud V, Paysant-Le Roux C, Delannoy E, Martin Magniette ML, Lepiniec L, Faure JD, Balzergue S, Dubreucq B (2018) Combining laser-assisted microdissection (LAM) and RNA-seq allows to perform a comprehensive transcriptomic analysis of epidermal cells of Arabidopsis embryo. Plant Methods 14:10
Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579:4666–4670
Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54:608–620
Schneider A, Aghamirzaie D, Elmarakeby H, Poudel AN, Koo AJ, Heath LS, Grene R, Collakova E (2016) Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J. 85:305–319
Seefried WF, Willmann MR, Clausen RL, Jenik PD (2014) Global regulation of embryonic patterning in arabidopsis by microRNAs. Plant Physiol 165:670–687
Serivichyaswat P, Ryu HS, Kim W, Kim S, Chung KS, Kim JJ, Ahn JH (2015) Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol Cells 38:259–266
Shen Y, Devic M, Lepiniec L, Zhou DX (2015) Chromodomain, Helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol J 13:811–820
Sijacic P, Bajic M, McKinney EC, Meagher RB, Deal RB (2018) Chromatin accessibility changes between Arabidopsis stem cells and mesophyll cells illuminate cell type-specific transcription factor networks. Plant J 94:215–231
Sreenivasulu N, Wobus U (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol 64:189–217
Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu J-K (2018) UTR-dependent control of gene expression in plants. Trends Plant Sci 23:248–259
Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. PNAS 98:11806–11811
Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci USA 105:3151–3156
Sugliani M, Brambilla V, Clerkx EJ, Koornneef M, Soppe WJ (2010) The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell 22:1936–1946
Sun F, Liu X, Wei Q, Liu J, Yang T, Jia L, Wang Y, Yang G, He G (2017) Functional characterization of TaFUSCA3, a B3-superfamily transcription factor gene in the wheat. Front Plant Sci 8:1133
Suzuki M, McCarty DR (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11:548–553
Suzuki M, Wang HHY, McCarty DR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143:902–911
Swain S, Myers ZA, Siriwardana CL, Holt BF 3rd (2017) The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. Biochim Biophys Acta 1860:636–644
Swaminathan K, Peterson K, Jack T (2008) The plant B3 superfamily. Trends Plant Sci 13:647–655
Tan T, Sun Y, Peng X, Wu G, Bao F, He Y, Zhou H, Lin H (2017) ABSCISIC ACID INSENSITIVE3 is involved in cold response and freezing tolerance regulation in Physcomitrella patens. Front Plant Sci 8:1599
Tanaka M, Kikuchi A, Kamada H (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146:149–161
Tang X, Hou A, Babu M, Nguyen V, Hurtado L, Lu Q, Reyes JC, Wang A, Keller WA, Harada JJ, Tsang EW, Cui Y (2008) The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol 147:1143–1157
Tang X, Lim MH, Pelletier J, Tang M, Nguyen V, Keller WA, Tsang EW, Wang A, Rothstein SJ, Harada JJ, Cui Y (2012a) Synergistic repression of the embryonic programme by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J Exp Bot 63:1391–1404
Tang X, Bian S, Tang M, Lu Q, Li S, Liu X, Tian G, Nguyen V, Tsang EW, Wang A, Rothstein SJ, Chen X, Cui Y (2012b) MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet 8:e1003091
Tang LP, Zhou C, Wang SS, Yuan J, Zhang XS, Su YH (2017) FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol 213:1740–1754
Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y (2017) Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551:124–128
To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18:1642–1651
Trindade I, Schubert D, Gaudin V (2017) Epigenetic regulation of phase transitions in Arabidopsis thaliana. In: Rajewsky N, Jurga S, Barciszewski J (eds) Plant epigenetics. Springer, Cham, pp 359–383
Troncoso-Ponce MA, Barthole G, Tremblais G, To A, Miquel M, Lepiniec L, Baud S (2016) Transcriptional activation of two delta-9 palmitoyl-ACP desaturase genes by MYB115 and MYB118 is critical for biosynthesis of omega-7 monounsaturated fatty acids in the endosperm of Arabidopsis seeds. Plant Cell 28:2666–2682
Tsai AY, Gazzarrini S (2012) AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J 69:809–821
Tsuchiya Y, Nambara E, Naito S, McCourt P (2004) The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37:73–81
Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K (2005) Analysis of a sugar response mutant of arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol 138:675–685
Tsukagoshi H, Morikami A, Nakamura K (2007) Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA 104:2543–2547
Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3:e86
van Zanten M, Zoll C, Wang Z, Philipp C, Carles A, Li Y, Kornet NG, Liu Y, Soppe WJ (2014) HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J 80:475–488
Vashisht D, Nodine MD (2014) MicroRNA functions in plant embryos. Biochem Soc Trans 42:352–357
Veerappan V, Wang J, Kang M, Lee J, Tang Y, Jha AK, Shi H, Palanivelu R, Allen RD (2012) A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to derepression of seed-specific gene expression. Planta 236:1–17
Veerappan V, Chen N, Reichert AI, Allen RD (2014) HSI2/VAL1 PHD-like domain promotes H3K27 trimethylation to repress the expression of seed maturation genes and complex transgenes in Arabidopsis seedlings. BMC Plant Biol 14:293
Verdier J, Thompson RD (2008) Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol 49:1263–1271
Vicente-Carbajosa J, Carbonero P (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49:645–651
Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161:1251–1264
Wang H, Caruso LV, Downie AB, Perry SE (2004) The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16:1206–1219
Wang X, Niu QW, Teng C, Li C, Mu J, Chua NH, Zuo J (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19:224–235
Wang Y, Deng D, Zhang R, Wang S, Bian Y, Yin Z (2012) Systematic analysis of plant-specific B3 domain-containing proteins based on the genome resources of 11 sequenced species. Mol Biol Rep 39:6267–6282
Wang H, Liu C, Cheng J, Liu J, Zhang L, He C, Shen WH, Jin H, Xu L, Zhang Y (2016) Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet 12:e1005771
Wang Y, Zhang T, Song X, Zhang J, Dang Z, Pei X, Long Y (2018) Identification and functional analysis of two alternatively spliced transcripts of ABSCISIC ACID INSENSITIVE3 (ABI3) in linseed flax (Linum usitatissimum L.). PLoS ONE 13:e0191910
Willmann MR, Mehalick AJ, Packer RL, Jenik PD (2011) MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol 155:1871–1884
Wójcik AM, Nodine MD, Gaj MD (2017) miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in arabidopsis. Front Plant Sci 8:2024
Wójcikowska B, Gaj M (2015) LEAFY COTYLEDON2-mediated control of the endogenous hormone content: implications for the induction of somatic embryogenesis in Arabidopsis. Plant Cell. Tissue Organ Cult (PCTOC) 121:255–258
Wojcikowska B, Jaskola K, Gasiorek P, Meus M, Nowak K, Gaj MD (2013) LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 238:425–440
Xiao J, Wagner D (2015) Polycomb repression in the regulation of growth and development in Arabidopsis. Curr Opin Plant Biol 23:15–24
Xiao J, Jin R, Yu X, Shen M, Wagner JD, Pai A, Song C, Zhuang M, Klasfeld S, He C, Santos AM, Helliwell C, Pruneda-Paz JL, Kay SA, Lin X, Cui S, Garcia MF, Clarenz O, Goodrich J, Zhang X, Austin RS, Bonasio R, Wagner D (2017) Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat Genet 49:1546–1552
Xu Y, Guo C, Zhou B, Li C, Wang H, Zheng B, Ding H, Zhu Z, Peragine A, Cui Y, Poethig S, Wu G (2016) Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol 172:2416–2428
Xu F, Kuo T, Rosli Y, Liu MS, Wu L, Oliver Chen LF, Fletcher JC, Sung ZR, Pu L (2018) Trithorax group proteins act together with a Polycomb group protein to maintain chromatin integrity for epigenetic silencing during seed germination in Arabidopsis. Mol Plant 11:659–677
Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58:843–856
Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol 51:2031–2046
Yamamoto A, Yoshii M, Murase S, Fujita M, Kurata N, Hobo T, Kagaya Y, Takeda S, Hattori T (2014) Cell-by-cell developmental transition from embryo to post-germination phase revealed by heterochronic gene expression and ER-body formation in Arabidopsis leafy cotyledon mutants. Plant Cell Physiol 55:2112–2125
Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M (2013) VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr Biol 23:1324–1329
Yang R, Zheng Z, Chen Q, Yang L, Huang H, Miki D, Wu W, Zeng L, Liu J, Zhou JX, Ogas J, Zhu JK, He XJ, Zhang H (2017a) The developmental regulator PKL is required to maintain correct DNA methylation patterns at RNA-directed DNA methylation loci. Genome Biol 18:103
Yang W, Zhang W, Wang X (2017b) Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol J 15:4–14
Yoshii M, Yamamoto A, Kagaya Y, Takeda S, Hattori T (2015) The Arabidopsis transcription factor NAI1 is required for enhancing the active histone mark but not for removing the repressive mark on PYK10, a seedling-specific gene upon embryonic-to-postgerminative developmental phase transition. Plant Signal Behav 10:e1105418
Yuan W, Luo X, Li Z, Yang W, Wang Y, Liu R, Du J, He Y (2016) A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat Genet 48:1527–1534
Zaret KS (2018) Pioneering the chromatin landscape. Nat Genet 50:167–169
Zhang X, Garreton V, Chua N-H (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19:1532–1543
Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE (2007) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14:869–871
Zhang H, Rider SD Jr, Henderson JT, Fountain M, Chuang K, Kandachar V, Simons A, Edenberg HJ, Romero-Severson J, Muir WM, Ogas J (2008) The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J Biol Chem 283:22637–22648
Zhang Y, Cao G, Qu LJ, Gu H (2009) Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep 28:337–346
Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J (2012) The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol 159:418–432
Zhang D, Jing Y, Jiang Z, Lin R (2014a) The chromatin-remodeling factor pickle integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26:2472–2485
Zhang Y, Clemens A, Maximova SN, Guiltinan MJ (2014b) The Theobroma cacao B3 domain transcription factor TcLEC2 plays a duel role in control of embryo development and maturation. BMC Plant Biol 14:106
Zhao M, Yang S, Liu X, Wu K (2015) Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front Plant Sci 6:159
Zhao H, Wu D, Kong F, Lin K, Zhang H, Li G (2016) The Arabidopsis thaliana nuclear factor Y transcription factors. Frontiers in Plant Science 7:2045
Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21:2563–2577
Zheng J, Chen F, Wang Z, Cao H, Li X, Deng X, Soppe WJ, Li Y, Liu Y (2012) A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol 193:605–616
Zhiguo E, Li T, Zhang H, Liu Z, Deng H, Sharma S, Wei X, Wang L, Niu B, Chen C (2018) A group of nuclear factor Y transcription factors are sub-functionalized during endosperm development in monocots. J Exp Bot 69:2495–2510
Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu CW, Yang S, Dong S, Ruan J, Yuan L, Zhang Z, Zhao L, Li C, Chen H, Cui Y, Wu K, Huang S (2013) HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25:134–148
Zhou Y, Romero-Campero FJ, Gomez-Zambrano A, Turck F, Calonje M (2017) H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol 18:69
Zhu C, Perry SE (2005) Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J 41:583–594
Zhu Y, Xie Z, Wang J, Liu Y, Wang J (2013) Cloning and characterization of two genes coding for the histone acetyltransferases, Elp3 and Mof, in brown planthopper (BPH), Nilaparvata lugens (Stal). Gene 513:63–70
Acknowledgments
We apologize to all our colleagues whose works and original articles could not be cited because of space limitations. The IJPB benefits from the support of the Labex Saclay Plant Sciences–SPS (ANR-10-LABX-0040-SPS). The collaboration between the authors was supported by the CERES project (ANR-10-BLAN-1238). We thank our colleagues D. De Vos, D. Grain, M. Miquel, J. Thévenin, and A. To for their valuable support to the work presented .
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Lepiniec, H. North, G. Ingram.
A contribution to the special issue ‘Seed Biology’.
Rights and permissions
About this article
Cite this article
Lepiniec, L., Devic, M., Roscoe, T.J. et al. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod 31, 291–307 (2018). https://doi.org/10.1007/s00497-018-0337-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-018-0337-2