Abstract
Main conclusion
Heterografting induced intraclonal epigenetic variations were detected among rubber plants. Interaction between genetically divergent root stock and scion tissues might have triggered these epigenetic changes which may eventually lead to intraclonal variability in rubber.
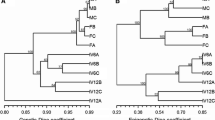
DNA methylation in response to stress may be associated with the alteration in gene transcription leading to morphological changes in plants. Rubber tree is commercially propagated by bud grafting where the scion of a high yielding variety is grafted on to a genetically divergent root stock. Still, significant levels of intraclonal variations exist among them. Epigenetic changes associated with heterografting may be partly responsible for this conundrum. In the present study, an attempt was made to identify the impact of divergent root stock on the epigenome of scion in grafted rubber plants. Heterografts were developed by grafting eye buds from a single polyembryony derived seedling on to genetically divergent root stocks of unknown parentage. The plants were uniformly maintained and their DNA was subjected to MSAP analysis. Polymorphic DNA methylation bands corresponding to CG as well as the plant-specific CHG types of methylation were observed. Cloning of selected polymorphic regions and bisulfite sequencing confirmed the presence of methylation in the promoter and coding region of important genes including an LRR receptor kinase gene. Since divergent root stock is the major factor differentiating the grafted plants, the changes in DNA methylation patterns might have been triggered by the interaction between the two genetically different tissues of stock and scion. The study assumes importance in Hevea, because accumulation and maintenance of epigenetic changes in functional genes and promoters during subsequent cycles of vegetative propagation may contribute towards intraclonal variability eventually leading to altered phenotypes.


Similar content being viewed by others
References
Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ (2007) Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA 104:14741–14746. https://doi.org/10.1073/pnas.0706701104
Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99:16491–16498
Cardinal ABB, Gonçalves PS, Mello MAL (2007) Stock–scion interactions on growth and rubber yield of Hevea brasiliensis. Sci Agric (Piracicaba, Braz) 64:235–240
Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6:351–360
Combe JC (1975) Demonstration of intraclonal variability in young graft trees. Revue Generaledes Caouth Plast 52:91–94
Combe JC, Gerner P (1977) Effect of the stock family on the growth and production of grafted Heveas. J Rubber Res Inst Sri Lanka 54:83–92
Cohen S, Naor A, Bennink J, Grava A, Tyree M (2007) Hydraulic resistance components of mature apple trees on rootstocks of different vigours. J Exp Bot 58(15–16):4213–4224
Cookson SJ, Clemente Moreno MJ, Hevin C, Nyamba Mendome LZ, Delrot S, Magnin N, Trossat-Magnin C, Ollat N (2014) Heterografting with nonself rootstocks induces genes involved in stress responses at the graft interface when compared with autografted controls. J Exp Bot 65:2473–2481. https://doi.org/10.1093/jxb/eru145
Dyachenko OV, Zakharchenko NS, Shevchuk TV, Bohnert HJ, Cushman JC, Buryanov YaI (2006) Effect of hypermethylation of CCWGG sequences in DNA of Mesembryanthemum crystallinum plants on their adaptation to salt stress. Biochemistry (Mosc) 71(4):461–465
Dorsett D, Ström L (2012) The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol 22:R240–R250
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107:8689–8694
Fulneček J, Kovařík A (2014) How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? BMC Genet 15:2
Gainza F, Opazo I, Muñoz C (2015) Graft incompatibility in plants: metabolic changes during formation and establishment of the rootstock/scion union with emphasis on Prunus species. Chilean J Agric Res. https://doi.org/10.4067/s0718-58392015000300004
Galaud J-P, Gaspar T, Boyer N (1993) Inhibition of internode growth due to mechanical stress in Bryonia dioica: relationship between changes in DNA methylation and ethylene metabolism. Physiol Plant 87(1):25–30
Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74:481–514
Gonçalves PS, Martins ALM (2002) Combining ability effects of clonal rootstocks and scions in rubber trees (Hevea). Crop Breed Appl Biotechnol 2:445–452
Haffani YZ, Silva NF, Goring DR (2004) Receptor kinase signalling in plants. Can J Bot 82:1–15
Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O (2007) CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulfite sequencing. Plant J 51(3):526–536
Kanehira A, Yamada K, Iwaya T, Tsuwamoto R, Kasai A, Nakazono M, Harada T (2010) Apple phloem cells contain some mRNAs transported over long distances. Tree Genet Genomes 6:635–642
Koepke T, Dhingra A (2013) Rootstock scion somatogenetic interactions in perennial composite plants. Plant Cell Rep 32:1321–1337
Kovařik A, Koukalová B, Bezděk M, Opatrný Z (1997) Hypermethylation of tobacco heterochromatic loci in response to osmotic stress. TAG Theor Appl Genet 95(1–2):301–306
Lämke J, Bäurle I (2017) Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol 18:124. https://doi.org/10.1186/s13059-017-1263-6
Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M (2002) Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol 4(6):694–699
Liang D, Zhang Z, Wu H, Huang C, Shuai P, Ye C-Y et al (2014) Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet 15(Suppl 1):S9. https://doi.org/10.1186/1471-2156-15-S1-S9
Maor GL, Yearim A, Ast G (2015) The alternative role of DNA methylation in splicing regulation. Trends Genet 5:274–280. https://doi.org/10.1016/j.tig.2015.03.002
Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328:872–875. https://doi.org/10.1126/science.1187959
Mun M, Yi H, Nou I, Hur Y (2015) Grafting-Induced gene expression change in Brassica rapa leaves is different from fruit trees. Plant Breed Biotechnol 3:67–76. https://doi.org/10.9787/PBB.2015.3.1.067
Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43:525–558
Nayanakantha NMC, Seneviratne P (2007) Tissue culture of rubber: past, present and future prospects. Cey J Sci (Bio Sci) 36:116–125
Ng AP, Ho CY, Sultan MO, Ooi CB, Lew HL, Yoon PK (1981) Influence of six rootstocks on growth and yield of six scion clones of Hevea brasiliensis. In: RRIM planters’s conference, London, Proceedings, pp 134–151
Omokhafe KO (2004) Clonal stability of tree dryness in Hevea brasiliensis Muell. Argic Genet Mol Biol 27:242–244
Pradillo M, Knoll A, Oliver Varas J, Corredor E, Puchta H, Santos JL (2015) Involvement of the cohesin cofactor PDS5 (SPO76) during meiosis and DNA repair in Arabidopsis thaliana. Front Plant Sci. https://doi.org/10.3389/fpls.2015.01034
Premakumari D, Nair RB, Soman TA, John A, Nazeer MA (2002) Genetic influence for intraclonal variations and associations of juvenile yield and girth in thirteen Hevea clones grown in Kanyakumari region of south India. Indian J Nat Rubber Res 15:28–32
Priyadarshan PM (2016) Genetic diversity and erosion in hevea rubber. In: Ahuja MR, Jain SM (eds) Genetic diversity and erosion in plants: case histories. Springer, Switzerland, pp 233–267
Rekha K, Jayashree R, Sushamakumari S, Sobha S, Gireesh T, Supriya R, Anita A, Saha T, Thulaseedharan A (2010) Induction of zygotic polyembryony and production of true-to-type seedlings of Hevea brasiliensis. In: Proceedings of the international conference on biotechnology: a global scenario. Kakatiya University, Warangal, pp 121–122
Rekha K, Thomas KU, Sobha S, Divya UK, Saha T, Sushamakumari S (2015) Genetic and epigenetic uniformity of polyembryony derived multiple seedlings of Hevea brasiliensis. Protoplasma. https://doi.org/10.1007/s00709-014-0713-1)
Sanabria N, Goring D, Nurnberger T, Dubery I (2008) Self/nonself perception and recognition mechanisms in plants: a comparison of self incompatibility and innate immunity. New Phytol 178:503–513
Saze H (2008) Epigenetic memory transmission through mitosis and meiosis in plants. Semin Cell Dev Biol 19:527–536
Schönberger B, Chen X, Mager S, Ludewig U (2016) Site-dependent differences in DNA methylation and their impact on plant establishment and phosphorus nutrition in Populus trichocarpa. PLoS One. https://doi.org/10.1371/journal.pone.0168623
Shang X, Cao Y, Ma L (2017) Alternative splicing in plant genes: a means of regulating the environmental fitness of plants. Int J Mol Sci. https://doi.org/10.3390/ijms18020432
Sobhana P, Jayasree G, Jacob J, Sethuraj MR (2001) Physiological and biochemical aspects of stock–scion interaction in Hevea brasiliensis. Indian J Nat Rubber Res 14:131–136
Souza LM et al (2015) Genetic diversity strategy for the management and use of rubber genetic resources: more than 1,000 wild and cultivated accessions in a 100-genotype core collection. PLoS One. https://doi.org/10.1371/journal.pone.0134607
Steward N, Kusano T, Sano H (2000) Expression of ZmMET1, a gene encoding a DNA methyltransferase from maize, is associated not only with DNA replication in actively proliferating cells, but also with altered DNA methylation status in cold-stressed quiescent cells. Nucleic Acids Res 28:3250–3259
Tang C et al (2016) The rubber tree genome reveals new insights into rubber production and species adaptation. Nat Plants. https://doi.org/10.1038/nplants.2016.73
Tsaftaris AS, Polidoros AN (2000) DNA methylation and plant breeding. Plant Breed Rev 18:87–176
Uthup TK, Ravindran M, Bini K, Saha T (2011) Divergent DNA methylation patterns associated with abiotic stress in Hevea brasiliensis. Mol Plant 4:996–1013
Wada Y, Miyamoto K, Kusano T et al (2004) Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol Genet Genom 271:658–666
Waidmann S, Kusenda B, Mayerhofer J, Mechtler K, Jonak C (2014) A DEK domain-containing protein modulates chromatin structure and function in Arabidopsis. Plant Cell 26:4328–4344
Wendt KS, Peters JM (2009) How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res 17:201–214
Wu R, Wang X, Lin Y, Ma Y, Liu G, Yu X et al (2013) Inter-species grafting caused extensive and heritable alterations of DNA methylation in Solanaceae plants. PLoS One 8:e61995. https://doi.org/10.1371/journal.pone.0061995
Yin H, Yan B, Sun J et al (2012) Graft-union development: a delicate process that involves cell-cell communication between scion and stock for local auxin accumulation. J Exp Bot 63:4219–4232
Zhang WN, Gong L, Ma C, Xu HY, Hu JF, Harada T, Li TZ (2012) Gibberellin acid-insensitive mRNA transport in Pyrus. Plant Mol Biol Rep 30:614–623
Zhang W, Spector TD, Deloukas P, Bell JT, Engelhardt BE (2015) Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol 16:14. https://doi.org/10.1186/s13059-015-0581-9
Zheng BS, Chu HL, Jin SH, Huang YJ, Wang ZJ, Chen M, Huang JQ (2010) cDNA-AFLP analysis of gene expression in hickory (Carya cathayensis) during graft process. Tree Physiol 30:297–303
Acknowledgements
We thank Dr. James Jacob, Director of Research, Rubber Research Institute of India for his support. We gratefully aknowledge the bioinformatic support rendered by Mr. Anantharamanan R. We also thank Mr. Madhusoodanan, Scientist, Rubber Technology division, RRII for the art work. This work was supported by the research funds of the Rubber Research Institute of India, Rubber Board (Ministry of Commerce and Industry, Government of India).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Uthup, T.K., Karumamkandathil, R., Ravindran, M. et al. Heterografting induced DNA methylation polymorphisms in Hevea brasiliensis. Planta 248, 579–589 (2018). https://doi.org/10.1007/s00425-018-2918-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-018-2918-6