Abstract
Main conclusion
Duplicated petunia clade-VI SPL genes differentially promote the timing of inflorescence and flower development, and leaf initiation rate.
The timing of plant reproduction relative to favorable environmental conditions is a critical component of plant fitness, and is often associated with variation in plant architecture and habit. Recent studies have shown that overexpression of the microRNA miR156 in distantly related annual species results in plants with perennial characteristics, including late flowering, weak apical dominance, and abundant leaf production. These phenotypes are largely mediated through the negative regulation of a subset of genes belonging to the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family of transcription factors. In order to determine how and to what extent paralogous SPL genes have partitioned their roles in plant growth and development, we functionally characterized petunia clade-VI SPL genes under different environmental conditions. Our results demonstrate that PhSBP1and PhSBP2 differentially promote discrete stages of the reproductive transition, and that PhSBP1, and possibly PhCNR, accelerates leaf initiation rate. In contrast to the closest homologs in annual Arabidopsis thaliana and Mimulus guttatus, PhSBP1 and PhSBP2 transcription is not mediated by the gibberellic acid pathway, but is positively correlated with photoperiod and developmental age. The developmental functions of clade-VI SPL genes have, thus, evolved following both gene duplication and speciation within the core eudicots, likely through differential regulation and incomplete sub-functionalization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Variation in plant form results largely from the differential timing of developmental phase transitions that can occur gradually (e.g., leaf size) or rapidly (e.g., flowering) in response to both endogenous and exogenous signals (Poethig 2003; Bäurle and Dean 2006). The ability of plants to match the timing of these transitions to favorable environmental conditions is a critical component of fitness, and is often associated with life history and architectural trait differences within and between populations (Hall and Willis 2006; Franks et al. 2007; Forrest and Miller-Rushing 2010). In some cases, genes involved in growth and differentiation are tightly synchronized, resulting in a predictable number of organs at a given stage of shoot growth. In contrast, some genes involved in differentiation are unaffected by shoot growth. An example of the latter are many genes involved in flowering time that have no effect on the rate of leaf initiation. Mutations in these genes result in early- or late flowering, and cause a concomitant decrease or increase of leaves, respectively (Koorneef et al. 1991; Haselhorst et al. 2011). Understanding to what extent genes involved in phase change can be uncoupled from genes involved in shoot growth under different environmental conditions is a key question in plant developmental biology (Poethig 2003).
Several genetic pathways that converge on floral integrator genes involved in floral competency and meristem identity tightly control phase change in angiosperms. In the annual rosid Arabidopsis (Arabidopsis thaliana, Brassicaceae), the recently duplicated clade-VI SQUAMOSA-PROMOTER BINDING PROTEIN (SBP)-LIKE (SPL) genes—AtSPL3, AtSPL4, and AtSPL5—integrate signals from the age, autonomous, photoperiod, and gibberellic acid signal transduction pathways to redundantly promote the formation of flowers within the inflorescence (Wu and Poethig 2006; Gandikota et al. 2007; Wang et al. 2009; Yamaguchi et al. 2009, 2014; Jung et al. 2012; Porri et al. 2012; Yu et al. 2012). Under short-day conditions, all three SPL genes are negatively regulated in an age-dependent manner by the microRNA miR156, and are positively regulated by SUPPRESSION OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Wu and Poethig 2006; Gandikota et al. 2007; Wang et al. 2009; Yamaguchi et al. 2009; Jung et al. 2012). Conversely, under long-day conditions, SOC1, FLOWERING LOCUS T (FT), and FLOWERING LOCUS D (FD) positively regulate SPL genes in leaves (Jung et al. 2012). In a positive feedback loop, SPL proteins then indirectly activate leaf FT expression, possibly through the direct binding of the inflorescence meristem gene FRUITFULL (FUL), and directly activate transcription of FUL, SOC1, APETALA1 (AP1) and LEAFY (LFY) in the shoot apex to promote flower production (Corbesier and Coupland 2006; Corbesier et al. 2007; Wang et al. 2009; Yamaguchi et al. 2009).
Although atspl3 mutants have no aberrant phenotypes, overexpression of AtSPL3 lacking the miR156-binding site accelerates juvenile to adult phase change, and results in precocious flowering without affecting the rate of leaf development (Wu and Poethig 2006; Gandikota et al. 2007; Schwarz et al. 2008; Wang et al. 2008, 2009; Yamaguchi et al. 2009). Precocious flowering is also evident in AtSPL4 and AtSPL5 overexpression lines (Wu and Poethig 2006). However, with the exception of abaxial leaf trichomes, overexpression of AtSPL4 and AtSPL5 does not decrease the number of leaves with juvenile characteristics (Wu and Poethig 2006). In accordance with the overexpression results, miR156-regulated silencing of multiple SPL genes (including AtSPL3/4/5) delays phase transitioning, but maintains apical dominance (Wu and Poethig 2006). Together these data suggest that Arabidopsis clade-VI SPL genes function redundantly in promoting reproductive, and possibly vegetative, phase change without affecting leaf or branch number. However, because of functional redundancy, further evidence is needed to determine the exact role of each gene in these developmental transitions.
Evidence from the perennial asterid species snapdragon (Antirrhinum majus, Plantaginaceae) and tomato (Solanum lycopersicum, Solanaceae) support conservation and diversification of core eudicot clade-VI SPL gene function following speciation (Klein et al. 1996; Manning et al. 2006; Preston and Hileman 2010). Similar to Arabidopsis, silencing of the AtSPL3/4/5 snapdragon homolog AmSBP1 has a negative effect on flowering time (Preston and Hileman 2010). Although inflorescence development is not delayed, flower production is completely abolished in AmSBP1-silenced plants. Furthermore, AmSBP1-silenced plants display abnormal vegetative phenotypes due to the loss of apical dominance, developing lateral vegetative branches after the main axis bearing an inflorescence fails to flower (Preston and Hileman 2010). Conversely, in tomato, epigenetic mutations in the AtSPL3/4/5 homolog LeSPL-COLORLESS NON-RIPENING (LeSPL-CNR) result in failed fruit ripening (Manning et al. 2006; Chen et al. 2015). Despite these functional insights, it is unclear how AmSBP1 and LeSPL-CNR are regulated during development compared to AtSPL3/4/5, and whether the clade-VI SPL paralogs of AmSBP1 and LeSPL-CNR also affect flowering time, branching, and fruit development.
In order to better understand the extent to which clade-VI SPL genes have functionally diversified following both gene duplication and speciation, and the underlying mechanism for these functional changes, we characterized the expression and function of two clade-VI SPL genes from the perennial asterid petunia (Petunia × hybrida, Solanaceae) under different growth conditions. Unlike Arabidopsis and snapdragon, which grow from a single dominant stem and have racemose inflorescences, petunia grows from multiple stems and has cymose inflorescences (Castel et al. 2010; Preston and Hileman 2010). Combining data from Arabidopsis, snapdragon, tomato, and petunia thus allows comparison of clade-VI SBP-box gene function across both phylogenetically and morphologically diverse species. Our results demonstrate that the petunia clade-VI SPL genes, PhSBP1, PhSBP2, and possibly PhCNR, have overlapping but divergent functions in the reproductive transition and plastochron length (leaf initiation rate). Furthermore, we show that gibberellic acid regulation of clade-VI SPL genes differs across annual and perennial species of core eudicots.
Materials and methods
Plant material and growth conditions
Petunia × hybrida (petunia) ‘Fantasy blue’ (2PET131), Mimulus guttatus IM767, and Antirrhinum majus (snapdragon) ANT11 seed was initially obtained from Seedman.com, J.K. Kelly at the University of Kansas, and the Gatersleben collection in Germany, respectively. For the photoperiod experiments, 20 wild type plants were grown under continuous light, long days (16 h light/8 h dark) or short days (8 h dark/16 h light) at 21–22 °C in a growth chamber until death or flowering with two experimental replicates. For the gibberellic acid experiments, 6-month-old non-flowering short-day grown plants were separated into two treatments. Plants in treatment one were sprayed twice weekly with 20 uM GA3, whereas plants in treatment two were sprayed twice weekly with water. Ice plant (Aizoaceae, Caryophyllales), carnation (Caryophyllaceae, Caryophyllales), Ruellia trittoniana (ruellia, Acanthaceae, Lamiales), Plumeria rubra (frangipani, Apocynaceae, Gentianales), Bidens torta (beggartick, Asteraceae, Asterales), Primula hortensis (Primulaceae, Ericales), and Penstemon barbatus (bear-tongue, Plantaginaceae, Lamiales) were grown under standard greenhouse conditions at the University of Kansas.
Gene isolation and phylogenetic analysis
In order to isolate all homologs of clade-VI SPL genes from petunia and other representative asterids, multiple degenerate primers were designed based on previously published and aligned SPL gene sequences from core eudicots (Supplemental Table S1). Total RNA was extracted from flower buds and leaves using TriReagent (Life Technologies), and contaminating DNA was removed with DNase I (Qiagen). One μg of RNA was used as a template for iScript cDNA synthesis (BioRad), the resulting cDNA was diluted 1:10, and 2 μl was used in PCR reactions. Amplicons derived from standard PCR reactions using different combinations of degenerate primers were cloned into pGEM-T (Promega), and 10–20 colonies per cloning reaction were sequenced. SPL genes were identified using BLAST searches and aligned with related genes from asterids and rosids in MacClade (Maddison and Maddison 2003). Phylogenetic relationships between SPL genes of petunia and other species were estimated using maximum likelihood methods in GARLI (Zwickl 2006). Analyses were run using the best-fitting model of molecular evolution (GTR + I + Γ), according to results of ModelTest 3.7 (Posada and Crandall 1998). The maximum likelihood analysis was run with 10 random additions, and bootstrap values were obtained using 500 bootstrap replicates. Isolation of putative petunia SPL target genes was accomplished using gene-specific primers designed from previously published sequences, and degenerate primers designed from aligned core eudicot AP1/FUL-like genes (Supplemental Table S2). Newly generated sequences longer than 200 bp in length were deposited in Genbank under the accession numbers KT717959–KT717966; sequences shorter than 200 bp in length can be found in Supplemental Table S3.
TRV2 plasmid construction
In order to control for any potentially adverse treatment effects of virus-induced gene silencing (VIGS), 194 bps of the petal pigment gene CHALCONE SYNTHASE (CHS) was PCR amplified from petunia floral cDNA, sequence verified, and cloned into the TRV2 vector as previously described (Chen et al. 2004). For SPL gene silencing, four constructs were made: PhSBP1code-TRV2 and PhSBP2code-TRV2, containing a 250 bp fragment of PhSBP1 or PhSBP2 just downstream of the SBP-box domain, and PhSBP1utr-TRV2 and PhSBP2utr-TRV2 containing a 200 bp fragment of the PhSBP1 or PhSBP2 3′-UTR. Target regions of the SPL genes were PCR amplified from petunia (Supplemental Table S1) and cloned into TRV2 using the restriction enzymes BamHI and XhoI. Each construct was sequence verified and transformed into Agrobacterium tumefaciens strain EHA105.
VIGS and phenotyping
Agrobacterium growth and plant infiltration methods followed Hileman et al. (2005) and Drea et al. (2007). Batches of 25 plants at the 4–6 leaf stage were infiltrated in half their leaves with a 1:1 ratio of TRV1:TRV2 using a needleless syringe, with at least three experimental replicates for each construct conducted at different times of the year. Following infiltration, plants were grown under long-day or short-day conditions for 2 weeks at which time RNA was extracted from the youngest (upper) leaf to screen for infection with TRV1 and TRV2 using the primers OYL195F and OYL198R for TRV1, and pYL156F and pYL156R for TRV2 (Hileman et al. 2005). Flowering time for each plant was determined as the production of the first visible floral bud, leaf and vegetative branch number at flowering were scored when the first flower was fully open, and days to transition was determined following emergence of the first inflorescence bracts. Leaf area was measured as the ratio of laminar width to laminar length. Since all dependent variables had skewed distributions, data was log-transformed prior to analysis. ANOVA was used to test for significant differences in days to the inflorescence transition, days to flowering, leaf number, branch number, and leaf ratio between treatments. When differences were significant (P < 0.05) comparisons were carried out between the PhCHS-VIGS control and each SPL treatment using a one-tailed Dunnett test in the multcomp package of R version 2.13.0 (R Development Core Team 2011). In cases where mean leaf number was opposite to predictions, i.e., lower in the SPL treatment relative to the control treatment, significance differences were determined using a two-tailed Dunnett test.
RNA extraction and quantitative real-time PCR
Expression analyses were carried out on wild type and VIGS plants under continuous light, 16 h long days, 8 h short days, and short days plus gibberellic acid conditions using quantitative reverse transcriptase (qRT)-PCR on a StepOne Plus machine (Life Technologies). Total RNA was extracted from leaves, shoot apical meristems, and dissected flowers at different developmental stages, and used to make cDNA as previously described. To determine times during the diurnal cycle when PhSBP1 and PhSBP2 expression would be high enough to compare expression levels across treatments, RNA was collected from fully expanded mid stage (fourth leaf from base) and upper leaves of two independent plants at the eight leaf stage every 4–5 h during the light period. All other material was collected between 9 and 10 am, which corresponds to 3–4 h after the zeitgeber in long-day and short-day grown plants. Upper leaf samples were taken when leaves were approximately 1 cm long.
Primer pairs for qRT-PCR were designed in Primer3 (Rozen and Skaletsky 2000) and tested for PCR efficiency using Fast SYBR Green Master Mix (Life Technologies) as previously described (Preston and Hileman 2010) (Supplemental Table S2). The housekeeping genes EF1alpha and UBQ5 were selected as internal controls based on high PCR efficiency and low transcriptional variation across different tissues in petunia (Mallona et al. 2010) and previous studies in M. guttatus and snapdragon (Preston and Hileman 2010; Scoville et al. 2011). After correcting for PCR efficiency, cycle threshold (cT) values in target tissues were normalized against the geomean of housekeeping gene expression (Scoville et al. 2011), and the mean was calculated for three to four technical replicates. For VIGS experiments, fold change was calculated in the youngest 1 cm long leaf or vegetative shoot apices by dividing the normalized values of the infected plants with that of control plants at the same developmental stage based on leaf number. For wild type gene expression experiments, biological replicates comprised similar sized tissues from different individuals at the same developmental stage or age as indicated.
Results
Isolation and phylogenetic analysis of core eudicot SPL genes and their putative targets
In order to identity all clade-VI SPL genes in petunia, and to verify their orthology relative to previously characterized genes, we cloned, sequenced, and phylogenetically analyzed SPL genes from multiple species of core eudicot. Sequencing of amplicons from both flower- and leaf-derived cDNA using multiple degenerate primer pairs identified three clade-VI SPL genes in petunia (Fig. 1), two of which (PhSBP1 and PhSBP2) were chosen to be the main focus of this study. Nucleotide comparison revealed that PhSBP1 shares 63 % identity with PhSBP2, and 83 % identity with PhCNR. Phylogenetic analysis based on the SBP-box domain of multiple core eudicot genes support orthology between petunia PhSBP1 and Solanum lycopersicum SlySBP3 (93 % ML bootstrap), PhCNR and SlyCNR (86 % ML bootstrap), and PhSBP2 and Solanum phujea SpSBP2 (94 % ML bootstrap) (Fig. 1). Although not well supported by ML bootstrapping, copy number and the most likely topology suggest at least two major lineages of SPL genes (hereafter the SBP1 and SBP2 clades), similar to that reported by Preston and Hileman (2010) (Fig. 1).
Maximum likelihood (ML) phylogeny of core eudicot clade-VI SPL genes. Genes fall into two major lineages labeled SBP1 and SBP2. The SBP1 lineage contains genes from both representative rosids (black labels) and asterids (colored labels), whereas the SBP2 lineage only contains genes from asterids. ML bootstrap values above 50 % are shown. Yellow Ericales; green Caryophyllales; red Solanales; blue Lamiales; purple Gentianales; orange Asterales. Petunia genes are in bold red with arrows. Outgroups are indicated
With the exception of tomato SlySBP4, crown group asterid genes in the SBP1 clade, including PhSBP1, PhCNR and AmSBP1, largely track the species phylogeny (Fig. 1). However, relationships among the crown asterid, early-diverging asterid Dianthus caryophyllus, Delosperma cooperi, and Primula hortensis, and rosid SBP1 clade genes are generally not well supported. The remaining asterid clade-VI SPL genes fall successively sister to the SBP1 lineage with little support among them (Fig. 1). Based on gene copy number, tree topology, and branch support, we infer that the two clade-VI SPL genes are derived from a duplication that predates diversification of the core eudicots. If the topology of the most likely tree is correct then we infer the loss of SBP2 clade genes, which include PhSBP2 and AmSBP2, from rosid eudicots. Alternatively, rosids genes forming a basal grade within the SBP1 clade, such as Medicago truncatula MtSBP1, are orthologous to asterid SBP2 clade genes (Fig. 1).
Sequencing of multiple clones from both flower- and leaf-derived petunia cDNA using gene-specific primers revealed one copy of previously uncharacterized FT, and one copy of ALF, FBP26, FBP29, PFG, UNSHAVEN, FBP21-SOC1, and FBP28-SOC1 as previously described (Gerats et al. 1988; Souer et al. 1998; Immink et al. 1999, 2003; Ferrándiz et al. 2000; Litt and Irish 2003; Vandenbussch et al. 2003). However, despite multiple attempts, both specific and degenerate primers failed to amplify PhFUL and PhFL (Litt and Irish 2003), and no SQUA orthologs containing the euAP1 transcriptional activation or farnesylation motif were isolated. Although the absence of other AP1/FUL-like genes from our search does not discount their presence in the genome, these results strongly suggest that FBP26, FBP29 and PFG are the only AP1/FUL-like genes expressed in leaf and floral tissues, concomitant with relatively high levels of PhSBP1 and PhSBP2 transcripts (Supplemental Fig. S1).
PhSBP1 and PhSBP2 expression in wild type petunia
In Arabidopsis and snapdragon, expression of SBP1 clade transcripts increase over developmental time, consistent with a role for these genes in collectively promoting the transition from juvenile to adult, vegetative to reproductive, and/or bract to flower growth (Wu and Poethig 2006; Gandikota et al. 2007; Schwarz et al. 2008; Wang et al. 2008, 2009; Yamaguchi et al. 2009; Preston and Hileman 2010). To determine if petunia PhSBP1 and PhSBP2 transcripts similarly increase during development, we conducted qRT-PCR analyses on different wild type tissues separated in time (leaves and shoot apices) and space (leaves and nodes) (Fig. 2a–d). Since expression was detectable, but did not significantly differ across 16 h long days (Fig. 2a, b), all experimental tissues were harvested at a fixed time, 3–4 h post-zeitgeber. In support of our developmental predictions, expression of PhSBP1 and PhSBP2 in plants grown under 16 h long days increased at least twofold in fully expanded upper leaves and shoot apices from day 19 to 40 (leaves) or 56 (apices) post-germination (Fig. 2c, d). Furthermore, expression of both genes was higher in late versus early emerging leaves on the same plant (Fig. 2a, b), and in older emerging nodes for PhSBP1 (box in Fig. 2c). The only expression pattern that failed to match our developmental prediction was PhSBP2 in nodes one (earliest) to eight (latest), with expression being similar across these tissues (box in Fig. 2d).
Relative PhSBP1 and PhSBP2 expression in wild type tissues. Leaf PhSBP1 (a) and PhSBP2 (b) expression levels are similar across 16 h long days, but increase from the middle (4th leaf) to the apex (8th leaf) of individual plants. n = 2 for bars and SDs. PhSBP1 (c) and PhSBP2 (d) expression increases with developmental age in leaves and shoot apical meristems (SAM), but only PhSBP1 transcripts become more abundant in axillary meristems (associated with numbered leaf nodes) from the base to the apex of individual plants. n = 4 for bars and SDs. e PhSBP1 is more abundant in early development with continuous versus short-day (SD) photoperiods. n = 4 for bars and SDs. f PhSBP1 expression does not response to gibberellic acid treatment under short days. n = 4 for bars and SDs. g PhSBP2 is also more abundant in early development with continuous versus short-day photoperiods. h PhSBP2 expression also does not response to gibberellic acid treatment under short days. n = 4 for bars and SDs
To test the hypothesis that photoperiod and gibberellic acid affect expression of PhSBP1 and PhSBP2, we assayed gene expression in plants grown under continuous light versus 8 h short days, and under short days with and without the addition of gibberellic acid. As expected, expression of both genes was higher under continuous relative to short-day light at early stages of development (leaf 4 to 11) (Fig. 2e, g). However, by leaf stage 18, expression was slightly higher in short-day versus continuous light grown plants, despite the fact that short-day plants failed to flower after 200 days. Furthermore, although it stimulated flowering in short-day plants (see next section), exogenous gibberellic acid addition had no effect on PhSBP1 or PhSBP2 expression (Fig. 2f, h). To determine if this lack of gibberellic acid response is common across asterids, similar experiments were conducted on perennial snapdragon and annual M. guttatus. Gibberellic acid had no effect on snapdragon AmSBP1 and AmSBP2, or M. guttatus MgSBP2, but expression of MgSBP1 increased over twofold relative to mock treated plants without stimulating flowering (Supplemental Fig. S2).
Petunia clade-VI SPL genes differentially control the timing of developmental phase change and leaf initiation
To identify any functional differences following duplication in clade-VI SPL genes, virus-induced gene silencing (VIGS) was conducted in petunia targeting PhSBP1, PhSBP2, and the experimental control anthocyanin pathway gene CHALCONE SYNTHASE (PhCHS). Since the most efficient VIGS vectors have previously been found to match the coding region of target genes (Lu and Page 2008), we designed PhSBP1 and PhSBP2 silencing vectors that spanned the 3′-end of the coding regions (hereafter PhSBP1code and PhSBP2code). Additionally, to confirm specificity of gene silencing, we repeated experiments with PhSBP1 and PhSBP2 silencing vectors spanning the 3′-UTRs (hereafter PhSBP1utr and PhSBP2utr). Over 250 plants screened positive for petunia VIGS constructs, with an infection-efficiency of around 80 %. qRT-PCR analyses also revealed that infection was strongly negatively correlated with target gene expression (P < 0.001 for coding region vectors, P < 0.05 for 3′-UTR vectors, one-tailed Tukey’s test) (Fig. 3e) or, in the case of PhCHS-TRV2 infected control plants, with loss of petal anthocyanin production (Fig. 3g, n).
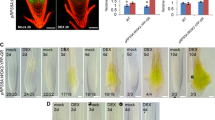
PhSBP1 and PhSBP2 VIGS phenotypes. a Days to flowering increase in PhSBP1code- relative to PhCHS- and PhSBP2code-silenced individuals under 16 h long days (LD) only (n = 29–53 for bars and SDs). Infected plants grown under 8 h short days (SD-GA) fail to flower even after 200 days unless treated with gibberellic acid (SD + GA) (n = 10 for bars and SDs). b PhSBP1utr-silenced plants take longer to transition to inflorescence development (Transition) in long days relative to PhCHS- and PhSBP2utr-silenced individuals, whereas both PhSBP1utr- and PhSBP2utr-silenced plants take longer to flower (Flowering) relative to PhCHS-infected controls. n = 45–52 for bars and SDs. c Leaves are initiated more quickly in PhSBP1code-silenced plants relative to PhCHS-silenced controls based on leaf counts at 22 and 33 days post-germination under long days. Increased leaf number in flowering (Flower.) PhSBP1code-infected plants correlate with their late-flowering phenotype. However, since PhSBP2code-silenced plants are not late-flowering, elevated leaf number suggests accelerated leaf initiation rate in long days. VIGS treatments do not significantly affect leaf number under short-day conditions. n = 29–53 for bars and SDs. d Leaves are also initiated more quickly in PhSBP1utr-silenced plants relative to PhSBP2utr- and PhCHS-silenced individuals based on leaf counts at 22 and 33 days post-germination under long days. Increased leaf number at flowering correlates with late flowering in PhSBP1utr-silenced plants. n = 30–52 for bars and SDs. e Infection with SBP1code and SBP1utr VIGS vectors significantly reduces expression of PhSBP1 (red bars) relative to PhCHS control plants (dashed line), but not PhSBP2 (orange bars) or PhCNR (purple bars). PhSBP2utr-TRV2 infection causes gene-specific silencing of PhSBP2, whereas infection with the PhSBP2code-TRV2 vector causes silencing of PhSBP2 and unexpected upregulation of PhCNR. n = 20 for bars and SDs. f PhCHS-silenced plant flowering at 68 days (left) versus PhSBP1code-silenced plant flowering at 151 days (right). g PhCHS-silenced (left) versus PhSBP2code-silenced (right) plant flowering at 75 days. Leaf node spacing is increased in PhSBP1code (i) and decreased in PhSBP2code (j) positive plants relative to PhCHS-silenced plants (h), consistent with leaf initiation rate. 33-day old PhCHS (k), PhSBP1code (l), and PhSBP2code (m) positive plants. PhCHS-silenced (n) flowering versus PhSBP2utr-silenced (o) late-flowering plant. Errors bars are standard deviations for multiple biological replicates. Asterisks denote significant differences at the P < 0.05 (asterisk), P < 0.01 (double asterisk), and P < 0.001 (triple asterisk) levels according to a Tukey’s (e) or Dunnett’s (a–d) test
For plants infected with SBP constructs, levels of off-target PhCNR and PhSBP1 (PhSBP2code-TRV2 and PhSBP2utr-TRV2) or PhSBP2 (PhSBP1code-TRV2 and PhSBP1utr-TRV2) transcripts were not significantly decreased relative to PhCHS-TRV2 infected plants (Fig. 3e). However, despite this evidence of no cross silencing among clade-VI SPL genes, PhCNR expression levels were significantly higher (P < 0.05, two-tailed Tukey’s test) in PhSBP2code- versus PhCHS-TRV2-infected plants (Fig. 3e). Based on these data, we infer that the PhSBP2code-TRV2 vector is less gene-specific than all the other VIGS vectors, resulting in the silencing of PhSBP2 and an unknown negative regulator of PhCNR. We nonetheless continued phenotypic characterization of PhSBP2code silenced plants to gain possible insight into the effect of elevated PhCNR levels on plant development.
Consistent with our SPL transcript level screen, silencing of PhSBP1 resulted in similar phenotypes for both the PhSBP1code- (Fig. 3a, c, f, i, l) and PhSBP1utr-TRV2 (Fig. 3b, d) vectors. PhSBP1 silencing resulted in significantly delayed transition to inflorescence and flower development relative to PhCHS control plants based on a Dunnett’s test (Fig. 3a–d, f). Delays in phenology were observed in post-germination days to inflorescence and flower development (Fig. 3a, b), as well as in increased numbers of leaves and branches during flower emergence of PhSBP1code- and PhSBP1utr-TRV2-infected plants (Fig. 3c, d; Supplemental Fig. S3a). Although leaves became narrower over time in early developing wild type petunias (Supplemental Fig. S3b), possibly reflecting the transition from juvenile to adult growth as in Arabidopsis (Poethig 2003; Wu and Poethig 2006), there was no difference in leaf shape or trichome density between developmentally comparable leaves of PhCHS-TRV2 and PhSBP1code-TRV2-infected plants (Supplemental Fig. S3c). However, while PhSBP1-silenced plants had increased leaf and branch numbers at the onset of flowering due to the significant delay in flowering time (Fig. 3c, d; Supplemental Fig. S3a), analysis of pre-flowering leaf number demonstrated that PhSBP1-silenced plants had a reduced rate of leaf initiation. Specifically, at 22 and 33 days post-germination, PhSBP1-silenced plants had significantly fewer leaves than PhCHS-silenced plants, suggesting that leaf initiation was suppressed (Fig. 3c, d).
Plants positive for the PhSBP2utr vector flowered significantly later than PhCHS-silenced plants under long days, despite not being delayed in the transition to inflorescence development (Fig. 3b, n, o). However, in contrast to PhSBP1, silencing of PhSBP2 using the PhSBP2utr-VIGS vector had no effect on the rate of leaf initiation under long days, at least during the initial 33 days post-germination (Fig. 3d). In plants infected with the PhSBP2code-TRV2 vector, where PhSBP2 is silenced and PhCNR expression is enhanced (Fig. 3e), there was no difference in flowering time relative to control plants, but leaf number and branch number at flowering were significantly increased (Fig. 3a, c, g; Supplemental Fig. S3a). Thus, because down-regulation of PhSBP2 alone (PhSBP2utr vector) leads to delayed flowering and no change in leaf or branch number, and down-regulation of PhSBP2 with increased PhCNR (PhSBP2code vector) has no effect on flowering and increases leaf number, these results suggest that PhCNR functions similarly to PhSBP1, accelerating flowering and leaf initiation rate in wild type petunia.
To determine if photoperiod has an effect on VIGS phenotypes, we conducted a subset of similar experiments using the PhCHS-, PhSBP1code-, and PhSBP2code vectors under 8 h short-day conditions. With this reduced photoperiod, PhCHS-, PhSBP1code-, and PhSBP2code-silenced plants failed to flower completely (Fig. 3a). However, addition of gibberellic acid to 6-month-old short-day grown plants promoted flowering, with no evidence of leaf number differences at flowering time between plants positive for either of the constructs compared to control plants (Fig. 3c).
Downstream targets of petunia clade-VI SPL genes are largely conserved
Differential regulation of downstream targets likely explains functional differences in phase change regulation and rate of leaf initiation between PhSBP1 and PhSBP2, and the phenotypic differences between PhSBP2utr- and PhSBP2code-TRV2 infected plants. In order to determine what genes might be differentially affected, we examined changes in expression of known target orthologs (Klein et al. 1996; Wang et al. 2009; Yamaguchi et al. 2009; Preston and Hileman 2010) in response to PhSBP1 and PhSBP2 silencing, and/or increased PhCHS expression (Fig. 4). The youngest (upper) leaves of plants from different treatments were matched by leaf number (Fig. 4a–c) and transitional shoot apical meristems were matched by developmental stage (Fig. 4d).
In all PhSBP1- and PhSBP2-silenced plants grown under long days, the flower development genes ALF, FBP26, FBP29, PFG, and FBP20 showed at least a mean 1.5-fold reduction in leaf and shoot apex expression relative to control plants (Fig. 4a, c, d). FT was also reduced at least 1.5-fold in long-day leaves of plants silenced with PhSBP1code and PhSBP2code constructs (Fig. 4a), and shoot apices of plants silenced with PhSBP1utr and PhSBP2utr constructs (Fig. 4d). However, silencing of either PhSBP1 or PhSBP2 with a 3′-UTR construct had little effect on FT expression in long-day grown leaves (Fig. 4c). Although expression of the SOC1-like genes, FBP21 and FBP28, was consistently reduced in leaves of PhSBP1-silenced plants, expression levels relative to control plants varied between leaves infected with PhSBP2utr and PhSBP2code (Fig. 4a versus 4c) vectors. Whereas PhSBP2utr-VIGS plants had reduced FBP21 and control-like FBP28 expression levels, PhSBP2code-VIGS plants had control-like FBP21 and elevated FBP28 expression levels (Fig. 4a versus c). Under short days, infection with PhSBP1code and PhSBP2code vectors caused at least 1.5-fold silencing of leaf FT, FBP29, and PFG; ALF and FBP26 only showed down-regulation when PhSBP1 was silenced, and FBP20 transcripts increased at least 1.5-fold in both PhSBP1- and PhSBP2-silenced plants (Fig. 4b). Taken together, these data suggest similar transcriptional targets for PhSBP1 and PhSBP2, and support the idea that the wild type flowering time of PhSBP2code-TRV2 plants is due to the coincident downregulation of FT, ALF, and AP1/FUL-like genes, and upregulation of a SOC1-like gene.
Discussion
Our study demonstrates that two petunia clade-VI SPL genes, PhSBP1 and PhSBP2, have evolved in function following their duplication at the base of core eudicots. The effects of gene silencing on development are consistent with PhSBP1 promoting inflorescence development, flower emergence, and leaf initiation in wild type petunias. In contrast, PhSBP2 has no obvious effect on the timing of inflorescence development or leaf development, but positively affects the onset of flower production. Although not a direct test of PhCNR function, comparison of plants infected with the PhSBP2utr and PhSBP2code vectors, the latter of which increases PhCNR levels, suggests that PhCNR in wild type plants functions similarly to PhSBP1 by promoting flowering and accelerating leaf initiation through decreased internode spacing. Unlike orthologs in Arabidopsis and M. guttatus, PhSBP1 and PhSBP2 transcript levels are unaffected by exogenous gibberellic acid treatment. However, increasing day length and developmental age positively regulates both genes, resulting in the transcriptional activation of a conserved set of target genes, including members of the FUL, SOC1, LEAFY, and FT clades of flowering time transcription factors (Klein et al. 1996; Corbesier and Coupland 2006; Corbesier et al. 2007; Wang et al. 2009; Yamaguchi et al. 2009; Preston and Hileman 2010; Preston et al. 2014).
Despite its efficacy in petunia, one important caveat to consider before making comparisons of clade-VI SPL gene function across species is that VIGS results in incomplete silencing that can vary spatially and temporally between plants. Thus, the silencing phenotypes described for PhSBP1 and PhSBP2 potentially underestimate the effects of these genes on development. Regardless of these limitations, we believe that the data presented here are meaningful in the sense that they can still reveal novel gene functions and, since several hundred plants were screened, distinguish the direction of developmental effects even for quantitative phenotypes. Similar levels of silencing between plants for PhSBP1 and PhSBP2 VIGS constructs also allow direct comparisons of petunia gene function following gene duplication. Ultimately, future studies exploiting powerful resources such as transposon-tagged petunia mutants or CRISPR-Cas site directed mutagenesis (Bortesi and Fischer 2015), will required to determine the overall effectiveness of VIGS approaches in petunia. These resources were unavailable to us at the start of this project.
Caveats notwithstanding, results of our study demonstrate a novel function for clade-VI genes in accelerating leaf initiation rate and a conserved function for these genes in promoting flower development. Specifically, whereas the Arabidopsis SBP1-like genes AtSPL3, AtSPL4, and AtSPL5 are likely functionally redundant in promoting vegetative phase change and flowering, petunia PhSBP1, PhSBP2, PhCNR and snapdragon AmSBP1 promote flowering, PhSBP1/2 and AmSBP1 control branching, PhSBP1 and PhCNR promote leaf initiation, PhSBP1 positively regulates late vegetative phase change, and tomato CNR promotes fruit ripening (Manning et al. 2006; Wu and Poethig 2006; Wang et al. 2009; Yamaguchi et al. 2009; Preston and Hileman 2010; this study). With the exception of fruit ripening and acceleration of leaf initiation, all of these functions have been described in analyses of more distantly related SPL homologs in Arabidopsis and rice (Schwarz et al. 2008; Shikata et al. 2009; Usami et al. 2009; Jiao et al. 2010; Miura et al. 2010). Thus, this evolutionary pattern is consistent with evolution mainly through differential sub-functionalization (Preston and Hileman 2013). Our data also demonstrate that clade-VI SPL genes differ in their response to the gibberellic acid signaling pathway, the lack of a response being correlated with perenniality. We suggest that future work focuses on the elucidation of downstream factors involved in functional diversification of clade-VI SPL genes following both speciation and duplication.
Author contribution statement
J.C.P., S.A.J., and R.O. conducted the experiments; J.C.P and S.A.J. analyzed the data; and J.C.P. and L.C.H. conceived of/designed the experiments and wrote the article. All authors read and approved the manuscript.
Abbreviations
- ALF :
-
ABERRANT LEAF AND FLOWER
- CHS :
-
CHALCONE SYNTHASE
- FBP :
-
FLORAL BINDING PROTEIN
- FUL :
-
FRUITFULL
- FT :
-
FLOWERING LOCUS T
- SOC1 :
-
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- SBP :
-
SQUAMOSA PROMOTER BINDING PROTEIN
- SPL :
-
SBP-like
- TRV:
-
Tobacco rattle virus
- VIGS:
-
Virus-induced gene silencing
References
Bäurle I, Dean C (2006) The timing of developmental transitions in plants. Cell 125:655–664
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J 12:367–377
Cardon G, Hohmann S, Klein J, Nettesheim K, Saedler H, Huijser P (1999) Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237:91–104
Castel R, Kusters E, Koes R (2010) Inflorescence development in petunia: through the maze of botanical terminology. J Exp Bot 61:2235–2246
Chen J-C, Jiang C-Z, Gookin TE, Hunter DA, Clark DG, Reid M (2004) Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol Biol 55:521–530
Chen W, Kong J, Lai T, Manning K, Wu C, Wang Y, Qin C, Li B, Yu Z, Zhang X, He M, Zhang P, Gu M, Yang X, Mahammed A, Li C, Osman T, Shi N, Wang H, Jackson S, Liu Y, Gallusci P, Hong Y (2015) Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci Rep 5:7852
Corbesier L, Coupland G (2006) The quest for florigen: a review of recent progress. J Exp Bot 57:3395–3403
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Drea S, Hileman LC, de Martino G, Irish VF (2007) Functional analyses of genetic pathways controlling petal specification in poppy. Development 134:4157–4166
Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127:725–734
Forrest J, Miller-Rushing AJ (2010) Toward a synthetic understanding of the role of phenology in ecology and evolution. Philos Trans R Soc Lond B Biol Sci 365:3101–3112
Franks SJ, Sim S, Weis AE (2007) Rapid evolution of flowering time by an annual plant in response to climate fluctuation. Proc Natl Acad Sci USA 104:1278–1282
Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP-box gene SPL3 prevents early flowering by translation inhibition in seedlings. Plant J 49:683–693
Gerats AGM, Kaye C, Collins C, Malmberg RL (1988) Polyamine levels in Petunia genotypes with normal and abnormal floral morphologies. Plant Physiol 86:390–393
Hall MC, Willis JH (2006) Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution 60:2466–2477
Haselhorst MS, Edwards CE, Rubin MJ, Weinig C (2011) Genetic architecture of life history traits and environment-specific trade-offs. Mol Ecol 20:4042–4058
Hileman LC, Drea S, de Martino G, Litt A, Irish VF (2005) Virus-induced gene silencing is an effective tools for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44:334–341
Immink RGH, Hannapel DJ, Ferrario S, Busscher M, Franken J, Campagne MML, Angenent GC (1999) A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 126:5117–5126
Immink RGH, Ferrario S, Busscher-Lange J, Kooiker M, Busscher M, Angenent GC (2003) Analysis of the petunia MADS-box transcription factor family. Mol Genet Genomics 268:598–606
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–545
Jung J-H, Ju Y, Seo PJ, Lee J-H, Park C-M (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69:577–588
Klein J, Saedler H, Huijser P (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250:7–16
Koorneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229:57–66
Litt A, Irish VF (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165:821–833
Lu E, Page JE (2008) Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods 4:5. doi:10.1186/1746-4811-4-5
Maddison DR, Maddison WP (2003) Macclade: analysis of phylogeny and character evolution. Sinauer Associates, Sunderland
Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower in Petunia hybrida. BMC Plant Biol 10:4
Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952
Miura K, Ikeda M, Matsubara A, Song X-J, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–550
Poethig RS (2003) Phase change and the regulation of developmental timing in plants. Science 301:334–336
Porri A, Tori S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory role for gibberellins in the promotion of flowering in Arabidopsis under long photoperiods. Development 138:2198–2209
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Preston JC, Hileman LC (2010) SQUAMOSA-PROMOTER BINDING PROTEIN1 initiates flowering in Antirrhinum majus through the activation of meristem identity genes. Plant J 62:704–712
Preston JC, Hileman LC (2013) Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Frontiers Plant Sci 4:80
Preston JC, Jorgensen SA, Jha SG (2014) Functional characterization of duplicated SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1-like genes in petunia. PLoS One 9(5):e96108. doi:10.1371/journal.pone.0096108
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://R-project.org/
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biological programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, NJ, USA, pp 365–386
Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67:183–195
Scoville AG, Barnett LL, Bodbyl-Roels S, Kelly JK, Hileman LC (2011) Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol 191:251–263
Shikata M, Koyama T, Mitsuda N, Ohe-Takagi M (2009) Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol 50:2133–2145
Souer E, van der Krol A, Kloos D, Spelt C, Bliek M, Mol J, Koes R (1998) Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development 125:733–742
Usami T, Horiguchi G, Yano S, Tsukaya H (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136:955–964
Vandenbussch M, Zethof J, Souer E, Koes R, Tornielli GB, Pezzotti M, Ferrario S, Angenenet GC, Gerats T (2003) Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15:2680–2693
Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20:1231–1243
Wang J-W, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133:3539–3547
Yamaguchi A, Wu M-F, Yang Li WuG, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17:268–278
Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D (2014) Gibberellin acts positively then negatively to control onset of flowering in Arabidopsis. Science 344:638–641
Yu S, Galvao VC, Zhang Y-C, Horer D, Zhang T-Q, Hao Y-H, Feng Y-Q, Wang S, Markus S, Wang J-W (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. Plant Cell 24:3320–3332
Zwickl D (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD Thesis, TX University of Texas at Austin
Acknowledgments
We thank several anonymous reviewers for their constructive comments on earlier versions of this manuscript. This work was supported by the University of Vermont College of Agriculture and Life Sciences, and a National Science Foundation grant (IOS-1051886) to L.C.H. and J.C.P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2015_2413_MOESM1_ESM.eps
Supplementary Supplemental Fig. S1 Expression of petunia clade-VI SPL genes and putative targets in floral organs and leaves. Black, white, and gray bars denote SPL, ALF, and AP1/FUL-like gene expression, respectively. n = 3 for bars and SDs (EPS 341 kb)
425_2015_2413_MOESM2_ESM.eps
Supplemental Fig. S2 Expression of clade-VI SPL genes from annual M. guttatus (a, b) and perennial snapdragon (c, d) in response to gibberellic acid (GA) treatment (white bars) under 8 h short days (SD). Only MgSBP1 increases in expression in response to GA. n = 4–5 for bars and SDs (EPS 450 kb)
425_2015_2413_MOESM3_ESM.eps
Supplemental Fig. S3 Effect of gene silencing and developmental time on branch number and leaf shape. a Boxplot showing significant differences in branch number at flowering between PhCHS-, PhSBP1code-, and PhSBP2code-VIGS infected plants. **P < 0.01; ***P < 0.001 according to a one-tailed Dunnett’s test. b Mean leaf shape (ratio of laminar length to width) in a series of leaves from wild type petunia plants grown under long days (black bars) and short days with gibberellic acid (GA) addition (gray bars). Stars denote flowering plants. A one-tailed Dunnett’s test comparing long-day leaf shapes was significant (P < 0.05) between leaf numbers one and 12 to 64, denoted a. c Mean leaf shape in a series of leaves from PhCHS-TRV2- (black bars), PhSBP1code-TRV2- (gray bars), and PhSBP2code-TRV2-infected (white bars) plants. No significant differences were found across treatments, suggesting that clade-VI SPL genes do not affect leaf shape as a proxy for juvenile-adult growth. n = 6–10 for bars and SDs (EPS 375 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Preston, J.C., Jorgensen, S.A., Orozco, R. et al. Paralogous SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes differentially regulate leaf initiation and reproductive phase change in petunia. Planta 243, 429–440 (2016). https://doi.org/10.1007/s00425-015-2413-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2413-2