Abstract
Main conclusion
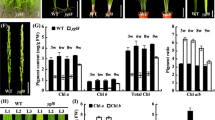
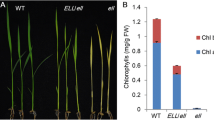
Rice heme oxygenase 2 (OsHO2) mutants are chlorophyll deficient with distinct tetrapyrrole metabolite and transcript profiles, suggesting a potential regulatory role of the stromal-localized OsHO2 in tetrapyrrole biosynthesis.
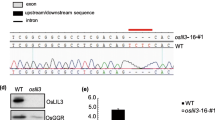
In plants, heme oxygenases (HOs) are classified into the subfamilies HO1 and HO2. HO1 are highly conserved plastid enzymes required for synthesizing the chromophore in phytochromes which mediate a number of light-regulated responses. However, the physiological and biochemical functions of HO2, which are distantly related to HO1, are not well understood, especially in crop plants. From a population of 60Coγ-irradiated rice mutants, we identified the ylc2 (young leaf chlorosis 2) mutant which displays a chlorosis phenotype in seedlings with substantially reduced chlorophyll content. Normal leaf pigmentation is gradually restored in older plants while newly emerged leaves remain yellow. Transmission electron microscopy further revealed defective chloroplast structures in the ylc2 seedlings. Map-based cloning located the OsYLC2 gene on chromosome 3 and it encodes the OsHO2 protein. The gene identification was confirmed by complementation and T-DNA mutant analyses. Subcellular localization and chloroplast fractionation experiments indicated that OsHO2 resides in the stroma. However, recombinant enzyme assay demonstrated that OsHO2 is not a functional HO enzyme. Analysis of tetrapyrrole metabolites revealed the reduced levels of most chlorophyll and phytochromobilin precursors in the ylc2 mutant. On the other hand, elevated accumulation of 5-aminolevulinic acid and Mg-protoporphyrin IX was observed. These unique metabolite changes are accompanied by consistent changes in the expression levels of the corresponding tetrapyrrole biosynthesis genes. Taken together, our work suggests that OsHO2 has a potential regulatory role for tetrapyrrole biosynthesis in rice.






Similar content being viewed by others
Abbreviations
- ALA:
-
5-Aminoleuvlinic acid
- BAC:
-
Bacterial artificial chromosome
- BV:
-
Biliverdin
- Car:
-
Carotenoid
- Chl:
-
Chlorophyll
- Chlide:
-
Chlorophyllide
- EYFP:
-
Enhanced yellow fluorescent protein
- His:
-
Histidine
- HM:
-
Homozygous
- HO:
-
Heme oxygenase
- MgCH:
-
Mg chelatase
- PΦB:
-
Phytochromobilin
- Pchlide:
-
Protochlorophyllide
- PG:
-
Plastoglobules
- Proto IX:
-
Protoporphyrin IX
- RT:
-
Reverse transcription
- STS:
-
Sequence-tagged site
- TEM:
-
Transmission electron microscopy
- Urogen III:
-
Uroporphyrinogen III
- WT:
-
Wild type
References
Adhikari ND, Froehlich JE, Strand DD, Buck SM, Kramer DM, Larkin RM (2011) GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23:1449–1467
Andrés F, Galbraith DW, Talón M, Domingo C (2009) Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol 151:681–690
Balestrasse KB, Yannarelli GG, Noriega GO, Batlle A, Tomaro ML (2008) Heme oxygenase and catalase gene expression in nodules and roots of soybean plants subjected to cadmium stress. Biometals 21:433–441
Beale SI (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60:43–73
Chen H, Cheng Z, Ma X, Wu H, Liu Y, Zhou K, Chen Y, Ma W, Bi J, Zhang X (2013) A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep 32:1855–1867
Davis SJ, Kurepa J, Vierstra RD (1999) The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci 96:6541–6546
Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD (2001) The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol 126:656–669
de Montellano PRO, Wilks A (2000) Heme oxygenase structure and mechanism. Adv Inorg Chem 51:359–408
Emborg TJ, Walker JM, Noh B, Vierstra RD (2006) Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol 140:856–868
Fu GQ, Jin QJ, Lin YT, Feng JF, Nie L, Shen WB, Zheng TQ (2011a) Cloning and characterization of a heme oxygenase-2 gene from alfalfa (Medicago sativa L.). Appl Biochem Biotech 165:1253–1263
Fu GQ, Xu S, Xie YJ, Han B, Nie L, Shen WB, Wang R (2011b) Molecular cloning, characterization, and expression of an alfalfa (Medicago sativa L.) heme oxygenase-1 gene, MsHO1, which is pro-oxidants-regulated. Plant Physiol Biochem 49:792–799
Gelly JC, Joseph AP, Srinivasan N, De Brevern AG (2011) iPBA: a tool for protein structure comparison using sequence alignment strategies. Nucleic Acids Res 39:18–23
Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinkel N (2010) Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochem J 425:425–434
Han B, Xu S, Xie YJ, Huang JJ, Wang LJ, Yang Z, Zhang CH, Sun Y, Shen WB, Xie GS (2012) ZmHO-1, a maize haem oxygenase-1 gene, plays a role in determining lateral root development. Plant Sci 184:63–74
Harel E, Klein S (1972) Light dependent formation of δ-aminolevulinic acid in etiolated leaves of higher plants. Biochem Biophys Res Commu 49:364–370
Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22:391–399
Jin QJ, Feng JF, Sun Y, Cui WT, Han B, Xu S, Shen WB, Cui J (2012a) Characterization of a heme oxygenase-1 from tobacco that is involved in some abiotic stress responses. Int J Plant Sci 173:113–123
Jin QJ, Yuan XX, Cui WT, Han B, Feng JF, Xu S, Shen WB (2012b) Isolation and characterization of a heme oxygenase-1 gene from Chinese cabbage. Mol Biotechnol 50:8–17
Katz JJ, Norris JR, Shipman LL, Thurnauer MC, Wasielewski MR (1978) Chlorophyll function in the photosynthetic reaction center. Annu Rev Biophys Bioeng 7:393–434
Lee S, Jeon JS, Jung KH, An G (1999) Binary vectors for efficient transformation of rice. J Plant Biol 42:310–316
Lee JY, Lee HS, Song JY, Jung YJ, Reinbothe S, Park YI, Lee SY, Pai HS (2013) Cell growth defect factor1/CHAPERONE-LIKE PROTEIN OF POR1 plays a role in stabilization of light-dependent protochlorophyllide oxidoreductase in Nicotiana benthamiana and Arabidopsis. Plant Cell 25:3944–3960
Li MY, Cao ZY, Shen WB, Cui J (2011) Molecular cloning and expression of a cucumber (Cucumis sativus L.) heme oxygenase-1 gene, CsHO1, which is involved in adventitious root formation. Gene 486:47–55
Linley PJ, Landsberger M, Kohchi T, Cooper JB, Terry MJ (2006) The molecular basis of heme oxygenase deficiency in the pcd1 mutant of pea. FEBS J 273:2594–2606
Liu H, Lau E, Lam MP, Chu H, Li S, Huang G, Guo P, Wang J, Jiang L, Chu IK (2010) OsNOA1/RIF1 is a functional homolog of AtNOA1/RIF1: implication for a highly conserved plant cGTPase essential for chloroplast function. New Phytol 187:83–105
Matera KM, Zhou H, Migita CT, Hobert SE, Ishikawa K, Katakura K, Maeshima H, Yoshida T, Ikeda-Saito M (1997) Histidine-132 does not stabilize a distal water ligand and is not an important residue for the enzyme activity in heme oxygenase-1. Biochemistry 36:4909–4915
Munro AW, Girvan HM, McLean KJ, Cheesman MR, Leys D (2009) Heme and hemoproteins. In tetrapyrroles: birth, life and death. Warren MJ, Smith AG (eds) pp 160–183
Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11:335–347
Muramoto T, Tsurui N, Terry MJ, Yokota A, Kohchi T (2002) Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol 130:1958–1966
Salvi D, Rolland N, Joyard J, Ferro M (2008) Purification and proteomic analysis of chloroplasts and their sub-organellar compartments. Methods Mol Biol 432:19–36
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Shi D, Zheng X, Li L, Lin W, Xie W, Yang J, Chen S, Jin W (2013) Chlorophyll deficiency in the maize elongated mesocotyl2 mutant is caused by a defective heme oxygenase and delaying grana stacking. PLoS One 8:e80107
Smith H (2000) Phytochromes and light signal perception by plants-an emerging synthesis. Nature 407:585–591
Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9:e0145
Terry MJ, Kendrick RE (1999) Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient Aurea and yellow-green-2 mutants of tomato. Plant Physiol 119:143–152
Tripathy BC, Sherameti I, Oelmüller R (2010) Siroheme: an essential component for life on earth. Plant Signal Behav 5:14–20
Wang L, Ma F, Xu S, Zheng T, Wang R, Chen H, Shen W (2014) Cloning and characterization of a heme oxygenase-2 gene from rice (Oryza sativa L.), and its expression analysis in response to some abiotic stresses. Acta Physiol Plant 36:893–902
Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21:1109–1128
Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145:29–40
Zhang H, Li J, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC (2006) Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62:325–337
Zhou K, Ren Y, Lv J, Wang Y, Liu F, Zhou F, Zhao S, Chen S, Peng C, Zhang X (2013) Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 237:279–292
Acknowledgments
This work is supported by the National Key Laboratory of Plant Molecular Genetics, the National Natural Science Foundation of China (grant no. 31171530), Zhejiang Provincial Natural Science Foundation of China (grant no. Y3100591), and the HKU Seed Funding for Basic Research (grant no. 201111159147).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Q. Li and F.-Y. Zhu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Q., Zhu, FY., Gao, X. et al. Young Leaf Chlorosis 2 encodes the stroma-localized heme oxygenase 2 which is required for normal tetrapyrrole biosynthesis in rice. Planta 240, 701–712 (2014). https://doi.org/10.1007/s00425-014-2116-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2116-0