Abstract
Fruits of persimmon (Diospyros kaki Thunb.) accumulate large amounts of proanthocyanidins (PAs) in the early stages of development. Astringent (A)-type fruits remain rich in soluble PAs even after they reach full-mature stage, whereas non-astringent (NA)-type fruits lose these compounds before full maturation. As a first step to elucidate the mechanism of PA accumulation in this non-model species, we used suppression subtractive hybridization to identify transcripts accumulating differently in young fruits of A- and NA-type. Interestingly, only a few clones involved in PA biosynthesis were identified in A–NA libraries. Represented by multiple clones were those encoding a novel 1-Cys peroxiredoxin and a new member of family 1 glycosyltransferases. Quantitative RT-PCR analyses confirmed correlation of the amount of PAs and accumulation of transcripts encoding these proteins in young persimmon fruits. Furthermore, the new family 1 glycosyltransferase was produced in Escherichia coli and shown to efficiently catalyze galactosylation at 3-hydroxyl groups of several anthocyanidins and flavonols. These findings suggest a complex mechanism of PA accumulation in persimmon fruits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proanthocyanidins (PAs) are colorless phenolic polymers that show brown coloration upon oxidation. They are present in fruits, bark, leaves, and seeds of many plants, and are postulated to play protective roles. These phenolic compounds are also recognized as useful agents for human health (Dixon et al. 2005). PAs consist of flavan-3-ol units that are synthesized via the pathway leading to various phenylpropanoid compounds such as flavonols and anthocyanins (glycosylated forms of anthocyanidins) (Fig. 1). Catalytic and regulatory mechanisms of phenylpropanoid metabolism have been elucidated well and a number of molecular components have been identified (Koes et al. 2005; Lepiniec et al. 2006; Yu and Jez 2008). Notably, two flavonoid reductases, leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR), have been shown to compete with the biosynthesis of anthocyanins, catalyzing steps committed for PA biosynthesis. Tanner et al. (2003) identified LAR, which catalyzes conversion of the immediate precursors of anthocyanidins to one of the PA subunits, 2,3-trans-flavan 3-ols [e.g., (+)-catechin], from the legume Desmodium uncinatum. Xie et al. (2003) demonstrated that anthocyanidins are common precursors to anthocyanins and PAs by identifying the BAN gene product as ANR, which converts anthocyanidins to the other PA subunits, 2,3-cis-flavan-3-ols [e.g., (−)-epicatechin], in Arabidopsis thaliana and Medicago truncatula. In addition to the biosynthetic enzymes, MYB transcription factors have been shown to regulate PA accumulation in several species, such as A. thaliana (Nesi et al. 2001) and grapes (Bogs et al. 2007; Deluc et al. 2008; Terrier et al. 2009).
One of the outstanding questions regarding the mechanism of PA metabolism is if and how flavan-3-ols are sorted from the cytoplasm to the vacuole, where they are postulated to be polymerized. Recently, several candidates involved in this process have been identified. One of them is a glucosyltransferase (GlcT), UGT72L1, from M. truncatula (Pang et al. 2008). Transcriptional profiling led to identification of the UGT72L1 gene, whose expression correlated well with massive accumulation of PAs and the presence of a low but significant amount of (−)-epicatechin glucoside in the seed coat of M. truncatula. A further biochemical study established the epicatechin 3′-O-GlcT activity of the UGT72L1 protein. Another component is a multidrug and toxic compound extrusion-type transporter, TRANSPARENT TESTA 12 (TT12), from A. thaliana. A genetic study showed that TT12 is necessary for PA accumulation in seeds (Debeaujon et al. 2001). A fusion of TT12 and green fluorescent protein was targeted to tonoplast in vivo, and yeast microsomes producing the TT12 protein were shown to have a cyanidin-3-O-glucoside/H+ antiporter activity in vitro (Marinova et al. 2007). Direct evidence for transport of flavan-3-O-glucosides through TT12 into the vacuole has not been provided yet. Nonetheless, these findings have led to a model for the critical step of PA biosynthesis, i.e., glycosylation and transport of flavan-3-ols from the cytoplasm to the vacuole where they are probably deglycosylated and polymerized (Pang et al. 2008).
A wide variety of plants is rich in PAs. However, only a handful of species has been used for biochemical and functional studies of these polyphenolic compounds. Among under-examined species is persimmon (Diospyros kaki Thunb.). Although it has not been mentioned in most review articles on PAs (e.g., Haslam 1977; Gu et al. 2004), D. kaki accumulates a large amount of high molecular weight PAs in leaves and fruits (Mallavadhani et al. 1998; Taira 1996). In Japan, this species provided the fifth most consumed fruit in 1996, and its immature fruits have been the source of kaki-shibu, the material rich in soluble PAs utilized in various industrial applications (Taira 1996). Persimmon cultivars can be divided into astringent (A)- and non-astringent (NA)-types based on the amount of soluble PAs in mature fruits, which cause astringency (drying and puckering sensation) in the mouth upon consumption. Soluble PAs in A-type fruits can be removed by ethanol treatment; this produces acetaldehyde, which in turn induces PA polymerization (Tanaka et al. 1994). Insolubilization of PAs during A. thaliana seed development was also shown to be caused by their oxidative polymerization (Pourcel et al. 2005). Hence, PA solubility in persimmon fruits may be negatively correlated with oxidation level as well as the degree of polymerization. Fruits of both A- and NA-types are rich in the soluble PAs in their early developmental stages. The NA-type loses its astringency during the development on the tree and becomes non-astringent with firm flesh. By contrast, the A-type fruit needs to be kept on the tree until it becomes over-ripe and extremely soft, or has to be subjected to pre- or post-harvest treatments, such as that with ethanol as described above, for removal of soluble PAs before human consumption (Taira 1996). A limited number of works has described the composition and structure of PAs in A-type fruits (Matsuo and Itoo 1978; Gu et al. 2008). The presence of soluble 2,3-cis- and -trans-flavan-3-ols in both A- and NA-type D. kaki fruits was also reported (Suzuki et al. 2005). In addition, accumulation of transcripts encoding putative homologs to flavonoid biosynthetic proteins was reported in PA-rich persimmon fruits (e.g., Ikegami et al. 2005a, 2005b, 2007). Overall, however, there has been no systematic study to clearly define compositions of PAs and other phenylpropanoid components of persimmon fruits. Furthermore, the molecular mechanism of PA accumulation in this non-model species remains largely unexplored. This situation may be mainly due to prolonged life cycle and genetic complexity of this hexaploid species.
As part of our horticultural program, we have crossed various cultivars of D. kaki and obtained their progenies segregating for fruit astringency. One of these crosses was made between two genetically distinct NA-types of D. kaki, cv. Luo Tian Tian Shi and cv. Taishu. Their F1 progenies were segregated into A- and NA-types, the latter of which appears to be determined by the presence of a dominant allele derived from cv. Luo Tian Tian Shi (Ikegami et al. 2005b, 2006). In this report, we made use of these materials, attempting to elucidate the molecular mechanism of soluble PA accumulation in young persimmon fruits. We employed suppression subtractive hybridization (SSH) to identify transcripts whose accumulation patterns were distinct between the segregated A- and NA-type fruits when their PA contents became distinct. Interestingly, only a few clones encoding flavonoid biosynthetic enzymes were identified. Among cDNAs represented by multiple clones were those encoding a new member of the 1-Cys peroxiredoxin (Prx) family and that of subgroup F of family 1 glycosyltransferases (GTs). A detailed sequence comparison and phylogenetic analysis revealed that the isolated 1-Cys Prx may be a novel type. In addition, UDP-galactose:anthocyanidin/flavonol 3-O-galactosyltransferase activity of the GT homolog was confirmed by using a bacterial expression system. These results may suggest complex mechanisms of PA accumulation in persimmon fruits.
Materials and methods
Plant materials and RNA preparation
Generation of F1 progenies between the two distinct NA-type persimmon cultivars (D. kaki cv. Luo Tian Tian Shi and cv. Taishu), their growth, and collection of their fruits were partly described by Ikegami et al. (2005b). Fruits usually become fully mature and ready to harvest for human consumption in the end of October to November. Immature green fruits used in this study were harvested considerably earlier, at three different dates (June 12, July 12, and August 3, 2001), during which PA accumulation started to decline in the NA-type but not in the A-type (Taira 1996; see Fig. 2). The A- and NA-types used in this study were defined previously based on the size of PA-accumulating cells and the soluble tannin content of fully mature fruits (Ikegami et al. 2005b). Fruit flesh was separated from seeds and peel, cut into small pieces (ca. 1 cm × 0.7 cm × 0.5 cm), frozen with liquid nitrogen, and stored at −80°C before further analysis. Total RNA was isolated from 5 g of the frozen sample using the hot borate method (Wan and Wilkins 1994).
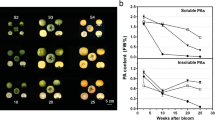
Estimation of PA contents in non-astringent- and astringent-type persimmon fruits at the early developmental stages. Fruits from three independent trees of non-astringent (NA)- and astringent (A)-type persimmon were collected on three different dates (June 12, July 12, and August 3, 2001) and the amount of 80% methanol-soluble and insoluble PAs in these fruits were measured as (+)-catechin equivalent by DMACA method (a) or cyanidin equivalent by n-butanol–HCl (b) method. The bar graphs indicate the average PA contents of NA- (white bars) or A- (black bars) type fruits. Error bars are standard deviations. Asterisks indicate that the differences are significant (P < 0.002)
PA estimation
Ten milligram of freeze-dried-powdered sample was resuspended in 25 ml of 80% methanol and was sonicated for 10 min three times. After centrifugation at 700×g for 15 min at 4°C, the supernatant containing the “soluble PAs” was collected. The fraction containing the “insoluble PAs” was prepared as described by Taira et al. (1998). Briefly, the residue after the 80%-methanol extraction was resuspended in 25 ml of 1% (v/v) HCl in methanol and incubated for 1 h at 60°C, followed by centrifugation at 700×g for 15 min at 4°C. The resultant supernatant was defined as the fraction containing the “insoluble PAs”. For quantifications of PAs, two methods were employed. The first method used p-dimethylaminocinnamaldehyde (DMACA; Li et al. 1996). Briefly, an aliquot of the extract (400 μl) was mixed with 1 ml of 0.1% (w/v) DMACA and 5% (v/v) HCl in methanol, incubated at room temperature for 15 min, and its absorbance at 643 nm was measured. The amount of PAs was calculated as equivalent to (+)-catechin. The second method used n-butanol–HCl as described by Porter et al. (1986). More specifically, 100 μl of the extract was mixed with 1 ml of 5% (v/v) HCl in n-butanol, boiled for 20 min, and its absorbance at 550 nm was measured. In this method, the amount of PAs was calculated as equivalent to cyanidin.
Suppression subtractive hybridization (SSH) and full-length cloning of selected cDNAs
For SSH analysis, RNA samples from eight trees for each type were combined and used as templates for cDNA synthesis with SMART PCR cDNA synthesis kit (Clontech, Mountain View, CA, USA) for the July samples (from 1 μg RNA), or with PCR-Select cDNA Subtraction Kit (Clontech) for the August fruits (from 2 μg RNA), according to the manufacturer’s instructions. Construction of the SSH library and differential screening were performed as previously described (Ikegami et al. 2007). DNA sequences of positive clones were determined with T7 or M13RV primers and DYEnamic ET terminator cycle sequence kit (GE Healthcare, Buckinghamshire, UK) by using an ABI 310 DNA sequencer (Applied Biosystems, Foster, CA, USA).
For full-length cloning of selected cDNAs, RNA of the immature fruit of an A-type persimmon (D. kaki cv. Kuramitsu), harvested on June 18, 2004, was used to generate a cDNA library with ZAP-cDNA synthesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer’s instruction. The library was screened with DIG-labeled probes, which were prepared using PCR DIG Probe synthesis kit (Roche Diagnostics, Basel, Switzerland) with sets of primers that were designed based on the sequences of SSH clones as shown in Supplementary Table S1. The cDNAs obtained from the screening were subcloned into a pBluescript KS vector and their sequences were determined as described above.
qRT-PCR analysis
cDNAs were synthesized with SuperScriptIII transcriptase (Invitrogen, Carlsbad, CA, USA) and an adapter primer from total RNA (1 μg as quantified by UV spectrometer) from fruits of each tree. Sequences of primer pairs used for amplification are listed in Supplementary Table S2. Primers for GST, Prx, and F3GT were designed based on the sequences of the full-coding cDNA clones, which were obtained as described above. Primers for SCPL were from the cDNA sequence reported previously (Ikegami et al. 2007). For DHD/SHD and CHS, primer pairs were designed based on cDNA clones isolated from young fruits of progenies derived from three cultivars (D. kaki cv. Taishu, cv. Aizu-mishirazu, and cv. Fuyu) harvested on July 30, 2004 (Akagi et al. 2009). Primers for PAL and DFR were based on partial clones obtained by another study (Ikegami et al. 2005a) using Primer Express Software (ver 2.1; Applied Biosystems). To design a primer set for ACTIN, a partial clone, which was isolated from young fruits of D. kaki cv. Kuramitsu harvested on July 17, 2007, by using a set of primers containing the consensus sequences (Ushijima et al. 2003), was used.
Each reaction was done with an aliquot of cDNA (0.1 μl) as a template using an ABI PRISM 7900HT (Applied Biosystem) and the SYBR Green system with SYBR Pre-mix Ex-Taq (Takara Bio, Otsu, Japan). The standard amplification protocol consisted of an initial denaturing step at 95°C for 30 s, followed by 40 cycles at 95°C for 10 s, 57°C for 5 s, and 72°C for 15 s. For each transcript, the average threshold cycle was determined according to the manufacturer’s instruction (Applied Biosystems). Standard curves for target genes and the housekeeping gene ACTIN were obtained by amplification of a serially diluted mixture of cDNA samples of six dilutions. For each measurement, independent standard curves were constructed for at least three replications of each sample and two biological replicates. The calculated cDNA copy numbers were normalized to those of ACTIN.
Phylogenetic analysis of Prx and GT
Amino acid sequences were aligned with ClustalX (Thompson et al. 1997). Phylogenetic analyses of the aligned amino acid sequences based on maximum parsimony were implemented in PAUP* (Swofford 2002) with heuristic searches using the TBR branch-swapping algorithm 1,000 random taxon addition replicates and no limit on the number of trees saved. Relative support for clades was assessed using 1,000 bootstrap replicates with 10 random taxon addition replicates per bootstrap replicate.
For the Prx sequences, 191 ambiguously aligned sites (131 from the amino terminus and 60 from the carboxyl terminus) were excluded from the original alignment of 357 sites. A final data set of 166 characters, of which 3 were constant, 6 were variable but parsimony-uninformative, and 157 were parsimony-informative, was subjected to phylogenetic analysis. The tree shown in Fig. 4a is one of four most parsimonious trees (length = 775, CI excluding uninformative characters = 0.8120, RI = 0.9124), arbitrarily rooted along a strongly supported internal branch.
For the GT sequences, the complete alignment of 610 amino acid characters, of which 61 characters were constant, 62 were variable but parsimony-uninformative, and 487 were parsimony-informative, was used in phylogenetic analysis. The unrooted phylogram shown in Fig. 5a is one of three most parsimonious trees (length = 6,700, CI excluding uninformative characters = 0.5902, RI = 0.4810).
Bacterial production and characterization of the DkFGT protein
The DkFGT-coding sequence in pBluescript KS vector was isolated by digesting the plasmid with EcoRI and XhoI, and transferred into a pGEX-KG vector (Guan and Dixon 1991). The resultant plasmid (pGEX-KG-DkFGT) encodes an in-frame fusion of the DkFGT protein with a glutathione S-transferase (GST) tag in the N-terminus, and was transformed into BL21(DE3)pLysS cells (Invitrogen). The transformants were pre-cultured at 37°C for 16 h in Luria Broth (LB) media containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol. Three millilitre of the preculture was transferred to the fresh LB media (300 ml) containing the two antibiotics, and incubated further at 37°C until its absorbance at 600 nm reached 0.2, when isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.3 mM. The induced culture was further incubated at 16°C for 48 h. The cells were harvested by centrifugation at 4,000 rpm at 4°C for 15 min, and stored at −80°C. Collected cells were lysed with PBS containing 1% Triton X-100 (PBST) and the insoluble bacterial debris was removed by centrifugation at 15,000 rpm for 15 min at 4°C. After dithiothreitol was added to the resulting supernatant to a final concentration of 1 mM, the soluble fraction was loaded onto a column containing 3 ml of glutathione–agarose (Sigma, St. Louis, MO, USA) that was pre-equilibrated with PBST. After the column was washed with 45 ml of PBST, the bound recombinant protein was eluted with 15 ml of the elution buffer (50 mM Tris–HCl, pH 9.5, and 50 mM glutathione). The eluted fraction was collected, its proteins were concentrated with Y30 membrane concentrator (Amicon, Billerica, MA, USA) and glycerol was added to a final concentration of 10% (v/v) before being stored at −80°C.
GT assay
Three anthocyanidins (cyanidin, delphinidin, and pelargonidin) were purchased from Indofine Chemicals (Hillsborough, NJ, USA). Three flavonols (quercetin, kaempferol, and myricetin), two flavan 3-ols [(+)-catechin and (−)-epicatechin], two flavonoid galactosides (cyanidin 3-galactoside and quercetin 3-galactoside), dihydroquercetin, and two UDP-sugars [UDP-galactose (UDP-Gal) and UDP-glucose (UDP-Glc)] were from Sigma-Aldrich (St. Louis, MO, USA). A standard reaction mixture (50 μl) contained 100 mM Tris–HCl (pH 8.35), 75 mM KCl, 100 μM MgCl2, 1 mM DTT, 5% glycerol, 1.56–400 μM of flavonoid substrates, 5 mM UDP-sugars, and an aliquot of the solution containing the recombinant protein (2.2–6.6 μg for flavonols or 0.53–1.6 μg for anthocyanidins). After incubation at 30°C (for anthocyanidins) or 50°C (for flavonols) for 3–30 min, the reaction was stopped by addition of 100 μl of 1% HCl/methanol (for anthocyanidins) or methanol (for flavonols). For analysis of temperature optima, the reaction mixture contained 0.5 mM quercetin and 2.5 mM UDP-Gal in 100 mM Tris-buffer (pH 8.0), which was incubated at 50°C for 30 min. For analysis of pH optima, the reaction was done with 0.5 mM quercetin and 2.5 mM UDP-Gal at 50°C for 3 min in following buffers of 100 mM: sodium acetate, pH 4.0, 4.5, 5.0, and 5.5; MES-KOH, pH 5.0, 5.5, 6.0. 6.5, and 7.0; potassium phosphate, pH 6.4, 7.0, 7.4, ad 8.0; and Tris–HCl, pH 7.5, 8.0, 8.5, 9.0, and 9.5.
For analysis of the reaction, the mixture was filtered through a PVDF membrane of 0.22 μm, and subjected to a high-performance liquid chromatography (HPLC) with a reversed-phase YMC-ODSA column (4.6 mm i.d.. × 150 mm; YMC, Kyoto, Japan) and 2% acetic acid (buffer A) and methanol (buffer B) as eluent with various gradient conditions at a flow rate of 1 ml/min, and the detection was done by photodiode array. The linear gradient condition used for anthocyanidins, anthocyanidin galactosides, and dihydroquercetin was: 0–5 min, 5% B; 5–30 min, 5–50% B; 30–32 min, 50–100% B; 32–37 min 100% B; and 37–39 min 100–5% B. The gradient used for flavonols and their galactosides was 0–5 min, 10% B; 5–30 min, 10–25% B; 30–35 min, 25–80% B, 35–36 min 80–100% B; 36–40 min 100% B; and 40–41 min 100–10% B. Flavan 3-ols were separated by using another linear gradient: 0–5 min, 5% B; 5–30 min, 5–25% B; 30–32 min, 25–100% B; 32–37 min 100% B; and 37–39 min 100–5% B. Detection of anthocyanidins and their galactosides was done at absorbance at 520 nm, flavonols at 360 nm, and flavan 3-ols and dihydroquercetin at 280 nm. Cyanidin 3-O-galactoside and quercetin 3-O-galactoside in the in vitro assay mixture were identified by co-elution with the standard samples and the absorption spectra of the peaks. To determine kinetic values (K m and V max), the reaction mixture containing 100 μM quercetin (for those for UDP-Gal) or 5 mM UDP-Gal (for those for acceptors) was incubated for 3 min (for anthocyanidins) or 5 min (for flavonols).
Results
SSH analyses between A- and NA-type persimmon fruits in their early developmental stages identify several transcripts encoding putative homologs to known proteins
The astringency of A-type fruits and that of NA-type fruits became distinct in those harvested in August. That is, the soluble PA content per dry weight of A-type fruits remained almost constant from June to August, whereas that of NA-type fruits dropped by more than 50% (Fig. 2). By contrast, during this period, (1) the concentrations of insoluble PAs remained largely unchanged and at comparable levels between the two fruit types (Fig. 2); and (2) fresh weight of a fruit of both types increased similarly up to five times (data not shown). These data suggest constant PA biosynthesis in A-type, but not in NA-type fruits. It may also be possible that the level of flavonoid oxidation, which has been shown to be negatively correlated with PA solubility (Pourcel et al. 2005), was lower in A-type than in NA-type. Hence, we decided to compare the transcript accumulation patterns in NA- and A-type fruits harvested in July and August. To identify differentially accumulating transcripts, we used suppression subtractive hybridization (SSH), which recently allowed us to isolate several transcripts including those encoding PA-biosynthetic enzymes whose accumulation levels were up- or down-regulated in persimmon fruits by ethanol treatment to remove astringency (Ikegami et al. 2007). We prepared RNAs from A- and NA-type fruits and generated reciprocal cDNA SSH libraries (A–NA and NA–A, respectively) for samples of each data point. Among 4,800 recombinant clones that were randomly selected from these libraries, 37 clones showed significantly different accumulation patterns between the two fruit types by RNA dot blot assays, and their nucleotide sequences were determined (Table 1). Based on the previous results (Ikegami et al. 2007), we had expected to find a number of PA-biosynthetic genes in A–NA libraries. Indeed, a total of nine independent clones involved in phenylpropanoid metabolism were isolated (Table 1). However, none of them encodes an enzyme catalyzing one of the committed steps for PA biosynthesis, i.e., LAR or ANR. Some other clones identified in these libraries encode homologs to known proteins that do not play a direct role for PA biosynthesis (Table 1). We also attempted to obtain full-length coding sequences of SSH clones isolated from A–NA libraries in order to facilitate further analysis. To this end, we screened a cDNA library from A-type persimmon fruits, and were able to obtain apparent full-length sequences for a subset of cDNAs (Table 1).
The presence of clones for the three flavonoid biosynthetic enzymes, phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), and dihydroflavonol 4-reductase (DFR), in A–NA libraries was consistent with the preliminary RNA-blotting data (Ikegami et al. 2005b). Also identified in A-NA libraries were clones for two other flavonoid biosynthetic enzymes, cinnamic acid 4-hydroxylase (C4H) and flavonoid 3′5′-hydroxylase (F3′5′H). Three truncated sequences (AB436539–436541) may derive from non-overlapping portions of a single mRNA encoding a C4H homolog (we could not obtain its full-length sequence). In addition, three SSH clones from the July sample were found to encode 3-dehydroquinate dehydratase/shikimate 5-dehydrogenase (DHD/SHD), which is involved in the biosynthesis of aromatic amino acids to fuel the phenylpropanoid pathway. Finally, represented by the highest number of clones for both the July and August samples were the transcripts encoding a protein with high sequence identities to various plant flavonoid GTs, such as UDP-Glc:anthocyanidin 3-O-GlcT from flower buds of Lobelia erinus L. (GenBank accession number: BAF49311), anthocyanin 3-O-GalT from cell suspension culture of Aralia cordata Thunb. (BAD06514; Nagashima et al. 1994), and kaempferol 3-O-GalT from pollen of Petunia x hybrida (Q9SBQ8; Miller et al. 1999).
The group of cDNAs not directly involved in phenylpropanoid metabolism consists of a total of eight independent clones. Three of them were found in A–NA libraries, encoding proteins similar to glucose acyltransferase (serine carboxylpeptidase-like protein = SCPL), 1-Cys peroxiredoxin (Prx), and glutathione S-transferase (GST), respectively. SCPL might be involved in PA accumulation, since correlation of its gene expression and accumulation of PAs has been reported at least twice, one in persimmon fruits (Ikegami et al. 2007) and another in hairy roots of grape overproducing MYB transcription factors (Terrier et al. 2009). The clone encoding a 1-Cys Prx homolog was represented by five SSH clones, whereas GST, which was shown to have multiple functions including conjugation to anthocyanidins in the cytoplasm for vacuole sorting (Dixon et al. 2002), was represented by a single clone (Table 1). Among the five cDNAs found in NA–A library, three clones (AB436544, AB436545, and AB46632) may encode non-overlapping portions of a single protein with a high sequence similarity to LATE BLOOMER 1 from pea, which plays roles in photoperiodic flowering, de-etiolation, and circadian regulation (Hecht et al. 2007). The other two (AB436630 and AP436631) appear to encode distinct chitinases, among which is one for a class II chitinase-like protein, which was represented by five SSH clones.
Differential accumulation of most but not all clones isolated by SSH analyses was confirmed by quantitative RT-PCR analysis
To validate the results obtained by A–NA SSH libraries, we performed quantitative reverse-transcriptase-mediated (qRT)-PCR to analyze accumulation patterns of a selected set of transcripts (Fig. 3). Although it was not identified by the SSH analyses, we included ANR for the assay because of its importance for PA biosynthesis (Xie et al. 2003). Consistent with the SSH results, transcript accumulation was found to be 10 times (for GST and DHD/SHD; Fig. 3c, d), 7.7 times (for FGT; Fig. 3e), 5 times (for SCPL and Prx; Fig. 3a, b), and 1.7 times (for CHS; Fig. 3 h) higher in A-type fruits than that in NA-type fruits harvested in July (P < 0.02). In fruits collected at the later stage (August), transcripts encoding FGT and PAL, which were identified in the A-NA SSH library (Table 1), were present in A-type fruits at levels 17 and 3 times higher, respectively, than in NA-type fruits (Fig. 3e, g; P < 0.01). Furthermore, transcripts of two additional clones that were not identified in the August A–NA SSH library, i.e., those for SCPL and Prx, were found to accumulate in A-type fruits to levels 7 and 26 times higher, respectively, than in NA-type fruits (Fig. 3a, b; P < 0.002). By contrast, in July and August, the levels of DFR transcript represented by one SSH clone at each of these time points (Table 1) were not significantly different between A- and NA-type fruits (Fig. 3f; P > 0.5). Thus, our current SSH analyses present both false negatives (for SCPL and Prx in August samples) and false positives (for DFR in both July and August samples). Finally, consistent with the results of the SSH analyses, we could not detect significant differences in the level of ANR transcript between A- and NA-type fruits examined in this study (Fig. 3i; P > 0.02).
Accumulation of a subset of transcripts in non-astringent and astringent-type persimmon at the early developmental stages. Accumulation of a subset of transcripts identified by SSH analysis (Table 1) as well as a that of ANR was examined by real time PCR with RT products from fruits of three independent non-astringent (NA)- and astringent (A)-type persimmon trees collected on three different time points (June 12, July 12, and August 3, 2001). The bar graphs indicate the average level of amplified cDNA, normalized to the amount in NA-type fruits of June 12, in NA- (white bars) or A- (black bars) type fruits. Error bars are standard deviations. An asterisk and double asterisks indicate that P values to indicate the significance of differences between the transcript levels in NA- and A-type are below 0.002 and 0.02, respectively
In addition to validating the SSH results, the qRT-PCR analysis revealed two interesting trends (Fig. 3). First, the level of all transcripts examined decreased by 68–99% during development of NA-type fruits (from June to August; P < 0.03). A similar tendency was found for A-type fruits although the rate of decrease was lower (29–91%) than that for the NA-type (P < 0.06), except for the FGT-coding transcript, which appeared to remain at comparable levels throughout fruit development (Fig. 3e; P > 0.22). Second, for the fruits harvested in June, a transcript encoding one of the flavonoid biosynthetic proteins, DFR, was found to be 100% higher in NA- than in A-type fruits (P < 0.005; Fig. 3f, h), whereas the amount of PAs in A-type fruits was comparable to or even higher than that in NA-type in June and July (Fig. 2). Hence, transcriptional level of DFR appears to have a negative correlation with soluble PA accumulation in the young persimmon fruits.
Among proteins encoded by identified clones, we decided to further examine Prx and FGT, because transcripts for both proteins were highly represented in A–NA SSH libraries (Table 1), and were also shown by qRT-PCR to accumulate to significantly higher levels in A-type fruits than in NA-type fruits (Fig. 3b, e). In particular, we wished to test if FGT acts on the PA precursor, flavan-3-ols, and plays a direct role in PA accumulation, similar to the recently identified GlcT, UGT72L1 (Pang et al. 2008). In the following sections, these two proteins are designated as DkPrx and DkFGT, respectively.
Comparative and phylogenetic analysis of the DkPrx amino acid sequence suggests that it is a novel member of the plant 1-Cys peroxiredoxin family
Prxs are ubiquitous antioxidant enzymes that use thiols to reduce various potentially toxic peroxides such as hydrogen peroxide, alkyl hydroperoxides, and peroxinitrite (Dietz 2003). In plants, Prxs consist of four groups (1-Cys Prx, 2-Cys Prx, PrxQ, and Type-II Prx), and have been implicated in antioxidation, cellular signaling pathways, and redox regulation (Dietz et al. 2006). DkPrx is apparently similar to 1-Cys Prxs, whose gene expression and/or protein accumulation was found in germinating seeds (Haslekas et al. 1998; Stacy et al. 1996; Lewis et al. 2000) and also in roots exposed to arsenic (Requejo and Tena 2005). As an initial approach to predict the function of DkPrx, we compared its deduced amino acid sequence with those of all the Prxs identified so far from three model plant species, A. thaliana, rice, and poplar (Dietz et al. 2006; Gama et al. 2007; See Supplementary Table S3). As shown in the phylogenetic tree (Fig. 4a), DkPrx was clearly grouped in the 1-Cys Prx family. The amino acid sequence alignment (Fig. 4b) revealed that in addition to the catalytic cysteine residue (Cys-47 in DkPrx) conserved in all the Prx proteins analyzed, the 1-Cys Prx proteins contain another cysteine residue (Cys-73 in DkPrx) unique to this group. Interestingly, DkPrx appears to lack the basic residues that were implicated in nuclear localization of barley 1-Cys Prx (Stacy et al. 1999) and which are also found in other homologs (Fig. 4c). Taken together, our results suggest that DkPrx represents a new and divergent member of the 1-Cys Prx proteins.
Phylogenetic analysis of peroxiredoxins from various plant species. a One of four most parsimonious trees from phylogenetic analysis of partial amino acid sequences of peroxiredoxins listed in Table S3. Numbers above branches are bootstrap support values >50%. Asterisks indicate branches that collapsed in the strict consensus tree. Six distinct groups are indicated on the right. b Alignment of amino acid sequences in the middle region of 1-Cys Prxs (top six) and one protein each from five other groups in A. thaliana as shown in A (bottom five). The catalytic cysteine residue conserved in all the Prxs is indicated with an asterisk, whereas the residue conserved only in 1-Cys Prx is indicated with a plus symbol. The second cysteine residues conserved in PrxQ and type II Prx (Rouhier and Jacquot 2002) are indicated as white characters on a black background. c Alignment of C-terminal region of 1-Cys Prxs. Basic residues, which may play roles in nucleus-targeting, are indicated as white characters on a black background
Amino acid sequence analysis shows that DkFGT belongs to group F of family 1 GT and may catalyze galactosylation of flavonoid at C-3 position
Based on their amino acid sequences, family 1 GT in plants can be divided into 14 subgroups, all of which contain a conserved 44-amino-acid-long stretch called a plant secondary product glycosyltransferase (PSPG) box (Bowles et al. 2005; Gachon et al. 2005), which was shown to interact with UDP-sugar by three-dimensional structure analyses (Shao et al. 2005; Offen et al. 2006; Li et al. 2007). In addition, a previous detailed sequence comparison and a site-directed mutagenesis study showed that the sugar-donor specificity would be partly determined by the last amino acid residue in the PSPG box, i.e., Gln for UDP-Glc and His for UDP-Gal (Kubo et al. 2004). To gain a first clue for the function of DkFGT, we aligned its deduced amino acid sequence with those of various plant GTs available in the database (Supplementary Table S5). An unrooted phylogenetic tree was generated (Fig. 5a), which showed that DkFGT belongs to a subclade F (Bowles et al. 2005; Gachon et al. 2005) with seven other sequences, six of which have already been shown to have anthocyanidin/flavonoid 3-O-GT activities, i.e., flavonol 3-O-rhamnosyltransferase (UGT78D1; Jones et al. 2003), flavonoid 3-O-GlcT (UGT78D2; Tohge et al. 2005) and flavonoid 3-O-arabinosyltransferase (UGT78D3; Yonekura-Sakakibara et al. 2008) from A. thaliana (Tohge et al. 2005), and three flavonoid 3-O-galactosyltransferase (F3GalT) from petunia (Miller et al. 1999), Aralia cordata (Nagashima et al. 2004), and Vigna mungo L. (Mato et al. 1998). This group is distinct from group E, which includes the enzyme involved in PA accumulation, UGT72L1 (Pang et al. 2008), and group L, which contains FGTs with less strict region-specificity (Tian et al. 2006).
Phylogenetic analysis and comparison of glycosyltransferases from various plant species. a One of three most parsimonious trees from phylogenetic analysis of complete amino acid sequences of GT listed in Table S4. Numbers along branches are bootstrap support values >50%. Asterisks indicate branches that collapsed in the strict consensus tree. Fourteen groups (A–N) defined by Bowles et al. (2005) are indicated. b Alignment of the regions near C-termini of GTs encompassing the Plant Secondary Product Glycosyltransferase (PSPG) box. The top panel figure was generated with Weblogo (Crooks et al. 2004), based on the sequences analyzed in a. The bottom panel shows the sequence alignment of group F proteins. Residues conserved in at least five proteins are shown as white characters on a black background. The histidine residue conserved among FGalTs is indicated with an asterisk in the bottom. Three conserved residues unique to this group are indicated with arrows
Various GTs were compared in the regions corresponding to the PSPG boxes. Figure 5b shows two sets of alignments. The first set (top panel) includes all the sequences used to make the unrooted tree in Fig. 5a, presented as a Weblogo format (Crooks et al. 2004), whereas the second set (bottom panel) lists sequences of group F proteins. DkFGT was found to contain the PSPG box, whose last residue is His (at residue 378), which is conserved among GalTs and also the recently identified arabinosyltransferase (UGT78D3; Yonekura-Sakakibara et al. 2008), instead of Gln in GlcTs. Finally, the persimmon protein contains three additional amino acids appear to be conserved between the F-group proteins, i.e., Val-349, Val-359, and Arg-372 (indicated with arrows in the bottom panel of Fig. 5b). These data indicate that DkFGT is the new member of subgroup F of family 1 GT, and it may catalyze galactosylation at 3-O-position of flavonoid substrates.
Biochemical characterization of DkFGT
To test catalytic activities of DkFGT, we produced it with an N-terminal GST-tag in E. coli. The soluble recombinant protein was further bound to and eluted from a glutathione–agarose column. The final fraction contained a protein of 77 kD, which corresponds well with the predicted size of GST-DkFGT (77.4 kD), as well as proteins of smaller sizes detectable after Coomassie Blue staining on SDS-PAGE (Fig. 6a, lane 2). Since all these proteins immunoreacted with the anti-GST antisera (data not shown), we assumed that the 77-kD protein was the intact GST-DkFGT protein, and the smaller ones were truncated forms containing GST. Using this fraction, we first tested whether the recombinant protein indeed has an UDP-hexose-dependent flavonoid GT activity. As shown in Fig. 6b, the fraction containing GST-DkFGT, but not the control prepared from E. coli cells containing the expression vector, converted a flavonol (quercetin) to a new compound with the presence of UDP-Gal (compare top and middle panels). The elution time and spectra of the new peak matched those of the authentic quercetin-3-O-galactoside. When UDP-Glc was used as a substrate, by contrast, no new peak was detected (Fig. 6b, bottom panel). The optimum temperature and pH of the quercetin 3-O-GalT activity of the recombinant protein were found to be 50°C (Fig. 6c) and 8.75 (Fig. 6d), respectively. We also tested the GalT activity of GST-DkFGT against a subset of potential acceptor substrates (see Fig. 1 for their structures). An anthocyanidin (cyanidin) was efficiently converted to its 3-O-galactoside (Fig. 6e), whereas the activity against other three substrates, dihydroquercetin, (−)-epicatechin, and (+)-catechin, was under the detection limit in the current assay (Fig. 6f).
Biochemical characterization of DkFGT. a Production of GST-DkFGT by E. coli. Proteins in the soluble fraction from E. coli transformed with pGEX-KG-DkFGT (lane 1) and those in the fraction bound to and eluted from glutathione agarose column (lane 2) were separated by SDS-PAGE and visualized by Coomassie blue staining. The 77-kD band, which cross-reacted with the αGST antibody by immunoblotting (data not shown), and thus corresponded to the intact GST-DkFGT, is indicated with an arrow. The smaller bands of 30–40 kD, which also cross-reacted with the αGST antibody, are indicated with an asterisk. The black line separates images from different portions of the same gel. b HPLC elution profiles of GT assay mixtures with various combinations of proteins and substrates. The reactions were done for 75 min at 30°C. GST protein extracts purified with a glutathione column from E. coli transformed with pGEX-KG, GST-DkFGT fraction 2 in a, Q quercetin, Q3Gal quercetin 3-O-galactoside, K (IC) kaempferol (as an internal control). c Temperature optimum of DkFGT activity. d pH optimum of DkFGT activity. e, f HPLC elution profiles of GT assay mixtures. The reactions were done with GST-DkFGT for 5 min (e), or with recombinant proteins indicated for 15 min (f)
Kinetic properties of GST-DkFGT were further examined for both the donor and acceptor substrates. By using quercetin (100 μM), the dependence of activity upon the concentration of UDP-Gal was analyzed between 0.125 and 8 mM. An apparent hyperbolic saturation curve was obtained, from which Lineweaver–Burk transformation gave a K m for UDP-Gal of 1.43 mM and V max for the formation of quercetin 3-O-galactoside of 17.5 nkat/mg protein. Similarly, when the UDP-Gal concentration was fixed at 5 mM, K m for quercetin was calculated to be 49 μM, and V max was 20 nkat/mg protein. As for the cyanidin-GalT activity of the recombinant protein, its K m and V max were estimated to be 55 μM and 6 nkat/mg protein, respectively. We also tested GalT activity of GST-DkFGT against two flavonols (kaempferol and myricetin) and two anthocyanidins (pelargonidin and delphinidin; see Fig. 1 for their structures). All the reactions resulted in a production of a single compound although we could not measure their amounts due to the lack of available authentic galactosides. Based on the area of the peak corresponding to the products, we could estimate the K m values for kaempferol, myricetin, pelargonidin and delphinidin as 143, 14, 186, and 19 μM, respectively. These data clearly indicate that DkFGT is a novel anthocyanidin/flavonol 3-O-GalT with a preference for substrates with a high number of hydroxyl groups in B-ring.
Discussion
PAs benefit various aspects of human life, most notably, health. Recent molecular genetic and biochemical studies with model plant species have greatly advanced our understanding of the biosynthesis of these important compounds at the molecular level (Dixon et al. 2005). Diospyros kaki is a PA-rich species, but it has been under-examined mainly due to its complex genetic background and prolonged life cycle. The biochemical and molecular mechanisms underlying PA-accumulation in D. kaki have remained unexplored, although astringency of certain fruits is known to be due to the presence of soluble PAs, whose formation appears to be negatively correlated with the degree of oxidative polymerization of flavan-3-ols. In this work, we attempted to shed some light on the mechanism of PA accumulation in this non-model species. Among possible mechanisms, underlying sustained accumulation of soluble PAs in A-type but not in NA-type persimmon fruits is transcriptional regulation of the biosynthesis of these flavonoid compounds. In this scenario, transcripts encoding PA biosynthetic enzymes, such as LAR and ANR, should be expressed more highly in A-type than in NA-type fruits. However, our initial transcriptome analysis by SSH failed to identify clones encoding these proteins. This might be due to false negative results, indicating the limitation of the current strategy. However, qRT-PCR analysis showed that the levels of transcripts for ANR were largely comparable between A- and NA-type fruits (Fig. 3i). Thus, PA biosynthesis in these persimmon fruits is more likely regulated at the posttranscriptional level. Rapid turnover of soluble PAs in NA-type fruits and/or slow turnover of soluble PAs in A-type fruits are another possible scenario.
The present work demonstrates that the abundance of transcripts encoding 1-Cys Prx (DkPrx) and flavonoid 3-O-GalT (DkFGT) correlates with the accumulation of soluble PAs in young persimmon fruits. DkPrx identified in this study is the first member of 1-Cys Prx isolated from fruits of any plant species. It is also unique in that it lacks the C-terminal nuclear-targeting domain, although its biochemical properties remain unexamined. DkFGT is the fourth group F GalT, whose catalytic activities have been examined. Its pH optimum (8.75) is slightly higher than those of GalTs from Vigna mungo (8.0; Mato et al. 1998) and petunia (7.5; Miller et al. 1999). Its affinities to UDP-Gal and flavonoid substrates are about one-third and one-tenth, respectively, of those of petunia GalT (Miller et al. 1999). Furthermore, the substrate preference of DkFGT for the three flavonols, which is in the order of myricetin > quercetin > kaempferol, is distinct from that of the petunia enzyme, which showed a preference of quercetin = kaempferol > myricetin. Thus, DkFGT may represent a novel class of flavonoid GalT. Since none of other GalT has been examined for its activities against anthocyanidins, however, it remains unknown whether the comparable activity towards the two distinct types of flavonoids (anthocyanidins and flavonols) is unique to DkFGT.
To our knowledge, involvement of either 1-Cys Prx or flavonoid 3-O-GalT in PA accumulation has never been shown in any plant systems. Do these proteins indeed play roles in the biosynthesis and/or accumulation of PAs in persimmon fruits? Or, conversely, does accumulation of PAs induce their gene expression? On one hand, Pourcel et al. (2005) demonstrated that oxidation of PAs by a laccase-type flavonoid oxidase leads to their insolubilization. As an antioxidant, DkPrx may prevent this reaction, and thus have a positive effect on accumulation of soluble PAs. On the other hand, it is known that persimmon fruits accumulate PAs in specialized cells (Taira 1996). Formation of these cells may somehow increase oxidative stress, and expression of the DkPRX gene may be induced to prevent the oxidative damage of the plant tissues.
As for DkFGT, we originally speculated that it might be directly involved in PA biosynthesis by acting on the PA precursor, flavan-3-ols, similar to the case with the recently identified GlcT UGT72L1 (Pang et al. 2008). However, our biochemical study revealed that it may be involved in the biosynthesis of galactosides of anthocyanidins and/or flavonols, instead of those of PA subunits. It has been suggested that flavonol galactosides play roles in pollen germination in petunia (Miller et al. 1999). The presence of such compounds has also been reported in leaves and calyx of D. kaki (Mallavadhani et al. 1998), but not from fruit flesh. Our preliminary attempts to detect flavonoid galactosides in D. kaki fruits failed (A. Ikegami, unpublished). Thus, it remains inconclusive if DkFGT plays a role in accumulation of anthocyanidin and/or flavonol galactosides in persimmon fruits. Another possibility is that DkFGT may be involved in feed-back regulation of PA biosynthesis. Previous studies showed that quercetin could inhibit key enzymes in PA biosynthesis, PAL and ANR, in vitro (Xie et al. 2004; Sarma and Sharma 1999). Similarly, lack of DkFGT in NA-type fruits may result in excess flavonol aglycones, inhibiting catalytic activity of the PA biosynthetic enzymes.
Because of the lack of molecular genetic tools, it will not be easy to test the above hypotheses with D. kaki. Nonetheless, the results of the current work should encourage further biochemical studies of this non-model species. It would also be interesting to test if and how 1-Cys Prx and FGalT homologs are involved in PA accumulation in model species, such as A. thaliana.
Abbreviations
- ANR:
-
Anthocyanidin reductase
- A-type:
-
Astringent-type
- C4H:
-
Cinnamic acid 4-hydroxylase
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol 4-reductase
- DHD/SHD:
-
Dehydratase/shikimate 5-dehydrogenase
- DMACA:
-
p-Dimethylaminocinnamaldehyde
- F3′5′H:
-
Flavonoid 3′5′-hydroxylase
- FGT:
-
Flavonoid glycosyltransferase
- GalT:
-
Galactosyltransferase
- GlcT:
-
Glucosyltransferase
- GST:
-
Glutathione S-transferase
- GT:
-
Glycosyltransferase
- LAR:
-
Leucoanthocyanidin reductase
- NA-type:
-
Non-astringent-type
- PA:
-
Proanthocyanidin
- PAL:
-
Phenylalanine ammonia lyase
- Prx:
-
Peroxiredoxin
- PSPG:
-
Plant secondary product glycosyltransferase
- qRT-PCR:
-
Quantitative reverse-transcriptase-mediated PCR
- SCPL:
-
Serine carboxylpeptidase-like protein
- SSH:
-
Suppression subtractive hybridization
References
Akagi T, Ikegami A, Suzuki Y, Yoshida J, Yamada M, Sato A, Yonemori K (2009) Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kaki Thunb.) fruit. Planta. doi:10.1007/s00425-009-0991-6
Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361
Bowles D, Isayenkova J, Lim EK, Poppenberger B (2005) Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8:254–263
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Debeaujon I, Peeters AJM, Leon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant J 13:853–871
Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147:2041–2053
Dietz KJ (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54:93–107
Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I (2006) The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57:1697–1709
Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol 3:REVIEWS3004
Dixon RA, Xie D-Y, Sharma SB (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165:9–28
Gachon CM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10:542–549
Gama F, Keech K, Eymery F, Finkemeier I, Gelhaye E, Gardeström P, Dietz KJ, Rey P, Jacquot J-P, Rouhier R (2007) The mitochondrial type II peroxiredoxin from poplar. Physiol Plant 129:196–206
Gu L, Kelm MA, Hammerstone JF, Beecher G, Holden J, Haytowitz D, Gebhardt S, Prior RL (2004) Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J Nutr 134:613–617
Gu H, Li C, Xu Y, Hu W, Chen M (2008) Structural features and antioxidant activity of tannin from persimmon pulp. Food Res Intern 41:208–217
Guan KL, Dixon JE (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem 192:262–267
Haslam E (1977) Symmetry and promiscuity in procyanidin biochemistry. Phytochemistry 16:1625–1640
Haslekas C, Stacy RA, Nygaard V, Culianez-Macia FA, Aalen RB (1998) The expression of a peroxiredoxin antioxidant gene, AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Mol Biol 36:833–845
Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJ, Weller JL (2007) Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 144:648–661
Ikegami A, Kitajima A, Yonemori K (2005a) Inhibition of flavonoid biosynthetic gene expression coincides with loss of astringency in pollination-constant, non-astringent (PCNA)-type persimmon fruit. J Hort Sci Biotech 80:225–228
Ikegami A, Sato A, Yamada M, Kitajima A, Yonemori K (2005b) Expression of genes involved in proanthocyanidin biosynthesis during fruit development in a Chinese pollination-constant, nonastringent (PCNA) persimmon, ‘Luo Tian Tian Shi’. J Am Soc Hort Sci 130:830–835
Ikegami A, Eguchi S, Sato A, Yamada M, Kitajima A, Mitani N, Yonemori K (2006) Segregations of astringent progenies in the F1 populations derived from crosses between a Chinese pollination-constant non-astringent (PCNA) ‘Luo Tian Tian Shi’, and Japanese PCNA and pollination-constant, astringent (PCA) cultivars. HortScience 41:561–563
Ikegami A, Eguchi S, Kitajima A, Inoue K, Yonemori K (2007) Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci 172:1037–1047
Jones P, Messner B, Nakajima J, Schäffner AR, Saito K (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278:43910–43918
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:237–242
Kubo A, Arai Y, Nagashima S, Yoshikawa T (2004) Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch Biochem Biophys 429:198–203
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonids. Annu Rev Plant Biol 57:405–430
Lewis ML, Miki K, Ueda T (2000) FePer 1, a gene encoding an evolutionarily conserved 1-Cys peroxiredoxin in buckwheat (Fagopyrum esculentum Moench), is expressed in a seed-specific manner and induced during seed germination. Gene 246:81–91
Li YG, Tanner G, Larkin P (1996) The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric 70:89–101
Li L, Modolo LV, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X (2007) Crystal structure of Medicago truncatula UGT85H2—insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferase. J Mol Biol 370:951–963
Mallavadhani UV, Panda AK, Rao YR (1998) Pharmacology and chemotaxonomy of Diospyros. Phytochemistry 49:901–951
Marinova K, Pourcel L, Lucille L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M (2007) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19:2023–2038
Mato M, Ozeki Y, Itoh Y, Higeta D, Yoshitama K, Teramoto S, Aida R, Ishikura N, Shibata M (1998) Isolation and characterization of a cDNA clone of UDP-galactose: flavonoid 3-O-galactosyltransferase (UF3GaT) expressed in Vigna mungo seedlings. Plant Cell Physiol 39:1145–1155
Matsuo T, Itoo S (1978) The chemical structure of kaki-tannin from immature fruit of the persimmon (Diospyros kaki L.). Agric Biol Chem 42:1637–1643
Miller KD, Guyon V, Evans JN, Shuttleworth WA, Taylor LP (1999) Purification, cloning, and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J Biol Chem 274:34011–34019
Nagashima S, Okamoto A, Suzuki H, Asada Y, Kondo T, Yoshikawa T (2004) Anthocyanin galactosyltransferase from Aralia cordata, cDNA cloning and characterization. Plant Biotech 21:191–195
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114
Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25:1396–1405
Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA (2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105:14210–14215
Porter LJ, Hrstich LN, Chan BG (1986) The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry 25:223–230
Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17:2966–2980
Requejo R, Tena M (2005) Proteome analysis of maize roots reveals that oxidative stress is a main contributing factor to plant arsenic toxicity. Phytochemistry 66:1519–1528
Rouhier N, Jacquot JP (2002) Plant peroxiredoxins: alternative hydroperoxide scavenging enzymes. Photosynth Res 74:259–268
Sarma AD, Sharma R (1999) Purification and characterization of UV-B induced phenylalanine ammonia-lyase from rice seedlings. Phytochemistry 50:729–737
Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17:3141–3154
Stacy RA, Munthe E, Steinum T, Sharma B, Aalen RB (1996) A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol 31:1205–1216
Stacy RAP, Nordeng TW, Culianez-Macia FA, Aalen RB (1999) The dormancyrelated peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. Plant J 19:1–8
Suzuki T, Someya S, Hu F, Tanokura M (2005) Comparative study of catechin compositions in five Japanese persimmons (Diospyros kaki). Food Chem 93:149–152
Swofford DL (2002) paup*: phylogenetic analysis using parsimony (*and Other Methods) ver4.0b10. Sinauer, Sunderland
Taira S (1996) Astringency in persimmon. In: Linskens HF, Jackson JF (eds) Fruit analysis. Springer, Berlin, pp 97–110
Taira S, Matsumoto N, Ono M (1998) Accumulation of soluble and insoluble tannins during fruit development in nonastringent and astringent persimmon. J Jpn Soc Hort Sci 67:572–576
Tanaka T, Takahashi R, Kouno I, Nonaka G (1994) Chemical evidence for the de-astringency (insolubilization of tannins) of persimmon fruit. J Chem Soc Perkin Trans 1:3013–3022
Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278:31647–31656
Terrier T, Torregrosa L, Ageorges A, Vialet S, Verries C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in Vitis vinifera L. and suggests additional targets in the pathway. Plant Physiol 149:1028–1041
Thompson JD, Gibson F, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian L, Blount JW, Dixon RA (2006) Phenylpropanoid glycosyltransferases from osage orange (Maclura pomifera) fruit. FEBS Lett 580:6915–6920
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito K (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15:771–781
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299:396–399
Xie DY, Sharma SB, Dixon RA (2004) Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch Biochem Biophys 422:91–102
Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20:2160–2176
Yu O, Jez JM (2008) Nature’s assembly line: biosynthesis of simple phenylpropanoids and polyketides. Plant J 54:750–762
Acknowledgments
We thank Professor L. Tian, all the members of the Inoue laboratory, and two anonymous reviewers for giving insightful and useful comments on this work. This project was partly supported by the Grant-in-Aid for Japan Society for the Promotion of Science Fellows, The Ministry of Education, Culture, Sports, Science and Technology, Japan (to AI) and United States National Science Foundation Award #DEB-0515431(to D.P.). The Inoue laboratory has been supported by Energy Biosciences Program at the United States Department of Energy (award #DE-FG02-08ER15963).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Contributions from Ayako Ikegami and Takashi Akagi to this work are considered equal.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ikegami, A., Akagi, T., Potter, D. et al. Molecular identification of 1-Cys peroxiredoxin and anthocyanidin/flavonol 3-O-galactosyltransferase from proanthocyanidin-rich young fruits of persimmon (Diospyros kaki Thunb.). Planta 230, 841–855 (2009). https://doi.org/10.1007/s00425-009-0989-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-0989-0