Abstract
Objective
Adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP) is caused by mutations in CSF1R. Pathogenic mutations in exons 12–22 including coding sequence of the tyrosine kinase domain (TKD) of CSF1R were previously identified. We aimed to identify CSF1R mutations in patients who were clinically suspected of having ALSP and to determine the pathogenicity of novel CSF1R variants.
Methods
Sixty-one patients who fulfilled the diagnostic criteria of ALSP were included in this study. Genetic analysis of CSF1R was performed for all the coding exons. The haploinsufficiency of CSF1R was examined for frameshift mutations by RT-PCR. Ligand-dependent autophosphorylation of CSF1R was examined in cells expressing CSF1R mutants.
Results
We identified ten variants in CSF1R including two novel frameshift, five novel missense, and two known missense mutations as well as one known missense variant. Eight mutations were located in TKD. One frameshift mutation (p.Pro104LeufsTer8) and one missense variant (p.His362Arg) were located in the extracellular domain. RT-PCR analysis revealed that the frameshift mutation of p.Pro104LeufsTer8 caused nonsense-mediated mRNA decay. Functional assay revealed that none of the mutations within TKD showed autophosphorylation of CSF1R. The p.His362Arg variant located in the extracellular domain showed comparable autophosphorylation of CSF1R to the wild type, suggesting that this variant is not likely pathogenic.
Conclusions
The detection of the CSF1R mutation outside of the region-encoding TKD may extend the genetic spectrum of ALSP with CSF1R mutations. Mutational analysis of all the coding exons of CSF1R should be considered for patients clinically suspected of having ALSP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP) is an autosomal dominant neurological disorder that predominantly affects the cerebral white matter [1]. ALSP encompasses two similar entities previously known as hereditary diffuse leukoencephalopathy with spheroids (HDLS) and pigmentary orthochromatic leukodystrophy (POLD) [1]. ALSP is clinically characterized by executive dysfunction, memory decline, personality changes, motor impairments, and seizures [2]. Frontal lobe syndrome in ALSP is characterized by loss of judgment, lack of social inhibitors, lack of insight, and motor persistence, which usually appears early in the disease course. The mean age at onset is 43 years ranging from 18 to 78 years [3]. Patients with ALSP eventually become bedridden with a mean disease duration of 6.8 years from onset to death [3].
Mutation in the colony stimulating factor 1 receptor (CSF1R) was identified as the cause of ALSP [4]. To date, 58 pathological mutations in CSF1R have been identified [3]. All reported mutations were found in exons 12−22 including the coding sequence of the tyrosine kinase domain (TKD) of CSF1R [1,2,3]. We previously reported that CSF1R mutation-mediated pathogenesis may be explained by haploinsufficiency or the loss of CSF1R-mediated signals [5].
In this study, we attempted to identify the CSF1R mutations in patients who were referred to our institute for genetic analysis. By this analysis, we found novel and previously reported CSF1R mutations including a novel frameshift in exon 4 outside of TKD and examined the pathogenicity of novel CSF1R variants.
Methods
Patients
Sixty-one patients from 59 pedigrees who fulfilled the diagnostic criteria of probable or possible ALSP were included in this study [6]. All the patients or their relatives provided their informed consent. This study was approved by the institutional review board committees of Niigata University and Mayo Clinic Florida.
Genetic and mRNA analyses
Genomic DNA and total RNA were isolated from peripheral leukocytes using standard methods. Mutational analysis of CSF1R was performed using sequences of both strands of all PCR-amplified coding exons and flanking intronic sequences as previously described [4, 5]. Briefly, all the exons and exon–intron boundaries of CSF1R were amplified by PCR, followed by Sanger sequence. We conducted in silico analysis using the PolyPhen-2, SIFT, PROVEAN, VEP and CADD algorithms to predict the pathogenicity of novel missense or indel mutations. Total RNA was extracted from peripheral leukocytes. Complementary DNA was synthesized using ReverTra Ace® (TOYOBO, Osaka, Japan).
Functional and immunoblot analyses
Mutagenesis was performed to generate cDNA of mutant CSF1R as previously described [5]. HEK293T cells were transiently transfected with wild-type or mutant CSF1R, and were cultured in a medium containing 10% fetal bovine serum (FBS). In another set of experiments, HEK293T cells were stimulated with the ligands of CSF1R, CSF1, and IL-34, in the absence of FBS to induce autophosphorylation of CSF1R. Detergent cell lysates were subjected to immunoblot analysis using antibodies including C-20 (C-terminus of CSF1R, Santa Cruz Biotechnology, Dallas, TX, USA) and B-8 (N-Terminus of CSF1R, Santa Cruz Biotechnology, Dallas, TX, USA) to detect total CSF1R. CSF1R phosphorylated at Tyr546, Tyr708, and Tyr723 was detected using specific anti-phosphorylated CSF1R antibodies (Cell Signaling Technology, Beverly, MA, USA).
Clinical and neuroimaging analyses
Clinical presentations and neuroimaging findings of the patients carrying CSF1R variants were retrospectively evaluated. MRI was conducted for routine diagnostic purposes using 1.5T systems in all the patients (n = 11). CT using thin slices was performed in seven patients.
Neuropathological examination
Neuropathological examination of brain samples from patient 10 was performed as previously described [7]. Briefly, tissue sections were embedded in paraffin, and 5-µm-thick sections were mounted on glass slides for histopathological and immunohistochemical analyses. The paraffin-embedded sections were stained with hematoxylin and eosin (H&E) and Luxol fast blue. Sections were also stained with antibodies against CD68, phosphorylated neurofilaments, and αB-crystallin.
Statistical analysis
The signal intensity of immunoblots was semiquantitatively analyzed using a LAS 4000 analyzer (GE Health Science, Piscataway, NJ, USA). The intensity of each autophosphorylation signal of CSF1R was normalized to the total amount of CSF1R. All statistical tests were run in GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA). Data are presented as mean ± SEM. Statistical analysis was performed by two-way ANOVA with Bonferroni correction.
Results
Identification of CSF1R mutations
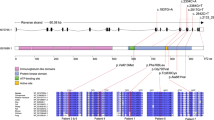
Eight missense variants including five novel mutations (p.Ile662Thr, p.Asp778Glu, p.Ile794Phe, p.Pro878Ser, and p.Pro878Ala), two known missense mutations (p.Gly765Asp and p.Ile794Thr), one known homozygote variant (p.His362Arg), and two frameshift mutations (p.Pro104LeufsTer8 and p.Tyr886GlnfsTer55) in CSF1R were identified (Fig. 1). Eight mutations were located in TKD. One frameshift mutation (p.Pro104LeufsTer8) was located in the extracellular domain and was predicted to cause nonsense-mediated RNA decay (NMD) [8]. Results of in silico analysis of CSF1R variants in ALSP patients revealed that these mutations within TKD are predicted to be pathogenic with high probability (Electronic Supplementary Table 1). Furthermore, these mutations were not found in the ExAC database, supporting the pathogenicity of these mutations, except for p.Ile794Thr, which is the most frequently reported CSF1R mutation (Electronic Supplementary Table 1). The p.His362Arg variant located outside TKD appeared to be nonpathogenic, because the allele frequency of p.His362Arg was 0.049 in the ExAC database.
Illustration of CSF1R structure and location of mutations. The positions of the variants identified in this study are indicated by arrows. The phospho-specific antibodies used in this study against CSF1R are also depicted. The position of each domain is shown on the basis of information from UniProt (http://www.uniprot.org/uniprot/P07333)
Analysis of mRNA expression of frameshift CSF1R mutations
We performed reverse transcription (RT)-PCR analysis using mRNA obtained from peripheral leukocytes to examine the mRNA expression of the frameshift mutations of p.Pro104LeufsTer8 and p.Tyr886GlnfsTer55. The analysis revealed that the expression level of the mutant allele derived from the p.Pro104LeufsTer8 mutation was markedly lower than that of the normal allele (Fig. 2a). This finding suggests that the p.Pro104LeufsTer8 frameshift undergoes NMD. On the other hand, the mutant allele derived from the p.Tyr886GlnfsTer55 frameshift mutation was expressed at a level comparable to that of the wild-type allele (Fig. 2b).
mRNA expression of frameshift CSF1R mutations. a Frameshift mutation in exon 4 (p.Pro104LeufsTer8), which was predicted to undergo NMD. The expression level of the mutant allele was markedly decreased, suggesting that this frameshift mutation results in NMD of mutant mRNA. b Frameshift mutation in exon 21 (p.Tyr886GlnfsTer55), which was predicted not to undergo NMD because the premature terminal codon is generated in exon 22, the last exon of CSF1R. The expression level of the mutant allele was comparable to that of the wild-type allele
Defective autophosphorylation of mutant CSF1R
To examine the pathogenicity of missense variants, we examined the autophosphorylation of CSF1R in HEK293T cells expressing wild-type or mutant CSF1R. CSF1R phosphorylated at Tyr546, Tyr708, and Tyr723 was detected in cells expressing wild-type CSF1R cultured in the presence of FBS (Fig. 3a). No autophosphorylation of CSF1R was observed in cells expressing missense variants including p.Ile662Thr, p.Gly765Asp, p.Asp778Glu, p.Ile794Phe, p.Pro878Ser, or p.Pro878Ala in TKD, and p.Tyr886GlnfsTer55 (Fig. 3a). The p.His362Arg variant located in the extracellular domain showed a comparable degree of autophosphorylation of CSF1R to the wild type (Fig. 3a).
Functional assay of mutant CSF1Rs. a HEK293T cells transfected with the wild type or mutant CSF1R were cultured in a medium containing 10% FBS. Western blot analysis using anti-CSF1R antibodies (B-8 and C-20) revealed comparable expression levels of total CSF1R between the wild-type and mutant CSF1Rs. Note that mutant CSF1R with Tyr886GlnfsTer55 lacking the C-terminal portion of CSF1R showed no band reactive to the anti-C-terminal antibody (C-20), but was recognized as a slightly smaller molecular weight by the anti-N-terminal antibody (B-8). None of the variants within TKD revealed autophosphorylation of CSF1R. The p.His362Arg variant showed a comparable degree of autophosphorylation of CSF1R to the wild type. The anti-β-actin antibody was used as a loading control. b Ligand-dependent autophosphorylation of CSF1R was examined in cells transiently transfected with the wild type or variant CSF1Rs. Detergent-extracted lysates were collected at the indicated time (min) after CSF1 stimulation. Increased levels of phosphorylated CSF1R were observed to be comparable in cells expressing the wild type and p.His362Arg. In contrast, neither of the mutants within TKD underwent autophosphorylation of CSF1R following CSF1 stimulation. c Results of semiquantification of immunoblot data (n = 3). There is no statistically significant difference in the degree of ligand-dependent autophosphorylation between the wild type and the p.His362Arg variant. Data are presented as mean ± SEM. d Ligand-dependent autophosphorylation of CSF1R. Cells were stimulated with CSF1 (left panel) or IL-34 (right panel). Detergent-extracted lysates were collected 20 min after ligand stimulation. Phosphorylated CSF1R levels were comparable between cells expressing wild-type CSF1R and p.His362Arg, whereas cells expressing mutant CSF1Rs within TKD showed no autophosphorylation
In another set of experiments, ligand-induced autophosphorylation of CSF1R was examined. None of the missense mutations within TKD or the frameshift mutation p.Tyr886GlnfsTer55 showed autophosphorylation upon the stimulation of CSF1 or IL-34 (Fig. 3b, d). The p.His362Arg variant showed a comparable degree of autophosphorylation of CSF1R to the wild type, suggesting that this variant does not cause the loss of CSF1R-mediated signals (Fig. 3b, c, d).
Clinical and neuroimaging characteristics
The clinical characteristics of 11 patients are shown in Table 1. Familial occurrence was observed in three patients. The case of a sibling of patient 4 with the p.Gly765Asp mutation was previously reported [5]. Eight patients apparently had no family history of the disease. The mean age at onset of the patients was 42.8 years ranging from 22 to 60 years. Dementia was observed in all the patients, and pyramidal signs were observed in 6 (55%) of the 11 patients.
Neuroimaging features are shown in Electronic Supplementary Table 2. The white matter lesions detected by MRI were usually distributed bilaterally, but could be distributed asymmetrically (Fig. 4, Electronic Supplementary Fig. 1). Hyperintensity lesions on diffusion-weighted MR images were observed in 8 of the 11 patients (73%). Calcifications were observed in six (86%) of the seven patients by CT (Electronic Supplementary Table 2, Electronic Supplementary Fig. 2).
Brain MRI and CT findings of ALSP patient with the frameshift mutation in exon 4. Characteristic hyperintensities in the white matter are observed in fluid-attenuated inversion recovery MR images (upper panels). Diffusion-weighted MR images revealed high-intensity lesions with left-side predominance (middle panels). CT scan revealed small spotty calcifications in the frontal and parietal white matter (lower panels)
Neuropathological findings
Patient 10 was a 64-year-old man from family 6510 and his depression and anxiety insidiously developed at the age of 57. Subsequently, he exhibited cognitive decline and personality/behavioral changes. At the age of 62 years, he developed parkinsonism characterized by rigidity, bradykinesia, and gait disturbance. He became totally dependent by the age of 63 years and died at 64.
The autopsy examination revealed marked frontal white matter pathology with enlargement of the frontal horn of the lateral ventricle and thinning of the corpus callosum as the gross findings (Fig. 5a). The cerebral white matter showed myelin loss on Luxol fast blue staining (Fig. 5b). There were pigment-laden macrophages visible on H&E staining and the pigment was autofluorescent (Fig. 5c, f, g). Immunohistochemical analysis using an anti-phosphorylated neurofilament antibody revealed axonal spheroids (Fig. 5d). Numerous ballooned neurons were detected in the frontal cortex (Fig. 5e). These neuropathological findings were consistent with ALSP [7].
Neuropathological findings of ALSP patient with p.Pro878Ala mutation. a Coronal section of cerebrum showing frontal white matter pathology. b Frontal white matter myelin loss on Luxol fast blue staining (note the relative sparing of subcortical arcuate fibers). c Macrophages in frontal white matter (CD68 immunohistochemistry). d Axonal spheroids in frontal white matter (phosphorylated neurofilament immunohistochemistry). e Ballooned neurons in frontal cortex (αB-crystallin immunohistochemistry). f Macrophages in white matter with brown granular pigment (arrows) (H&E stain). g Pigment in macrophages in white matter showing autofluorescence. Bar in a = 1 cm; bar in b = 200 µm; bars in c, d, and e = 50 µm; bars in f and g = 20 µm
Discussion
In this study, we identified seven novel and two previously reported mutations in CSF1R among patients who fulfilled the diagnostic criteria of ALSP [6]. This study provides several noteworthy insights relevant to the role of CSF1R mutations in ALSP.
First, we identified a novel frameshift mutation caused by single-nucleotide deletion (c.310delC) in exon 4 resulting in p.Pro104LeufsTer8 located outside of TKD. All the previously reported mutations in ALSP were found within exons 12−22 including the coding sequence of TKD [3,4,5]. The frameshift mutation p.Pro104LeufsTer8 generates a premature stop codon, which was predicted to cause NMD [8]. Our expression analysis revealed that the level of mRNA expression derived from the mutant allele was substantially decreased as predicted (Fig. 2a). These findings suggest that the p.Pro104LeufsTer8 mutation causes ALSP owing to the haploinsufficiency of CSF1R.
Second, we identified another novel frameshift mutation caused by two-nucleotide deletion (c.2655_2656delAT) in exon 21 resulting in p.Tyr886GlnfsTer55. This frameshift mutation does not fulfill the criterion of NMD because the premature stop codon is generated within exon 22, the last exon of CSF1R. Our expression analysis revealed that the mRNA expression level of the mutant allele was comparable to that of the wild-type allele (Fig. 2b). These findings suggest that this frameshift mutation does not cause CSF1R haploinsufficiency. Because this mutation is located within TKD, we examined whether the mutant CSF1R of p.Tyr886GlnfsTer55 affects autophosphorylation upon ligand stimulation. Our examination revealed that the mutant CSF1R of p.Tyr886GlnfsTer55 did not show autophosphorylation (Fig. 3a, b, d). Thus, the pathogenic mechanism caused by p.Tyr886GlnfsTer55 mutation appears to be the loss of CSF1R-mediated signals.
Third, we showed that the autophosphorylation of novel missense variants (p.Ile662Thr, p.Asp778Glu, p.Ile794Phe, p.Pro878Ser, and p.Pro878Ala) within TKD was impaired. These findings suggest that these mutants within TKD are the causal factors [4, 5]. As previously reported [3], pathogenic mutations identified in this study occur more frequently in the distal part of TKD than in the proximal part. There is apparently no significant difference in the degree of autophosphorylation impairment between the p.Ile662Thr mutant within the proximal part of TKD and other mutants within the distal part of TKD.
Finally, we identified the homozygous CSF1R variant of p.His362Arg located in the extracellular domain in patient 2 who fulfilled the possible criteria of ALSP. We considered that the p.His362Arg missense variant is not the causative factor for the following reasons. First, the degree of autophosphorylation of this variant was comparable to that of the wild type. Second, this variant was reported in the ExAC database to have a frequency of 0.049 in the general population and 0.368 in the East Asian population. Thus, the cause of leukoencephalopathy in this patient is not likely to be this CSF1R variant. Recently, mutations in AARS2 have been reported in ALSP patients who lack CSF1R mutations [9]. We performed genetic analysis of AARS2 and found no causative mutation in this patient.
The clinical and neuropathological findings with CSF1R mutations in this study were consistent with those reported previously [1,2,3, 7]. Indeed, all the patients in this study fulfilled possible or probable diagnostic criteria for ALSP [6]. The MRI findings of the patients were similar to those reported previously (Supplementary Table 2) [10]. Attention should be paid to patient 1 in whom signal changes detected by MRI were observed with marked left-side predominance (Fig. 4). Brain calcifications were observed in all the patients with CSF1R mutation, supporting the diagnostic value of this finding for ALSP [11].
In conclusion, the detection of the CSF1R mutation outside of exons 12−22 may extend the genetic spectrum of ALSP with CSF1R mutations. Mutational analysis of all the exons of CSF1R should be considered for patients clinically suspected of having ALSP.
References
Nicholson AM, Baker MC, Finch NA et al (2013) CSF1R mutations link POLD and HDLS as a single disease entity. Neurology 80:1033–1040
Lynch DS, Jaunmuktane Z, Sheerin UM et al (2016) Hereditary leukoencephalopathy with axonal spheroids: a spectrum of phenotypes from CNS vasculitis to parkinsonism in an adult onset leukodystrophy series. J Neurol Neurosurg Psychiatry 87:512–519
Konno T, Yoshida K, Mizuno T et al (2017) Clinical and genetic characterization of adult-onset leukoencephalopathy with axonal spheroids and pigmented glia associated with CSF1R mutation. Eur J Neurol 24:37–45
Rademakers R, Baker M, Nicholson AM et al (2012) Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet 44:200–205
Konno T, Tada M, Tada M et al (2014) Haploinsufficiency of CSF-1R and clinicopathologic characterization in patients with HDLS. Neurology 82:139–148
Konno T, Yoshida K, Mizuta I et al (2018) Diagnostic criteria for adult-onset leukoencephalopathy with axonal spheroids and pigmented glia due to CSF1R mutation. Eur J Neurol 25:142–147
Baba Y, Ghetti B, Baker MC et al (2006) Hereditary diffuse leukoencephalopathy with spheroids: clinical, pathologic and genetic studies of a new kindred. Acta Neuropahol 111:300–311
Kuzmiak HA, Maquat LE (2006) Applying nonsense-mediated mRNA decay research to the clinic: progress and chalenges. Trends Mol Med 12:306–316
Lynch DS, Zhang WJ, Lakshmanan R et al (2016) Analysis of mutations in AARS2 in a series of CSF1R-negative patients with adult-onset leukoencephalopathy with axonal spheroids and pigmented glia. JAMA Neurol 73:1433–1439
Sundal C, Van Gerpen JA, Nicholson AM et al (2012) MRI characteristics and scoring in HDLS due to CSF1R gene mutations. Neurology. 79:566–574
Konno T, Broderick DF, Mezaki N et al (2017) Diagnostic value of brain calcifications in adult-onset leukoencephalopathy with axonal spheroids and pigmented glia. AJNR Am J Neuroradiol 38:77–83
Acknowledgements
We would like to thank the patients and their relatives for participating in this study and Ms. Reiko Kawai for her technical assistance. Dr. Konno is supported by JSPS Overseas Research Fellowships and the gift from Carl Edward Bolch, Jr., and Susan Bass Bolch. Dr. Wszolek is partially supported by the NIH/NINDS P50 NS072187, NIH/NIA (primary) and NIH/NINDS (secondary) 1U01AG045390-01A1, Mayo Clinic Center for Regenerative Medicine, Mayo Clinic Neuroscience Focused Research Team (Cecilia and Dan Carmichael Family Foundation, and the James C. and Sarah K. Kennedy Fund for Neurodegenerative Disease Research at Mayo Clinic in Florida), the gift from The Sol Goldman Charitable Trust, and Donald G. and Jodi P. Heeringa. Dr. Ikeuchi is supported by JSPS KAKENHI Grant number JP16H01331 and 26117506, AMED under Grant number JP18kk0205009, and MHLW Grant number 18062640.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical standard
The study was approved by the ethics committee of Niigata University.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Miura, T., Mezaki, N., Konno, T. et al. Identification and functional characterization of novel mutations including frameshift mutation in exon 4 of CSF1R in patients with adult-onset leukoencephalopathy with axonal spheroids and pigmented glia. J Neurol 265, 2415–2424 (2018). https://doi.org/10.1007/s00415-018-9017-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-018-9017-2