Abstract
Background
The out-of-hospital cardiac arrest (OHCA) in the young may be associated with a genetic predisposition which is relevant even for genetic counseling of relatives. The identification of genetic variants depends on the availability of intact genomic DNA. DNA from autopsy may be not available due to low autopsy frequencies or not suitable for high-throughput DNA sequencing (NGS). The emergency medical service (EMS) plays an important role to save biomaterial for subsequent molecular autopsy. It is not known whether the DNA integrity of samples collected by the EMS is better suited for NGS than autopsy specimens.
Material and methods
DNA integrity was analyzed by standardized protocols. Fourteen blood samples collected by the EMS and biomaterials from autopsy were compared. We collected 172 autopsy samples from different tissues and blood with postmortem intervals of 14–168 h. For comparison, DNA integrity derived from blood stored under experimental conditions was checked against autopsy blood after different time intervals.
Results
DNA integrity and extraction yield were higher in EMS blood compared to any autopsy tissue. DNA stability in autopsy specimens was highly variable and had unpredictable quality. In contrast, collecting blood samples by the EMS is feasible and delivered comparably the highest DNA integrity.
Conclusions
Isolation yield and DNA integrity from blood samples collected by the EMS is superior in comparison to autopsy specimens. DNA from blood samples collected by the EMS on scene is stable at room temperature or even for days at 4 °C. We conclude that the EMS personnel should always save a blood sample of young fatal OHCA cases died on scene to enable subsequent genetic analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Every year in the European population, 67–170 out-of-hospital cardiac arrest (OHCA) cases per 100,000 are reported. Of those, 19–97 per 100,000 receive out-of-hospital resuscitation attempts (OHRAs) by the emergency medical services (EMS) [1]. Among these patients of all age groups unrecognized cardiac conditions such as coronary heart disease, cardiomyopathies or different arrhythmogenic diseases may lead to sudden cardiac death (SCD) [2]. However, the incidence of these conditions is not precisely known for several reasons [3].
Especially in the young, SCD is associated with higher proportion of arrhythmogenic or structural heart diseases, which may be related to genetic predispositions [4, 5]. However, the identification of genetic forms of SCD needs next-generation sequencing technologies (NGSs) which can only be applied if non-degraded biological material for extraction of high-quality DNA is available [6, 7]. The combination of autopsy and genetic analysis of fatal OHCA cases allows the identification of genetic forms of SCD in the young [5]. Due to low autopsy frequencies in several countries, a high proportion of these cases remain unexamined (see for an overview [4, 8]). This may have consequences on health of others in affected families [2, 4, 8]. Moreover, biological materials for the isolation of genomic DNA (gDNA) may be not suitable for high-throughput DNA sequencing because of postmortem degradation processes [9, 10] leading to fragmented gDNA at autopsy [11–14].
The recent European Resuscitation Council guidelines from 2021 (ERC-2021) do not consider the collection of blood samples from fatal OHCA cases by the EMS on scene. Instead, it is recommended to collect biomaterial from autopsy [15]. However, the minority of OHCA cases are autopsied, and autopsy biomaterial may be not suitable for NGS. Especially formalin-fixed paraffin-embedded (FFPE) tissue will result in comparably low-quality sequencing results. For these reasons, blood samples from fatal OHCA cases collected by the EMS on scene may play a deciding role to allow at least molecular autopsy by specialized cardiogenetic services if autopsies are not performed [16].
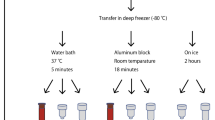
In this study, we determined the DNA integrity number (DIN) of the gDNA isolated from autopsy specimens and compared the data to gDNA extracted from blood collected by the EMS (Fig. 1). Moreover, we investigated different storage conditions for the blood samples with later gDNA extraction to simulate real-life conditions for handling blood samples by the EMS.
Study overview (compare also Table 1 for details)
Material and methods
Study cohort
We included in total 42 hospital and forensic autopsy cases of deceased individuals aged 2–76 years. In total, 172 samples from hospital and forensic autopsies derived from heart (n = 40), lung (n = 15), kidney (n = 17), cartilage (n = 17), fibrous connective tissue (n = 16), bone (n = 16), skeletal muscle (n = 25), and blood (n = 26) were collected. Samples were taken at postmortem intervals (PMIs) between 14 and 168 h and stored at − 80 °C until gDNA extraction. All but one autopsy case was cooled until examination. For comparison, the EMS collected blood samples from 14 fatal OHCA cases at scene who were later autopsied were included (for baseline data, see Table 1, Figs. 1 and 2). For experiments simulating real-life conditions of blood collection by the EMS, we have also included samples of 20 anonymous blood donors.
Inclusion criteria for autopsy samples
We included individuals who died, the time of death was known, and an autopsy was performed. As an autopsy subgroup fatal OHCA cases were included to compare biomaterial collected by the EMS at scene with samples collected during autopsy.
Blood samples for experimental testing
We also analyzed under experimental conditions blood samples isolated from 20 anonymous blood donors to reveal the effects of different storage conditions on the DNA stability. Blood was incubated up to 2 weeks at different temperatures and time intervals. Incubation was carried out at 4 °C, room temperate (RT), 32 °C, and 42 °C, and aliquots for gDNA isolation were frozen at − 22 °C at days 0, 1, 2, 3, 4, 5, 7, 10, and 14. For each time point, five biological replicates were analyzed. Five samples of the blood donors were also analyzed for DNA stability after repeated freeze-thaw cycles (0, 3×, 6×, 9×, 12×; see Supplements sFig. 1). All blood samples were collected in ethylenediaminetetraacetic acid (EDTA) containing plasma tubes (S-Monovette®, Sarstedt, Nümbrecht, Germany). Autopsy specimens were directly frozen on dry ice during examination. Samples included here were not used for genotyping in this study.
Extraction of gDNA from biomaterials
The gDNA was extracted using the High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. To isolate gDNA from tissues, 30 mg (w/w) of specimens were incubated overnight with lysis buffer and Proteinase K at 55 °C. The gDNA was eluted in 50 μl elution buffer and stored at − 20 °C for further analysis. The concentration was measured with the NanoDrop 2000. The technical scatter of gDNA isolation on the DNA integrity was analyzed in triplicate.
Determination of DNA integrity
DNA integrity was analyzed using the TAPEStation2200 (Agilent Technologies, Santa Clara, CA, USA). DNA stability was given as DNA integrity number (DIN) ranging from 0 to 10 with 10 representing no degraded gDNA. Approximately 5–150 ng/μl of gDNA was applied on a Genomic DNA ScreenTape (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer’s instructions. The DIN is calculated by an automated algorithm, which determines the fragmentation of a gDNA sample by assessing the distribution of signal across the size range [17]. We regarded a DIN of 7 as a cutoff value for reliable NGS as recommended by the manufacturer [17].
To estimate the technical and biological scatter, we isolated gDNA in triplicates of three tissue specimens and determined the DINs by TapeStation analysis.
PCR amplification of specific target sites in selected cardiac genes
We further validated the DIN analysis by PCR amplification of gDNA with different integrity values [18]. For PCR fragment analysis, single amplicons of four different genes were used (NEXN, TTN, TNNI3, and MYL2). These genes may carry DNA variants in genetic forms of cardiomyopathies and are located at chromosomal sites known to be sensitive for hydrolytic DNA cleavage [19]. PCR amplicons of these chromosomal sites were designed with different amplicon sizes (for MYL2-Ex.6 it was 206bp, TNNI3-Ex.7 381bp, TTN-Ex.27.1 430bp, and NEXN-Ex.12 529bp) for parallel analysis in one lane (primer sequences are available upon request).
Statistics
For statistical analysis GraphPad Prism v9.4.1 for Mac was used (GraphPad Software, San Diego, CA, USA; www.graphpad.com). Medians are given within the manuscript including the upper and lower quartiles. In box and whiskers plots, boxes represent the quartiles and whiskers extend from 10 to 90 percentiles. Statistical significance was analyzed for multiple comparisons by Kruskal-Wallis or pairwise by Mann-Whitney test for non-Gaussian distributed data, where appropriate.
Results
Extraction of gDNA
We included biomaterial for DNA integrity measurements of 42 autopsy cases with a median age at death of 39 years (range 2–77 years; Fig. 2; Table 1). The median PMI at autopsy was 64 h (range 14–168 h; Fig. 2). The yield for gDNA extraction was highest in samples from blood (median 44; 26.8–74.9 μg/mL) and lowest in fibrous connective tissue (median 61; 29–94 ng/mg tissue) given that blood has a specific weight of 1.05 mg/mL [20] (Fig. 3; Table 2). Repeated freeze-thaw cycles had no influence on the DNA integrity (see sFig. 1).
Violin plot of gDNA extraction yields of solid tissues (left Y-axis) and blood (red, right Y-axis) derived from autopsies. gDNA isolated from autopsy blood provides by far the highest yields of gDNA (Kruskal-Wallis test for multiple comparisons; medians are given as solid, quartiles as dotted lines)
The DNA integrity does not directly depend on the postmortem interval
The mean technical scatter for TapeStation analysis was 3.6% and 4.5% for combined gDNA isolation and TapeStation analysis (Supplements, sFig. 2).
We correlated the DNA integrity against the PMI in different tissues from 42 autopsies. We measured the DIN in specimens from 40 hearts, 15 lungs, 17 kidneys, 17 cartilage tissues, 16 fibrous connective tissues, 16 bones, 25 skeletal muscles, and 26 blood samples. As a reference, we measured the DIN also in blood samples of 14 EMS cases (for details, see Table 2). We found that the DNA integrity is not directly dependent on the PMI but differs considerably between autopsy cases. However, samples derived from skeletal or cardiac muscle appeared to have the lowest scatter of DIN, as compared to all other tissues (Fig. 4). We did not find a correlation between PMI and DIN in autopsy samples by linear regression analysis (Fig. 4). We also compared the DIN of different tissues within the same autopsy case with considerable differences between tissues and PMI. Thus, the DIN depends in an unpredictable manner on the case, the tissue, and the PMI (Supplements, sFig. 3).
DNA integrity plotted against the postmortem interval (PMI) for different tissues from autopsy biomaterial. There is no linear correlation between DNA integrity and PMI (maximum 6 days). Results from linear regression analysis are given as dotted lines (slopes are not significant from zero). Dashed lines indicate cutoff of DIN = 7. A Heart; B skeletal muscle; C lung; D kidney; E cartilage; F bone; G fibrous connective tissue; H blood. DIN relative DNA integrity number
gDNA isolated from blood samples collected by the EMS has the highest integrity
We compared the DNA integrity of all biomaterials collected during autopsy and by the EMS (Table 2; Fig. 5). The DIN of blood samples from autopsy (BL-A; median 6.6; 5.3–7.1) and from the EMS (BL-EMS; median 7.6; 7–8.1) were significantly different (Kruskal-Wallis test, p = 0.005). The 25% percentile of the BL-EMS DIN was 7.0 and above the level recommended for NGS analysis. Vice versa 75% of BL-A samples were at or below the cutoff recommended for NGS sequencing (Fig. 5) [17].
Comparison of the DNA integrity number (DIN) derived from autopsy samples including blood and blood samples collected by the emergency medical services (EMS). The DIN of the EMS blood samples from 14 fatal OHCA cases is significantly higher (p = 0.005) as compared to any other tissue except cartilage, bone, and skeletal muscle from autopsy. For the statistical evaluation, the Kruskal-Wallis test for multiple comparisons was used (box and whiskers plots: boxes represent the quartiles, whiskers extend from 10 to 90 percentiles, outliers are given as dots, medians as solid lines and means as crosses)
Notably, among the autopsy biomaterials tested in this study skeletal muscle revealed the highest levels of DIN, which was significantly higher as compared to samples from cardiac, renal, and fibrous connective tissues (Fig. 5).
EDTA blood samples from blood donors provide intact gDNA even for several days under robust storage conditions
We further compared blood samples collected during autopsy and from blood donors under experimental conditions. DINs of samples collected from blood donors plotted against the storage time could be fit by linear regression analysis independently of the storage conditions, revealing degradation of DNA integrity over time (slope significantly different from zero, p < 0.001; see Supplements sFig. 4). In contrast, DINs of blood samples from autopsy plotted against the PMI could not be fitted by regression analysis (slope different from zero in linear regression analysis, p = 0.967; see Supplements sFig. 4). This reveals that the stability of gDNA derived from autopsy blood samples cannot be predicted from the postmortem time.
We observed that the DIN of samples stored at room temperature (RT) remained above the cutoff of 7 for 2 days (Fig. 6). Even after storage of blood samples for 5 days at RT, the DIN was above the median of blood samples collected during autopsy (compare Fig. 5 and Fig. 6). The quality of gDNA could be improved storing the blood samples at 4 °C, yielding a median DIN of 7.6 (7.1–8.2; Fig. 6). The incubation of blood samples at 32 °C accelerated the degradation process, whereas the DNA integrity measured in samples stored at 42 °C was comparable to samples kept at 4 °C (Fig. 6), but the yield dropped considerably (Supplements sFig. 6).
Experimental comparison of the influence of different storage conditions (4 °C, room temperature (RT), 32 °C and 42 °C) on EDTA blood samples from donors or of blood collected during autopsy on DNA integrity, respectively. The DNA integrity number (DIN) is plotted against the blood sample incubation time or postmortem interval (PMI). Blood samples from autopsy are shown as means ± standard error. For donor blood samples stored at different temperatures, the means ± standard deviation is given (see also Supplements sFig. 4)
PCR works also with gDNA of low integrity
We found for all targets PCR amplicons, indicating that targeted Sanger sequencing may work even with fragmented gDNA (DIN range 1–7.7; see Supplements sFig. 5).
Discussion
Sudden cardiac death in the young is a devastating event for relatives. For the EMS, it remains challenging to identify non-ischemic cardiac failure in patient with need for OHRA. It is known that the etiology for cardiac failure differs considerably with patient age. In a Danish nationwide study, coronary artery disease was the main cause of SCD in patients between 1 and 49 years [4]. In young cases of OHCA, a major cause of cardiac arrest may be associated with a considerable burden of genetic etiologies [21]. It is known that in fatal OHRA, the cause of death may be unraveled by autopsy. Autopsy without molecular analysis may fail to identify the cause of death in a considerable high number of cases, i.e., in patients with sudden arrhythmic death [22]. In addition, low DNA integrity due to postmortem decomposition processes can impact the option to perform routine high-throughput NGS. However, the parallel DNA sequencing of a high number of genes or even the whole exome in cases of young fatal OHCA is mandatory [23]. We found that DNA isolated from frozen tissue and even with low DIN can provide PCR amplicons suitable for Sanger sequencing. In families with known genetic predispositions, PCR-based Sanger sequencing may be performed in autopsy material even with low-quality DNA [24]. Whereas in fatal OHCA cases of unknown etiology, hundreds of genes must be sequenced and therefore Sanger sequencing will not be appropriate for molecular autopsy.
The ERC-2021 guidelines recommend consistently to consider an advanced post-resuscitation care including autopsy and molecular analysis of cardiovascular diseases in not precisely selected fatal OHCA cases [1]. In an ideal setting and according to the ERC guidelines, this combined procedure will have an impact on unraveling even rare forms of genetic diseases, which not necessarily have to be inherited in cases of de novo variants [25] or homozygous genotypes [26].
Unfortunately, the rate of autopsies in many countries is notoriously low for several reasons and the PMI may lead to gDNA degradation limiting the extent of possible molecular analysis. In a recent EMS study, about 40% of fatal OHCA cases were not transported to a hospital and about 43% of cases were not autopsied [16]. This shows that the ERC-2021 guidelines do not consider fatal cases who are not transported to a hospital and/or are not transferred to autopsy.
We recently suggested that the EMS has the unique opportunity in young fatal cases of OHCA to collect a blood sample at scene [8]. We showed that the number of fatal OHCA cases with an identified predisposition for cardiac arrest was remarkably higher when the EMS collects a blood sample on scene as compared to those cases identified by autopsy only [16]. For these reasons, the EMS should always save a blood sample for later molecular autopsy in young (1–50 years of age) fatal OHCA cases [16], which is currently not recommended by the ERC guidelines [1].
In EMS practice, it is important to know how stable a blood sample will be for a possible subsequent NGS analysis. Therefore, we analyzed in parallel samples from different tissues derived from autopsies and compared the data to the biomaterial, which was collected by the EMS. We found that gDNA from autopsy had significantly lower DNA integrities depending on tissue and PMI as compared to EDTA blood samples collected by the EMS. We also found that the yields and the DNA integrity differed among autopsy cases in an unpredictable manner, which is in line with problems to calculate the PMI from gDNA fragmentation in forensic medicine (for a review, see [27]). The best DNA integrity but with a low extraction yield was found in samples of the skeletal muscle, which is in concordance with previous work [9, 12, 28], whereas the blood samples collected during autopsy delivered quite heterogenous results.
We also measured the DNA integrity of EDTA blood under challenging experimental storage conditions, simulating real-life conditions of blood sample collection by the EMS. The integrity of the DNA is not influenced by repeated freezing-thawing revealing stability of the genomic DNA under robust handling conditions (see sFig. 1). The gDNA degradation is enzyme dependent, which is reflected by the temperature dependency of its storage conditions. At experimental temperatures of 4 °C or 42 °C, the enzymatic degradation of gDNA is low in comparison to RT or 32 °C, respectively. Since DNA degradation is an enzyme catalyzed process, gDNA remains stable at higher temperature, but the extraction yield at 42 °C was considerably low (Supplements sFig. 6). However, EDTA blood samples incubated for even up to 10 days at 4 °C delivered gDNA with a DIN at 7. Even storage at RT will provide DNA stability for up to 2 days revealing the feasibility of sample handling by the EMS personnel. This further indicates that blood sample collection by the EMS will provide excellent biomaterial for later molecular autopsy—especially in fatal cases of OHCA.
An interdisciplinary network of pathologists, molecular biologists, cardiologists, and geneticists is most effective for unraveling genetic causes of fatal cardiac arrest. However, this network will be most effective by integrating the EMS. The EMS is essential to unravel the medical circumstances of death in fatal OHCA, who are not transported to a hospital and/or not autopsied. Thus, with little efforts, the identification of genetic forms of severe cardiovascular diseases with fatal outcome can be improved.
Conclusions
-
1.
Our data show that in this study, the integrity of gDNA in blood samples collected by the EMS is higher as in any biomaterial from autopsy. We recommend changing future ERC guidelines accordingly: in fatal cases of young OHCAs, the collection of a sample of 2 mL EDTA blood is a safe and reliable way to obtain gDNA for later molecular autopsy, especially of those who are not transported to hospital. This requires no special storage or handling conditions. The collection of a 2 mL blood sample by the EMS should be mandatory in fatal OHCA cases younger than 50 years of age. An adequate blood sample ensures the option of a later targeted consultation of survivors and the relatives, when compared with clinical or pathological findings.
-
2.
If no blood sample collected by the EMS is available and an autopsy is performed, specimen of the skeletal muscle should be considered for gDNA extraction. Since the yield of gDNA extraction is limited, in skeletal muscle, we recommend to isolate 1 cm3 tissue for immediate freezing (− 20 °C or lower). In any case, tissue must not be fixed by formalin if NGS is considered for subsequent investigation.
Limitations
For the determination of DNA stability, we determined the DNA integrity number (DIN) using an established semiautomated chromatography method [17]. We did not perform NGS genotyping in parallel. DINs above 7 are recommended by the NGS manufacturer to generate reproducible sequencing data and therefore used as a technical cutoff in this study. Since the correlation of DIN and NGS data quality has a high scatter, DINs below 7 may nevertheless deliver acceptable sequencing results in selected cases.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BL-A:
-
Blood sample collected during autopsy
- BL-EMS:
-
Blood sample-collected by the emergency medical service
- DIN:
-
DNA integrity number
- DNA:
-
Desoxyribonucleic acid
- EMS:
-
Emergency medical service
- EDTA:
-
Ethylendiaminetetraacetic acid
- ERC:
-
European Resuscitation Council
- FFPE:
-
Formalin-fixed paraffin-embedded tissue
- gDNA:
-
Genomic DNA
- NGS:
-
Next-generation sequencing
- OHCA:
-
Out-of-hospital cardiac arrest
- OHRA:
-
Out-of-hospital resuscitation attempt
- PCR:
-
Polymerase chain reaction
- PMI:
-
Postmortem interval
- RT:
-
Room temperature
- SCD:
-
Sudden cardiac death
References
Grasner JT, Herlitz J, Tjelmeland IBM et al (2021) European Resuscitation Council Guidelines 2021: Epidemiology of cardiac arrest in Europe. Resuscitation 161:61–79. https://doi.org/10.1016/j.resuscitation.2021.02.007
Markwerth P, Bajanowski T, Tzimas I, Dettmeyer R (2021) Sudden cardiac death-update. Int J Legal Med 135:483–495. https://doi.org/10.1007/s00414-020-02481-z
Tseng ZH, Olgin JE, Vittinghoff E et al (2018) Prospective countywide surveillance and autopsy characterization of sudden cardiac death: POST SCD study. Circulation 137:2689–2700. https://doi.org/10.1161/CIRCULATIONAHA.117.033427
Risgaard B, Winkel BG, Jabbari R et al (2014) Burden of sudden cardiac death in persons aged 1 to 49 years: nationwide study in Denmark. Circ Arrhythm Electrophysiol 7:205–211. https://doi.org/10.1161/CIRCEP.113.001421
Bagnall RD, Weintraub RG, Ingles J et al (2016) A prospective study of sudden cardiac death among children and young adults. N Engl J Med 374:2441–2452. https://doi.org/10.1056/NEJMoa1510687
Hertz CL, Christiansen SL, Ferrero-Miliani L et al (2016) Next-generation sequencing of 100 candidate genes in young victims of suspected sudden cardiac death with structural abnormalities of the heart. Int J Legal Med 130:91–102. https://doi.org/10.1007/s00414-015-1261-8
Hellenthal N, Gaertner-Rommel A, Klauke B et al (2017) Molecular autopsy of sudden unexplained deaths reveals genetic predispositions for cardiac diseases among young forensic cases. Europace 19:1881–1890. https://doi.org/10.1093/europace/euw247
Dalgaard CV, Hansen BL, Jacobsen EM et al (2022) Sudden unexplained death versus nonautopsied possible sudden cardiac death: findings in relatives. J Cardiovasc Electrophysiol 33:254–261. https://doi.org/10.1111/jce.15333
Bar W, Kratzer A, Machler M, Schmid W (1988) Postmortem stability of DNA. Forensic Sci Int 39:59–70. https://doi.org/10.1016/0379-0738(88)90118-1
Itani M, Yamamoto Y, Doi Y, Miyaishi S (2011) Quantitative analysis of DNA degradation in the dead body. Acta Med Okayama 65:299–306. https://doi.org/10.18926/AMO/47011
Alaeddini R, Walsh SJ, Abbas A (2010) Forensic implications of genetic analyses from degraded DNA--a review. Forensic Sci Int Genet 4:148–157. https://doi.org/10.1016/j.fsigen.2009.09.007
Hansen J, Lesnikova I, Funder AM, Banner J (2014) DNA and RNA analysis of blood and muscle from bodies with variable postmortem intervals. Forensic Sci Med Pathol 10:322–328. https://doi.org/10.1007/s12024-014-9567-2
Endrullat C, Glokler J, Franke P, Frohme M (2016) Standardization and quality management in next-generation sequencing. Appl Transl Genom 10:2–9. https://doi.org/10.1016/j.atg.2016.06.001
Morlote D, Janowski KM, Siniard RC et al (2019) Effects of improved DNA integrity by punch from tissue blocks as compared to pinpoint extraction from unstained slides on next-generation sequencing quality metrics. Am J Clin Pathol 152:27–35. https://doi.org/10.1093/ajcp/aqz014
Nolan JP, Sandroni C, Bottiger BW et al (2021) European Resuscitation Council and European Society of Intensive Care Medicine Guidelines 2021: post-resuscitation care. Resuscitation 161:220–269. https://doi.org/10.1016/j.resuscitation.2021.02.012
Tiesmeier J, Gaertner A, Homm S et al (2021) The emergency medical service has a crucial role to unravel the genetics of sudden cardiac arrest in young, out of hospital resuscitated patients: Interim data from the MAP-IT study. Resuscitation 168:176–185. https://doi.org/10.1016/j.resuscitation.2021.07.042
Diessl N, Ernst U, Schulz A, Wolf S (2018) Quality control in Illumina sequencing workflows using the TapeStation system. Agilent Technologies, Application Note Genomics
Preuner S, Danzer M, Proll J et al (2014) High-quality DNA from fingernails for genetic analysis. J Mol Diagn 16:459–466. https://doi.org/10.1016/j.jmoldx.2014.02.004
Kumar R, Nagpal G, Kumar V, Usmani SS, Agrawal P, Raghava GPS (2019) HumCFS: a database of fragile sites in human chromosomes. BMC Genomics 19:985. https://doi.org/10.1186/s12864-018-5330-5
Oestreich C (1931) Über das spezifische Gewicht des Blutes und seiner Bestandteile. Klin Wochenschr 10:160–162
Isbister JC, Nowak N, Butters A et al (2021) "Concealed cardiomyopathy" as a cause of previously unexplained sudden cardiac arrest. Int J Cardiol 324:96–101. https://doi.org/10.1016/j.ijcard.2020.09.031
Campuzano O, Allegue C, Partemi S, Iglesias A, Oliva A, Brugada R (2014) Negative autopsy and sudden cardiac death. Int J Legal Med 128:599–606. https://doi.org/10.1007/s00414-014-0966-4
Ingles J, Bagnall RD, Semsarian C (2018) Genetic testing for cardiomyopathies in clinical practice. Heart Fail Clin 14:129–137. https://doi.org/10.1016/j.hfc.2017.12.001
Milting H, Klauke B, Christensen AH et al (2015) The TMEM43 Newfoundland mutation p.S358L causing ARVC-5 was imported from Europe and increases the stiffness of the cell nucleus. Eur Heart J 36:872–881. https://doi.org/10.1093/eurheartj/ehu077
Gaertner-Rommel A, Tiesmeier J, Jakob T et al (2019) Molecular autopsy and family screening in a young case of sudden cardiac death reveals an unusually severe case of FHL1 related hypertrophic cardiomyopathy. Mol Genet Genomic Med 7:e841. https://doi.org/10.1002/mgg3.841
Brodehl A, Meshkov A, Myasnikov R et al (2021) Hemi- and homozygous loss-of-function mutations in DSG2 (Desmoglein-2) cause recessive arrhythmogenic cardiomyopathy with an early onset. Int J Mol Sci 22. https://doi.org/10.3390/ijms22073786
Tozzo P, Scrivano S, Sanavio M, Caenazzo L (2020) The role of DNA degradation in the estimation of post-mortem interval: a systematic review of the current literature. Int J Mol Sci 21. https://doi.org/10.3390/ijms21103540
Fan J, Khanin R, Sakamoto H et al (2016) Quantification of nucleic acid quality in postmortem tissues from a cancer research autopsy program. Oncotarget 7:66906
Acknowledgements
First and most of all, we would like to thank the relatives who gave their permission to include biomaterials of their deceased family members in our study. We also thank the local prosecutors and the criminal police for their continuous support. Thanks also to Zaher Elbeck, Karolinska Institute, Stockholm, Sweden, for his excellent technical support. For her continuous excellent support, we would like to thank Martina Lindemann who contacted relatives, coordinated, and organized the—sometimes challenging—logistics in this study. Finally, we would also like to show our appreciation to all colleagues, too numerous to be listed here, at all participating hospitals and institutes, who generously supported the goals of this study.
Funding
Open Access funding enabled and organized by Projekt DEAL. The study was kindly supported by the Erich and Hanna Klessmann-Foundation, Gütersloh, Germany.
Author information
Authors and Affiliations
Contributions
Study conception and design: CS, HM, AG, JT. Data collection: CS, SH, LH, AW, RK, GMP, HF, TL, LP, HM, JT, KK, AG. Biomaterial collection and investigation: CS, LH, UK, BK, SS, HP, JG, SG, TJ, GV, DM, EH, JP, BBM, DH, JT. Experimental work: CS, GMP. Interpretation of the results and writing of the manuscript: CS, HM, JT. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ethics committees of the Ruhr-University Bochum, Germany, located in Bad Oeynhausen and of the local Medical Association in Münster, Germany, gave approvals (file numbers: 2017-232, 2017-232_2, or 2017-514-b-S, respectively). All samples analyzed in this study were anonymized.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1:
Supplements. SFigs. 1–6 (DOCX 345 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stanasiuk, C., Milting, H., Homm, S. et al. Blood taken immediately after fatal resuscitation attempts yields higher quality DNA for genetic studies as compared to autopsy samples. Int J Legal Med 137, 1569–1581 (2023). https://doi.org/10.1007/s00414-023-02966-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-023-02966-7