Abstract
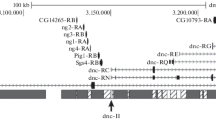
As part of our effort to induce and identify mutations in all genes on chromosome 4 of Drosophila melanogaster, we have mapped the breakpoints of eight chromosome 4 deficiencies relative to the predicted genes along this chromosome. Although the approximate locations of Df(4)G, Df(4)C3, Df(4)M101-62f, Df(4)M101-63a, Df(4)J2, Df(4)O2, Df(4)C1-10AT, and Df(4)B2-2D are known (some from cytological observations and others predicted from P element locations), the extents of these deletions have not been mapped with respect to the predicted genes identified by the Drosophila Genome Project. Polymerase chain reaction primers were designed to amplify the predicted exons of all chromosome 4 genes, and homozygous embryos for each deficiency were identified and their DNA used to test for the presence or absence of these exons. By testing for the inability to amplify various exons along the length of the chromosome, we were able to determine which predicted genes are missing in each deficiency. The five deficiencies, Df(4)G, Df(4)C3, Df(4)C1-10AT, and Df(4)B2-20 (all terminal deletions), and Df(4)M101-62f (a proximal interstitial deletion), enabled us to partition the gene-containing, right arm of chromosome 4 into five regions. Region A [uncovered by Df(4)M101-62f] contains the proximal-most 21 genes; region B [uncovered by Df(4)B2-2D] contains the next 12 genes; region C [uncovered by Df(4)B2-2D and Df(4)C1-10AT] contains the next 17 genes; region D [uncovered by Df(4)B2-2D, Df(4)C1-10AT, and Df(4)C3] contains the next 21 genes; and region E [uncovered by Df(4)B2-2D, Df(4)C1-10AT, Df(4)C3, and Df(4)G] contains the distal-most ten genes. By using Df(4)M101-62f, Df(4)B2-2D, Df(4)C1-10AT, Df(4)C3, and Df(4)G in complementation tests, we can assign newly induced recessive lethal mutations to one of the five regions on chromosome 4. This will substantially reduce the amount of DHPLC analysis required to match each mutation to a predicted transcript on chromosome 4.



Similar content being viewed by others
References
Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195
Ahmad K, Golic K (1998) The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148:775–792
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bridges CB (1935) The mutants and linkage data of chromosome four of Drosophila melanogaster. Biol Zh (Moscow) 4:401–420
Carmena M, Gonzalez C (1995) Transposable elements map in a conserved pattern of distribution extending from beta-heterochromatin to centromeres in Drosophila melanogaster. Chromosoma 103:676–684
Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A et al (2002) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3:RESEARCH0079. Epub
Hochman B (1976) The fourth chromosome of Drosophila melanogaster. In: Ashburner M, Novitski E (eds) The genetics and biology of Drosophila. Academic, London, pp 903–928
Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA et al (2002) Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol 3:RESEARCH0083. Epub
James TC, Eissenberg JC, Craig C, Deitrich V, Hobson A, Elgin SC (1989) Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol 50:170–180
Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, Ashburner M, Celniker SE (2002) The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3:RESEARCH0084. Epub
Kammermeier L, Leemans R, Hirth F, Flister S, Wenger U, Walldorf U, Gehring W, Reichert H (2001) Differential expression and function of the Drosophila Pax6 genes eyeless and twin of eyeless in embryonic central nervous system development. Mech Dev 103:71–78
Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Züllig S, Basler K (2002) Wnt/Wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear β-catenin-TCF complex. Cell 109:47–60
Larsson J, Chen D, Rasheva V, Rasmuson-Lestander A, Pirrotta V (2001) Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci U S A 98:6273–6278
Locke J, McDermid H (1993) Analysis of Drosophila chromosome four using pulsed field gel electrophoresis. Chromosoma 102:718–723
Locke J, Howard LT, Aippersbach N, Podemski L, Hodgetts RB (1999) The characterization of DINE-1, a short, interspersed repetitive element present on chromosome 4 and in the centric heterochromatin of Drosophila melanogaster. Chromosoma 108:356–366
Locke J, Podemski L, Aippersbach N, Kemp H, Hodgetts R (2000) A physical map of the polytenized region (101EF–102F) of chromosome 4 in Drosophila melanogaster. Genetics 155:1175–1183
Miklos GL, Yamamoto MT, Davies J, Pirrotta V (1988) Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the beta-heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci U S A 85:2051–2055
Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS et al (2002) Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol 3:RESEARCH0083. Epub
Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M (1995) Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci U S A 92:3804–3808
Sun F-L, Cuaycong M, Craig C, Wallrath L, Locke J, Elgin SCR (2000) The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci U S A 97:5340–5345
Wallrath LL, Elgin SC (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9:1263–1277
Wallrath LL, Guntur VP, Rosman LE, Elgin SC (1996) DNA representation of variegating heterochromatic P-element inserts in diploid and polytene tissues of Drosophila melanogaster. Chromosoma 104:519–527
Acknowledgements
We thank Scott Hanna for excellent technical assistance and advice, W. Gehring for providing the Df(4)O2 and Df(4)J2 stocks, Travis Thompson for providing the D(4)C3 and D(4)G stocks and the Bloomington Stock Center for providing the Df(4)M101-63a stock. This work was funded with grants from the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Podemski, L., Sousa-Neves, R., Marsh, J.L. et al. Molecular mapping of deletion breakpoints on chromosome 4 of Drosophila melanogaster . Chromosoma 112, 381–388 (2004). https://doi.org/10.1007/s00412-004-0286-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-004-0286-4