Abstract
Introduction
Maximising alternative sample types for genomics in advanced lung cancer is important because bronchoscopic samples may sometimes be insufficient for this purpose. Further, the clinical applications of comprehensive molecular analysis such as whole genome sequencing (WGS) are rapidly developing. Diff-Quik cytology smears from EBUS TBNA is an alternative source of DNA, but its feasibility for WGS has not been previously demonstrated.
Methods
Diff-Quik smears were collected along with research cell pellets.
Results
Tumour content of smears were compared to research cell pellets from 42 patients, which showed good correlation (Spearman correlation 0.85, P < 0.0001). A subset of eight smears underwent WGS, which presented similar mutation profiles to WGS of the matched cell pellet. DNA yield was predicted using a regression equation of the smears cytology features, which correctly predicted DNA yield > 1500 ng in 7 out of 8 smears.
Conclusions
WGS of commonly collected Diff-Quik slides is feasible and their DNA yield can be predicted.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
When a tissue diagnosis of lung cancer is made the samples must be simultaneously used for immunohistochemical subtyping and molecular genetic testing to assess for the presence of actionable mutations. Endobronchial ultrasound-guided, transbronchial needle aspiration (EBUS TBNA) is a common procedure to make the tissue diagnosis of advanced lung cancer [1, 2]. Small amounts of material from fine needle aspirates are deposited on smears for microscopy in the procedure room (Diff-Quik smears) and for formal microscopy (pap smears), while the majority of the sample is collected to make formalin-fixed paraffin-embedded (FFPE) cell blocks [3, 4]. FFPE cell blocks from EBUS TBNA may have adequate tumour content for molecular analysis in as few as 43% of samples [5]. Regarding next generation panel sequencing success rates for EBUS TBNA acquired FFPE cell blocks range from 60 to 93% [6,7,8,9,10]. Conversely Diff-Quik smears contain cancer cells in over 90% of lung cancer patients [11] and are typically never required after their use in the procedure room [12]. These smears have great potential for molecular testing. They allow a rapid estimation of tumour cell content and avoid the impact of formalin on DNA [12]. We and others have shown > 95% success of sequencing of these smears, including with large sequencing panels [12,13,14]. Using smears for sequencing preserves the FFPE cellblock for immunohistochemistry and other developing spatial techniques [15].
In this brief report we take Diff-Quik smears further by exploring their potential for whole genome sequencing (WGS). WGS is not standard of care at this time, however with progressively falling costs it could be more widely used in the clinic [16,17,18]. WGS can detect all forms of molecular abnormalities including point mutations, fusion genes, and chromosomal damage [19], indicating it can detect all actionable mutation types in lung cancer. Further, WGS future-proofs the need to incrementally expand the size of molecular panels.
Successful performance of WGS testing of Diff-Quik smears is highly dependent on the tumour content of the sample, with the proportion of malignant cells preferably to be > 40% and the slide to yield > 1500 ng DNA. We recently showed that over a third of smears had > 1500 ng DNA and all of which had > 40% tumour cellularity [12], suggesting WGS is potential feasibility for a significant number of Diff-Quik samples. Further, it is important to show that the Diff-Quick smear is representative of the FFPE cell block, such that they demonstrate similar tumour content, especially when considering the cell blocks are representative of multiple needle aspirates from the same lymph node, whereas each smear represents just one needle pass.
In demonstrating feasibility of Diff-Quik smears for WGS, we therefore sought to determine (i) whether Diff-Quik smears could yield sufficient tumour content compared with matched research cell pellets; (ii) whether we can predict the amount of DNA on a smear using a simple set of microscopy criteria [12] prior to attempting WGS; and (iii) whether Diff-Quik smears could represent the sole source of WGS material when cell pellets do not yield sufficient material for sequencing.
Methods
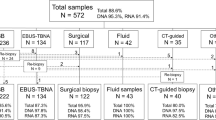
The samples were part of a large prospective study exploring the benefits of Diff-Quik smears (Institutional Review Board approval (HREC/17/QRBW/301); QIMR P2404) [20]. Patients with suspected lung cancer undergoing EBUS TBNA sampling had standard of care testing including Diff-Quik smears, PAP smears and FFPE cell blocks. Research samples were collected frozen or in RNALater for creation of cell pellets. Diff-Quik smears were evaluated by two cyto-pathologists to estimate the percent of malignant cells and overall malignant cell count on the slide. The smear evaluation process by the two cyto-pathologists took approximately 10 min per smear. In addition, smear area was measured from digital slide scans [15]. These 3 parameters were included in a lognormal regression [12] model to estimate DNA yield, as follows:
EXP([1.6 if malignant cells are ≥ 50% malignant cells] + [ 1.2 for malignant cell count estimate ≥ score 2 or 1000 cells] + [0.255* % slide area covered by smear] + 4.14) = ng DNA for that smear.
DNA was extracted from research cell pellets using the AllPrep DNA/RNA Mini Kit, from matched blood samples using the QiAmp DNA Blood Mini Kit, and from smears using the QiAmp DNA Micro Kit (Qiagen). DNA quantity and integrity were measured by Qubit Assay (Thermo Fischer) and TapeStation (Agilent). DNA from research pellets and Diff-Quik slides, with matched normal DNA, were analysed by Infinium Global Screening SNP arrays (Illumina) to determine the tumour content (% tumour) of the samples [21]. Eight smears with > 1ug of DNA and > 40% tumour content [22,23,24] estimated by SNP arrays underwent WGS: four smears had concomitant WGS from fresh cell pellet to allow comparison of genome data and four smears were selected to see if we could “rescue” cases where the fresh cell pellet was inadequate for WGS.
DNA from Diff-Quik smear, cell pellet and matched blood were subjected to WGS. Samples were sequenced to an average read depth of 36.9x (range 31.5 to 44.1) for blood and 70.3x (range of 50 to 77.5x) for tumour samples (smear or cell pellet). The detection of somatic mutations was performed as previously described [25, 26]. Results of standard of care pathology and molecular testing were recorded.
Results
The tumour content was estimated by SNP arrays for 55 Diff-Quik smears and 44 fresh samples from 42 patients. Figure 1 shows comparisons of tumour content and sequencing of fresh tissue and Diff-Quik smears. Median DNA yield of these smears was 1,965 ng (range: 216–12,690 ng) and the median DNA integrity (DIN) was 4.3 (range: 1.9–6.6). Fresh sample results were 17,600 ng (range 434–19,6200 ng) and 6.8 DIN (range 1.8–8.8), respectively. Tumour content estimated by SNP arrays for the two sample types (research cell pellet and Diff-Quik) showed good correlation (Spearman correlation 0.85, p < 0.0001) (Fig. 1A).
Comparison of Diff-Quik smears and fresh cell pellet samples obtained from the same EBUS TBNA procedures. A Tumour content estimated using SNP arrays for Diff-Quik samples and fresh cell pellets collected during the same EBUS TBNA procedure of advanced lung cancers. Spearman correlation = 0.85 (p < 0.0001). Orange dots are samples that both Diff-Quik smears and fresh cell pellet were subjected to whole genome sequencing. Green dots represent samples where only Diff-Quik smears were sequenced by WGS, as fresh samples had insufficient tumour content (< 40%) or DNA yield. B Number and distribution of tumour specific mutations, including point mutations, copy number changes and structural rearrangements detected in Diff-Quik smears and fresh cell pellets. C Circos plots for fresh cell pellets and Diff-Quik samples. Each plot shows chromosomes in the outer ring, followed by copy number alterations (green = loss, red = gain), inner ring represents B-allele frequency data which can be used to identify regions of loss of heterozygosity, lines in the middle of the circos plot indicate structural rearrangements. D Venn-diagrams show the overlap in somatic point mutations detected in Diff-Quik samples and fresh cell pellet from each patient
Table 1 shows the clinical and sample information of the eight cases that underwent WGS, including the extracted DNA yield and quality, as well as the DNA yield predicted by cytopathology scores. In these 8 patients which underwent WGS the median number of TBNA needle passes overall was 4.0 ± 1.0. All the smears with actual DNA content > 1500 ng had predicted DNA values also of > 1500 ng. This value was selected as an approximate value of DNA required to perform DNA quality control by SNP array and then WGS. For case D01_18_034, the smear had a DNA yield of 1452 ng and a predicted yield of 252 ng; the discrepancy likely due to microscopy raw scores being at the low margin of the algorithm, in particular the smear surface area was only 7% of the slide area.
Both smears and the research cell pellets obtained from the same procedure demonstrated good agreement in the number and type of somatic single nucleotide variants, copy number alterations, and structural rearrangements detected by WGS (Fig. 1B). Circos plots (Fig. 1C) illustrate the concordance in the pattern of structural alterations and Venn diagrams (Fig. 1D) indicates that the majority (67–91%) of point mutations detected in the freshly collected specimens were also detected in the diagnostic Diff-Quik smears.
In four samples where the fresh cell pellet did not provide appropriate material for WGS, the smear samples were successfully sequenced (Fig. 2). The somatic mutations identified included reportable mutations revealed by standard of care testing, i.e. KRAS: c. 35G > C p. (Gly12Ala) in case D01_19_046 and a fusion between ROS1 and CD74 in case D01_21_090, which was reported diagnostically as a ROS1 rearrangement by immunohistochemistry, and fluorescence in-situ hybridisation (Fig. 2C).
Somatic alterations detected in Diff-Quik smears from cases where the matched fresh cell pellet was not suitable for whole genome sequencing. A Number of point mutations and indels, percentage of the genomes affected by copy number, and the number and types of structural rearrangements. B Circos plots for individual cases, showing chromosomes in the outer ring, followed by copy number alterations (green = loss, red = gain), inner ring represents B-allele frequency data which can be used to identify regions of loss of heterozygosity, lines in the middle of the circos plot indicate structural rearrangements. C Images from diagnostic testing for case D01_21_090 (adenocarcinoma, CT core biopsy prior to EBUS TBNA staging), showing haematoxylin and eosin, ROS1 immunohistochemistry (H-score of 220) and ROS1 FISH (Zytolight SPEC ROS1 (6q22.1) Dual Colour, Break Apart Rearrangement Probe). ROS1 fusion positive cells were indicated by single red and green signals, as observed here
In three cases, extra mutations in KRAS and STK11 (Tier 4 mutations) were detected by WGS (Table 1). Considering the added overall benefit, from the 8 smears there was added molecular information from this WGS single test in 4 patients (50%): 3 with tier 4 mutations and 1 with a ROS 1 fusion. These 4 include 1 patient where the standard of care cell block was “insufficient for testing.”
Discussion
Others have demonstrated that WGS is feasible from EBUS-TBNA specimens [27]. Here we advance this knowledge to demonstrate for the first time the feasibility of conducting WGS on Diff-Quick smears. A common clinical problem arises when the FFPE cell block yields insufficient material for genomic testing; here we show that not only can Diff-Quik smears negate this problem, but also that they can yield comprehensive genomic data capturing multiple types of somatic mutations in one molecular test.
To demonstrate the utility of WGS, we observed good agreement between smear and fresh tissue WGS for point mutations (67 to 91% agreement), copy number alterations and structural rearrangements. Importantly, WGS identified Tier 1 somatic alterations reported by accredited methodology and Tier 4 mutations with potential relevance to future genomic based therapies. Further, smears permitted WGS for cases where the fresh tissue and/or FFPE cell pellets had insufficient material. In three NSCLC cases, smears provided sufficient material for comprehensive genomic testing where SOC testing was insufficient or had no reportable mutations.
Good correlation of tumour content between fresh cell pellets and smears gives confidence that a carefully selected Diff-Quik smear can represent an average of all needle passes made into the node.
Not all smears will have sufficient DNA (> 1500 ng) for WGS or have sufficient malignant cell content (> 40%). We suggest the use of our cytology-based prediction algorithm can assist in selecting smears that will yield sufficient DNA. Further this prediction could allow selection of the best of all the smears from a procedure for sequencing. The algorithm will continue to be improved with further samples from the clinic. Generally, our predictions under-estimated the actual yield of DNA obtained from the smears, however all smears with > 1500 ng DNA were correctly predicted above that threshold. Two cases significantly underestimated DNA content, as scores for estimating cell counts are arbitrarily set at a maximum of 4000 cells some smears clearly have many more cells than this. Some samples at the lower marginal end of the prediction algorithm may be excluded, but conversely the correct identification of samples with > 1500 ng can rule them in.
In our study, Diff-Quik smears were one or two years old, which might have impacted the DNA quality obtained. We would expect freshy collected smears to have DIN approaching that of fresh cell pellets.
Two smears were from patients with SCLC and were chosen for their tumour content to contribute to this proof-of-concept study. Presently there are no specific molecular targets for SCLC however WGS could have a future role in aiding treatment decision making in the future [28].
References
Gomez M, Silvestri GA (2009) Endobronchial ultrasound for the diagnosis and staging of lung cancer. Proc Am Thorac Soc 6(2):180–186
Torii A, Oki M, Yamada A, Kogure Y, Kitagawa C, Saka H (2022) EUS-B-FNA Enhances the diagnostic yield of ebus bronchoscope for intrathoracic lesions. Lung 200(5):643–648
Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH et al (2018) Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the college of american pathologists, the international association for the study of lung cancer, and the association for molecular pathology. Arch Pathol Lab Med 142(3):321–346
Magnini D, Fuso L, Varone F, D’Argento E, Martini M, Pecoriello A et al (2018) Molecular testing in EBUS-TBNA specimens of lung adenocarcinoma: a study of concordance between cell block method and liquid-based cytology in appraising sample cellularity and egfr mutations. Mol Diagn Ther 22(6):723–728
Knoepp SM, Roh MH (2013) Ancillary techniques on direct-smear aspirate slides: a significant evolution for cytopathology techniques. Cancer Cytopathol 121(3):120–128
Martin-Deleon R, Teixido C, Lucena CM, Martinez D, Fontana A, Reyes R et al (2021) EBUS-TBNA Cytological samples for comprehensive molecular testing in non-small cell lung cancer. Cancers 13(9):2084
Turner SR, Buonocore D, Desmeules P, Rekhtman N, Dogan S, Lin O et al (2018) Feasibility of endobronchial ultrasound transbronchial needle aspiration for massively parallel next-generation sequencing in thoracic cancer patients. Lung Cancer 119:85–90
Xie F, Zheng X, Mao X, Zhao R, Ye J, Zhang Y et al (2019) Next-generation sequencing for genotyping of endobronchial ultrasound-guided transbronchial needle aspiration samples in lung cancer. Ann Thorac Surg 108(1):219–226
Fielding D, Dalley AJ, Bashirzadeh F, Singh M, Nandakumar L, McCart Reed AE et al (2019) Diff-Quik cytology smears from endobronchial ultrasound transbronchial needle aspiration lymph node specimens as a source of DNA for next-generation sequencing instead of cell blocks. Res Int Rev Thoracic Dis 97(6):525–539
Royo BUCC, Sanz JL, Costa EC, Mitja PS, Terzi MA, Mezalek RT, Gratacos AT, Garcia FA (2021) Non-small-cell lung cancer molecular typification using next-generation sequencing in cytological samples from EBUS-TBNA. Eur Resp J. https://doi.org/10.1183/13993003.congress-2021.PA2181
Jain D, Allen TC, Aisner DL, Beasley MB, Cagle PT, Capelozzi VL et al (2018) Rapid on-site evaluation of endobronchial ultrasound-guided transbronchial needle aspirations for the diagnosis of lung cancer: a perspective from members of the pulmonary pathology society. Arch Pathol Lab Med 142(2):253–262
Fielding DI, Dalley AJ, Singh M, Nandakumar L, Lakis V, Chittoory H et al (2023) Evaluating diff-quik cytology smears for large-panel mutation testing in lung cancer-predicting DNA content and success with low-malignant-cellularity samples. Cancer cytopathol 131(6):373–382
Roy-Chowdhuri S (2019) Molecular testing of residual cytology samples: rethink, reclaim, repurpose. Cancer Cytopathol 127(1):15–17
Stoy SP, Segal JP, Mueller J, Furtado LV, Vokes EE, Patel JD et al (2018) Feasibility of EBUS TBNA cytology specimens for next generation sequencing in non-small-cell lung cancer. Clin Lung Cancer 19(3):230
Pourmaleki M, Socci ND, Hollmann TJ, Mellinghoff IK (2023) Moving spatially resolved multiplexed protein profiling toward clinical oncology. Cancer Discov 13(4):824–828
Samsom KG, Schipper LJ, Roepman P, Bosch LJ, Lalezari F, Klompenhouwer EG et al (2022) Feasibility of whole-genome sequencing-based tumor diagnostics in routine pathology practice. J Pathol 258(2):179–188
Simons MJHG, Retèl VP, Ramaekers BLT, Butter R, Mankor JM, Paats MS et al (2021) Early cost effectiveness of whole-genome sequencing as a clinical diagnostic test for patients with inoperable stage IIIB. C/IV Non-squamous Non-small-Cell Lung Cancer PharmacoEconomics 39(12):1429–1442
Wang C, Dai J, Qin N, Fan J, Ma H, Chen C et al (2022) Analyses of rare predisposing variants of lung cancer in 6004 whole genomes in Chinese. Cancer Cell 40(10):1223
Ramarao-Milne P, Kondrashova O, Patch AM, Nones K, Koufariotis LT, Newell F et al (2022) Comparison of actionable events detected in cancer genomes by whole-genome sequencing, in silico whole-exome and mutation panels. ESMO open 7(4):100540
Fielding D, Dalley AJ, Singh M, Nandakumar L, Nones K, Lakis V, Chittoory H, Ferguson K, Bashirzadeh F, Bint M, Pahoff C, Son JH, Hodgson A, Sharma S, Godbolt D, Coleman K, Whitfield L, Waddell N, Lakhani SR, Hartel G, Simpson PT (2022) Prospective optimisation of EBUS TBNA lymph node assessment for lung cancer: 3 needle agitations is non-inferior to 10 agitations for adequate for tumour cell and DNA yield. JTO CRR 3(10):100403
Song S, Nones K, Miller D, Harliwong I, Kassahn KS, Pinese M et al (2012) qpure: A tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS ONE 7(9):e45835
Nones K, Johnson J, Newell F, Patch AM, Thorne H, Kazakoff SH et al (2019) Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Annals Oncol Off J Eur Soci Med Oncol 30(7):1071–1079
Nones K, Waddell N, Wayte N, Patch A-M, Bailey P, Newell F et al (2014) Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun 5:5224
Scarpa A, Chang DK, Nones K, Corbo V, Patch A-M, Bailey P et al (2017) Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543(7643):65–71
Newell F, Kong Y, Wilmott JS, Johansson PA, Ferguson PM, Cui C et al (2019) Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun 10(1):3163
Patch A-M, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S et al (2015) Whole-genome characterization of chemoresistant ovarian cancer. Nature 521(7553):489–494
Leong TL, Aloe C, Aujla S, Wang H, Gayevskiy V, Asselin-Labat M-L et al (2023) Heterogeneity of tumour mutational burden in metastatic NSCLC demonstrated by endobronchial ultrasound sampling. Front Oncol 13:1150349
Pongor LS, Schultz CW, Rinaldi L, Wangsa D, Redon CE, Takahashi N et al (2023) Extrachromosomal DNA amplification contributes to small cell lung cancer heterogeneity and is associated with worse outcomes. Cancer Discov 13(4):928–949
Acknowledgements
We thank the patients who consented to be involved in our study, without whom this work would not be possible, and all nursing staff at The Royal Brisbane & Women’s Hospital, Gold Coast University Hospital, and Sunshine Coast University Hospital Bronchoscopy departments.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. Research was supported by funding from the following sources: The 2017 Priority-driven Collaborative Cancer Research Scheme, funded by Cancer Australia (Grant #1147067); Cancer Council of Queensland (Grant #1147067); Australian New Zealand Interventional Pulmonology Group (supported by Olympus Australia); Royal Brisbane and Women’s Hospital Foundation; and Australian Genomics (NHMRC grants GNT1113531 and GNT2000001), Cooperative Research Centres Project/Australian Government.
Author information
Authors and Affiliations
Contributions
Conceptualised the study and attracted funding: DFi, SRL, KN, PTS; patient recruitment and EBUS TBNA sampling and clinical data curation: DFi, DFa, FB, MB, CP, JHS; cytology sample preparation and analysis: MS, LN, AH; sample processing: AJD, HC, KF, KN; bioinformatics and data curation: VL, NW, JVP, KN; statistical analysis: GH; wrote the first draft: DFi, KN, PTS.
Corresponding author
Ethics declarations
Conflict of interest
No author has any conflict of interest for the material presented in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fielding, D., Dalley, A.J., Singh, M. et al. Whole Genome Sequencing in Advanced Lung Cancer can be Performed Using Diff-Quik Cytology Smears Derived from Endobronchial Ultrasound, Transbronchial Needle Aspiration (EBUS TBNA). Lung 201, 407–413 (2023). https://doi.org/10.1007/s00408-023-00631-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00408-023-00631-9