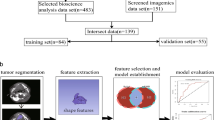
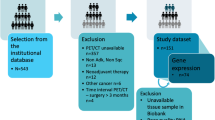
Abstract
Purpose
Classical prognostic indicators of head and neck squamous cell carcinoma (HNSCC) can no longer meet the clinical needs of precision medicine. This study aimed to establish a radiomics model to predict Granzyme A (GZMA) expression in patients with HNSCC.
Methods
We downloaded transcriptomic data of HNSCC patients from The Cancer Genome Atlas for prognosis analysis and then used corresponding enhanced computed tomography (CT) images from The Cancer Imaging Archive for feature extraction and model construction. We explored the influence of differences in GZMA expression on signaling pathways and analyzed the potential molecular mechanism and its relationship with immune cell infiltration. Subsequently, non-invasive CT radiomics models were established to predict the expression of GZMA mRNA and evaluate the correlation with the radiomics-score (Rad-score), related genes, and prognosis.
Results
We found that GZMA was highly expressed in tumor tissues, and high GZMA expression was a protective factor for overall survival. The degree of B and T lymphocyte and natural killer cell infiltration was significantly correlated with GZMA expression. The receiver operating characteristic curve showed that the Relief GBM and RFE_GBM radiomics models had good predictive ability, and there were significant differences in the Rad-score distribution between the high- and low-GZMA-expression groups.
Conclusions
GZMA expression can significantly affect the prognosis of patients with HNSCC. Enhanced CT radiomics models can effectively predict the expression of GZMA mRNA.




Similar content being viewed by others
Data availability
The data were downloaded from The Cancer Genome Atlas and The Cancer Imaging Archive.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249
Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR (2020) Head and neck squamous cell carcinoma. Nat Rev Dis Primers 6:92
Kitamura N, Sento S, Yoshizawa Y, Sasabe E, Kudo Y, Yamamoto T (2020) Current trends and future prospects of molecular targeted therapy in head and neck squamous cell carcinoma. Int J Mol Sci 22:240
Solomon B, Young RJ, Rischin D (2018) Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol 52:228–240
Chen D, Wang CY (2019) Targeting cancer stem cells in squamous cell carcinoma. Precis Clin Med 2:152–165
Cramer JD, Burtness B, Le QT, Ferris RL (2019) The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol 16:669–683
Kiselevsky DB (2020) Granzymes and mitochondria. Biochemistry (Mosc) 85:131–139
Zeglinski MR, Granville DJ (2020) Granzymes in cardiovascular injury and disease. Cell Signal 76:109804
Zhu J, Xiao J, Wang M, Hu D (2021) Pan-cancer molecular characterization of m6A regulators and immunogenomic perspective on the tumor microenvironment. Front Oncol 10:618374
Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y et al (2020) Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 368:eaaz7548
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Yang WS, Jiang HY, Liu C, Wei JW, Zhou Y, Gong PY et al (2021) Multi-omics and its clinical application in hepatocellular carcinoma: current progress and future opportunities. Chin Med Sci J 36:173–186
Moussa AM, Ziv E (2021) Radiogenomics in interventional oncology. Curr Oncol Rep 23:9
Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W, Beets-Tan R (2019) Radiogenomics: bridging imaging and genomics. Abdom Radiol (NY) 44:1960–1984
Garzón-Tituaña M, Sierra-Monzón JL, Comas L, Santiago L, Khaliulina-Ushakova T, Uranga-Murillo I et al (2021) Granzyme A inhibition reduces inflammation and increases survival during abdominal sepsis. Theranostics 11:3781–3795
Zhu P, Zhang D, Chowdhury D, Martinvalet D, Keefe D, Shi L et al (2006) Granzyme A, which causes single-stranded DNA damage, targets the double-strand break repair protein Ku70. EMBO Rep 7:431–437
Lieberman J (2010) Granzyme A activates another way to die. Immunol Rev 235:93–104
Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J (2003) Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112:659–672
Romero JM, Grünwald B, Jang GH, Bavi PP, Jhaveri A, Masoomian M et al (2020) A four-chemokine signature is associated with a T-cell-Inflamed phenotype in primary and metastatic pancreatic cancer. Clin Cancer Res 26:1997–2010
Gu X, Boldrup L, Coates PJ, Fahraeus R, Wang L, Wilms T et al (2019) High immune cytolytic activity in tumor-free tongue tissue confers better prognosis in patients with squamous cell carcinoma of the oral tongue. J Pathol Clin Res 5:240–247
Zhu Z, Li G, Li Z, Wu Y, Yang Y, Wang M et al (2022) Core immune cell infiltration signatures identify molecular subtypes and promote precise checkpoint immunotherapy in cutaneous melanoma. Front Immunol 13:914612
Mei Z, Huang J, Qiao B, Lam AK (2020) Immune checkpoint pathways in immunotherapy for head and neck squamous cell carcinoma. Int J Oral Sci 12:16
Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR et al (2018) A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 19:1180–1191
Staal FCR, van der Reijd DJ, Taghavi M, Lambregts DMJ, Beets-Tan RGH, Maas M (2021) Radiomics for the prediction of treatment outcome and survival in patients with colorectal cancer: a systematic review. Clin Colorectal Cancer 20:52–71
Kocher M (2020) Artificial intelligence and radiomics for radiation oncology. Strahlenther Onkol 196:847
Gillies RJ, Schabath MB (2020) Radiomics improves cancer screening and early detection. Cancer Epidemiol Biomark Prev 29:2556–2567
Chetan MR, Gleeson FV (2021) Radiomics in predicting treatment response in non-small-cell lung cancer: current status, challenges and future perspectives. Eur Radiol 31:1049–1058
Singh G, Manjila S, Sakla N, True A, Wardeh AH, Beig N et al (2021) Radiomics and radiogenomics in gliomas: a contemporary update. Br J Cancer 125:641–657
Kaur RP, Vasudeva K, Kumar R, Munshi A (2018) Role of p53 gene in breast cancer: focus on mutation spectrum and therapeutic strategies. Curr Pharm Des 24:3566–3575
Dang M, Lysack JT, Wu T, Matthews TW, Chandarana SP, Brockton NT et al (2015) MRI texture analysis predicts p53 status in head and neck squamous cell carcinoma. AJNR Am J Neuroradiol 36:166–170
Funding
This study was supported by the Fundamental Research Program of the Ninth People’s Hospital affiliated to the Shanghai Jiao Tong University School of Medicine (grant number, JYZZ158), Shanghai Jiaotong University Medical-Engineering Cross Research Fund (YG2022QN050), and the Science and Technology Commission of Shanghai Funding/Supporting (16411960900).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hang, R., Bai, G., Sun, B. et al. Construction of an enhanced computed tomography radiomics model for non-invasively predicting granzyme A in head and neck squamous cell carcinoma by machine learning. Eur Arch Otorhinolaryngol 280, 3353–3364 (2023). https://doi.org/10.1007/s00405-023-07909-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-023-07909-x