Abstract
Purpose
It is of obvious interest to identify clinical prognosis-related oncogenes in HNSCC (head and neck squamous cell carcinoma).
Methods
Based on the available datasets within the TCGA (The Cancer Genome Atlas) and the GEO (Gene Expression Omnibus) databases, the potential mechanism of action of the SEC61G (SEC61 translocon subunit gamma) gene in HNSCC tumorigenesis was explored by several bioinformatics approaches.
Results
There was a higher expression level of SEC61G in primary HNSCC tumor tissues than in normal tissues. Moreover, highly expressed SEC61G was statistically associated with the poor survival prognosis of HNSCC patients. When HPV (human papilloma virus) was considered, we also observed a relatively lower proportion of “arm-level gain” and “high amplification” types of CNA (copy-number alteration) in the HNSCC-HPV (+) group than in the HNSCC-HPV (−) group. Additionally, we identified SEC61G CAN-correlated genes, such as CCT6A (chaperonin-containing TCP1 subunit 6A) and HUS1 (HUS1 checkpoint clamp component), and found a correlation between SEC61G copy-number segments and prognosis related to overall and progression-free survival intervals of HNSCC patients. Moreover, the molecular regulation mechanisms of the spliceosome, ribosome, proteasome degradation, cell adhesion, and immune infiltration of B and CD8+ T cells may contribute to the involvement of SEC61G in the pathogenesis of HNSCC.
Conclusions
The SEC61G gene was identified for the first time as a prognostic biomarker of HNSCC. The detailed underlying mechanism merits further research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SEC61G is the main member of the Sec61 complex, which is a transmembrane channel across which proteins are translocated and integrated into the ER (endoplasmic reticulum) membrane [1, 2]. SEC61G is essential for protein translocation into the mammalian endoplasmic reticulum [2]. The expression of SEC61G was reportedly associated with IFN-K immunosuppressive therapy for lupus nephritis [3], while the methylation modification of SEC61G was related to Balkan endemic nephropathy [4, 5].
Several publications have reported the correlation between SEC61G expression and the pathogenesis of certain cancers and the prognosis of patients with these cancers. For example, the binding of lncRNA LINC02418 and miR-4677-3p (microRNA-4677-3p) was associated with the expression of SEC61G in NSCLC (non-small cell lung cancer) cells [6]. The SEC61G-EGFR fusion gene was detected in the MI (mitogen-independent) stem-like cells of pediatric ependymomas [7]. It was also reported that high expression of the SEC61G gene was correlated with a poor prognosis of glioblastoma patients in a study that used the TCGA and CGGA (Chinese Glioma Genome Atlas) datasets [8]. The SEC61G gene was identified as a hub molecule of parathyroid adenoma development [9].
In the present study, we first collected the relevant available datasets from the TCGA and GEO databases to explore the oncogenic role of SEC61G in HNSCC. A series of analyses, including gene expression, survival prognosis, genetic alteration, gene enrichment, and immune infiltration, were carried out.
Methods
Expression feature analysis
After logging into the UALCAN website (http://ualcan.path.uab.edu/analysis-prot.html), entering the gene information, and selecting the cancer type, we obtained the differential expression data of the SEC61G gene in normal and HNSCC tissues. The transcript per million was used for the expression level of SEC61G. Two factors, namely, human papilloma virus (HPV) infection status and sex, were also considered. In addition, we logged into the Oncomine database (https://www.oncomine.org/resource/main.html) and entered the word “SEC61G”. Differential expression data of SEC61G between the normal and different tumor tissues were obtained from the GEO database with the threshold of p value = 0.05 and fold change = 1.5. A pooled analysis across five analyses was also performed.
Survival prognosis analysis
We applied the “survival map” module of GEPIA2 [10] to obtain the OS (overall survival) and DFS (disease-free survival) rate significance map data of SEC61G for all tumor cases from the TCGA database. A cutoff point of 50% was set to split the high-expression and low-expression cohorts. “Significance Level” was set at 0.01, and “p value adjustment” of FDR was used. The survival plots with a log-rank test were also yielded via the “survival analysis” module of GEPIA2. We also considered three factors, race, histological grade, and sex, for the survival analyses. In addition, we utilized the interactive operation interface of the PrognoScan database [11] to conduct an RFS (relapse-free survival) rate analysis of HNSCC patients in the GSE2837 dataset.
Genetic alteration analysis
We then studied the association between SEC61G expression and CNA (copy-number alteration) across all TCGA cancers using the “sCNA” module of TIMER2.0 (http://timer.cistrome.org/). After logging into the cBioPortal web (https://www.cbioportal.org/) [12, 13], we chose the “TCGA PanCancer Atlas studies” in the “Quick select” section and entered “SEC61G” to obtain the association between log2 copy-number values and the mRNA expression Z scores of SEC61G. Using a LinkedOmics approach (http://www.linkedomics.org/login.php) [14], we identified the potential SEC61G CNA-associated genes. Spearman’s rho statistical analysis was conducted. We also utilized UCSC-Xena (https://xena.ucsc.edu/) to investigate the potential correlation between the copy-number segments of SEC61G and the overall survival, disease-specific survival, progression-free interval, and disease-free interval status of the patients. Kaplan–Meier plots with log-rank p values were generated.
Gene enrichment analysis
Next, we performed cluster analysis of the SEC61G-correlated significant genes through the LinkedOmics approach [14]. Heat maps and association plots covering the significant genes positively/negatively correlated with SEC61G expression are presented. We also probed the expression correlation between SEC61G and the selected genes through a Spearman correlation test. Additionally, the GO (gene ontology), KEGG (Kyoto encyclopedia of genes and genomes) pathway, WikiPathway, and GSEA (gene set enrichment analysis) profiles were obtained.
Immune infiltration analysis
Finally, based on the HNSCC dataset of the TCGA, we investigated the potential association between SEC61G expression and the infiltration level of different immune cells using the “immune-gene” module of TIMER2.0 with the algorithms TIMER, EPIC, QUANTISEQ, XCELL, MCPCOUNTER, CIBERSORT, and CIBERSORT-ABS [15].
Results
Gene expression
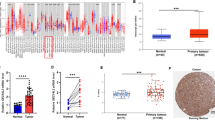
SEC61G structure is conserved across different species (Fig. S1). In the present study, we investigated the possible oncogenic role of human SEC61G in HNSCC. We first compared the differential expression of SEC61G between normal tissues and HNSCC tissues using two factors (HPV and sex). As shown in Fig. 1a, we observed high SEC61G expression in primary HNSCC tumor tissues (n = 520) compared with normal tissues (n = 44) (p = 1.62e-12). Additionally, there was a higher expression level of SEC61G in both HNSCC-HPV (+) (Fig. 1b, p = 5.04e-10) and HNSCC-HPV (−) (p = 5.97e-05) tissues than in controls. The expression of SEC61G was higher in the HNSCC-HPV (−) group than in the HNSCC-HPV (−) group (Fig. 1b, p = 2.86e-02). Likewise, compared with the normal tissues, we observed a higher expression level of SEC61G in both the male (Fig. 1c, p = 1.62e-12) and female (p = 1.71e-06) groups. However, there was no difference between the male and female groups (Fig. 1c).
The expression level of the SEC61G gene. a Expression differences in the SEC61G gene in normal and HNSCC tissues were detected based on the HNSCC dataset from the TCGA. Two factors, including, b HPV and c sex, were considered. d Through the Oncomine database, differential expression data of SEC61G between normal and different tumor tissues were provided. e Pooled analysis of SEC61G across five GEO studies was conducted
Based on the ONCOMINE database analysis, we found that the SEC61G gene was highly expressed in many types of tumors, such as brain/CNS cancer, head and neck cancer, and kidney cancer, compared with its expression in normal control tissues (Fig. 1d). We further determined the high expression pattern of SEC61G in head and neck cancer using pooled analysis (Fig. 1e, p = 9.16e-08).
Survival analysis
Subsequently, we divided the HNSCC samples into high-expression and low-expression groups according to the expression level of SEC61G and investigated the correlation between SEC61G expression and the prognosis of patients with different types of cancers, mainly using datasets from the TCGA and GEO databases. As shown in the survival maps in Fig. 2a, we observed that high expression of SEC61G was related to a poor OS (overall survival) rate among HNSCC, BRCA (breast invasive carcinoma), LGG (brain lower-grade glioma), and LIHC (liver hepatocellular carcinoma) patients in the TCGA database. DFS (disease-free survival) rate analysis data (Fig. 2a) indicated a correlation between high expression of SEC61G and poor prognosis of TCGA HNSCC patients. The detailed OS and DFS rate survival plots for the HNSCC patients are shown in Fig. 2b. We also observed positive correlations (Fig. S2) when considering the other three factors, race (p = 0.00079), histological grade (p = 0.033), and sex (p = 0.0018). Additionally, we detected a poor relapse-free survival rate in HNSCC patients with high SEC61G expression in the GSE2837 dataset (Fig. 2c). These data indicate a correlation between high expression of SEC61G and poor survival prognosis in HNSCC patients.
Genetic alteration analysis
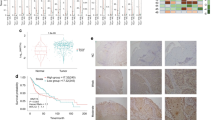
The CNA status of SEC61G in different tumors in the TCGA database was also analyzed. As shown in Fig. 3a, the “arm-level gain” variant of SEC61G showed a higher proportion in TGCT (testicular germ cell tumors), READ (rectum adenocarcinoma), KIRP (kidney renal papillary cell carcinoma), and other tumors. The “high amplification” variant was most common in GBM (glioblastoma multiforme) tumors. In contrast, in the HNSCC, HNSCC-HPV (+), and HNSCC-HPV (−) groups, “arm-level gain” and “high amplification” variants were present at moderate levels. We observed a relatively lower proportion of the two types of CNAs in the HNSCC-HPV (+) group compared with the HNSCC-HPV (−) group. Moreover, we observed a positive correlation between SEC11G log2 copy-number values and the mRNA expression Z scores (Fig. 3b, p = 9.24e-80, R = 0.72 for a Pearson test, p = 2.50e-34, R = 0.51 for Spearman’s test). Based on the HNSCC dataset of the TCGA, the potential SEC61G CNA-correlated genes identified using Spearman’s rho statistical analysis are shown in Fig. 3c. There was a positive correlation between SEC61G CNA and the expression level of CCT6A (Fig. 3d, p = 5.51e-52, Spearman correlation = 0.6019) and HUS1 (p = 3.59e-44, Spearman correlation = 0.5622). In addition, we explored the potential association between the genetic alteration of SEC61G and the clinical survival prognosis of TCGA HNSCC patients. We observed a correlation between SEC61G copy-number segments and overall survival (Fig. 3e, p = 0.0003) and progression-free intervals (p = 0.0199) but not with disease-specific survival rates or disease-free intervals in HNSCC patients.
Genetic alteration analysis of SEC61G. a Association between SEC61G expression and CNA across all TCGA tumors was analyzed via TIMER2.0. b Correlation analysis between SEC11G log2 copy-number values and the mRNA expression Z score was performed via cBioPortal. c We identified potential SEC61G CNA-associated genes via LinkedOmics. d Potential correlations between the copy-number segments of SEC61G and the status of overall survival, disease-specific survival, progression-free interval, and disease-free interval rates were determined via UCSC-Xena
Enrichment analysis of SEC61G-related partners
To further explore the molecular mechanism of the SEC61G gene in tumorigenesis, we sought to identify SEC61G-correlated genes for pathway enrichment analysis using the LinkedOmics approach. The heat map in Fig. 4a shows the top ten significant genes positively (e.g., MRPS17, CHCHD2, etc.) or negatively (e.g., IKZF3, NCOA2, KIAA0240, etc.) correlated with SEC61G expression. Figure 4b further shows the association analysis results and Spearman’s correlation analysis results for SEC61G and MRPS17 (p = 6.624e-90, R = 0.7363), CHCHD2 (p = 3.572e-88, R = 0.7314), and IKXF3 (p = 1.658e-58, R = 0.6285).
Additionally, GO enrichment, KEGG pathway, WikiPathway, and GSEA profile analyses were performed. As shown in Fig. 4c, we detected enriched cellular components (e.g., mitochondrial inner membrane or receptor complex), molecular functions (e.g., structural constituent of the ribosome or DNA-binding transcription activator activity, RNA polymerase II-specific), and biological processes (e.g., ribonucleoprotein complex biogenesis or regulation of small GTPase-mediated signal transduction). The KEGG pathway and GSEA data shown in Fig. 5 further revealed that the pathways “cell adhesion molecules (CAMs)”, “Spliceosome”, or “Ribosome”. Figure 6 of WikiPathway data also presents the enrichment plots of “B cell receptor signaling pathway”, “proteasome degradation”, and “cytoplasmic ribosomal proteins” were enriched. These results suggest that SEC61G participates in the pathogenesis of HNSCC, mainly through playing potential regulatory roles in the spliceosome, ribosome, proteasome degradation, immune cell regulation, and cell adhesion processes.
Immune infiltration analysis
Finally, we utilized the TIMER, EPIC, QUANTISEQ, XCELL, MCPCOUNTER, CIBERSORT, and CIBERSORT-ABS algorithms to evaluate the potential association between SEC61G expression and the infiltration of different immune cells based on the HNSCC dataset in the TCGA database. Following several analyses, we observed a significant negative correlation between the immune infiltration of B cells and CD8+ T cells and SEC61G expression in all HNSCC tumors, especially HNSCC-HPV (+) tumors (Fig. 7a). The scatter plot data showing B cell infiltration with the XCELL algorithm is presented in Fig. 7b (p = 1.72e-05, cor = − 0.439), while the scatter plot data showing CD8+ T cell infiltration with the CIBERSORT-ABS algorithm is shown in Fig. 7c (p = 3.18e-05, cor = − 0.426). These results indicated a potential relationship between SEC61G expression and the infiltration of B and CD8+ T cells in the HNSCC tissues.
Correlation analysis between SEC61G expression and immune cell infiltration in HNSCC. a Based on the HNSCC dataset of TCGA, the potential association between SEC61G expression and the infiltration status of B and T immune cells was studied via TIMER2.0. b One scatter plot of B cells or CD8+ T cells is shown as an example
Discussion
High expression of SEC61G
It has been reported that there is a higher level of SEC61G in the tissues of glioblastoma multiforme than in those of lower-grade gliomas [16, 17]. In the current study, we observed an increased level of SEC61G in brain or central nervous system (CNS) cancer tissues across ten studies compared with its expression level in the corresponding normal tissues. In addition to in brain tumors, we also detected a high expression level of SEC61G in other types of tumors. For instance, we observed high expression of the SEC61G gene in kidney cancer tissues in six studies. Using the TCGA and Oncomine datasets, we analyzed the expression levels of SEC61G in normal and HNSCC tissues. In the TCGA database, compared with the 40 normal tissues samples, SEC61G was highly expressed in 520 primary HNSCC tissue samples, mainly including 133 tongue tumor samples and 117 larynx tumor samples. In the Oncomine database, only five datasets containing four tongue carcinomas and one oral cavity carcinoma were included. We also observed high SEC61G expression in these tumor tissues compared with that in normal tissues. Thus, these results suggest that SEC61G is not a tumor-specific highly expressed gene.
CNA and SEC61G
SEC61G expression was not closely related to DNA copy number in 7p11.2 amplified oral squamous cell carcinoma (OSCC) [18]. However, genomic copy-number aberrations of SEC61G seem to be associated with the expression of SEC61G in gastric cancer (R = 0.93, p < 0.01) [19]. The data from a GWAS (genome-wide association study) indicated that single-nucleotide polymorphisms (SNPs) in SEC61G are associated with susceptibility to clinical SLE (systemic lupus erythematosus) [20]. In the present study, CNA of SEC61G was found to be linked to its mRNA expression level and patient overall survival rates and progression-free intervals. Furthermore, we observed a potential correlation between SEC61G CNA and the expression of CCT6A or HUS1. CCT6A is a member of the chaperonin-containing TCP1 complex and was reported to be associated with transcriptome reprogramming by HNSCC-derived exosomes [21]. HUS1, an important DNA damage checkpoint, participates in the regulation of the cell cycle [22, 23]. HUS1 can act as a potential tumor suppressor of hepatocellular carcinoma [24]. However, there is still no report regarding the potential correlation between HUS1 expression and oncogenesis in HNSCC. SEC61G CNA may be the downstream functional target of CCT6A or HUS1. This potential mechanism deserves future explorations.
Cell biological events and SEC61G
As SEC61G is located in the membrane structure of the ER [1, 2], regulation of the biology of the ER is the most critical mechanism of cellular SEC61G. Nevertheless, there is limited evidence to suggest that there is a correlation between SEC61G-mediated ER function alterations and the occurrence of clinical tumor diseases. It was reported that the response to ER stress might contribute to the oncogenic role of SEC61G in glioblastoma [17]. Herein, we carried out a series of SEC61G-correlated gene enrichment analyses and identified a group of biological structures or processes associated with SEC61G, such as the mitochondrial inner membrane or receptor complex, regulation of small GTPase-mediated signal transduction, CAMs, spliceosome, and proteasome degradation, apart from ER-associated ribosome aberrations. Tumor-infiltrating immune cells, as prominent components of the tumor microenvironment, are closely linked to the initiation, progression, or metastasis of cancer [25, 26]. In the present study, we also utilized different algorithms to observe the potential association between SEC61G expression and the infiltration of B cells and CD8+ T cells in HNSCC. These findings offer a reference or possible future investigation directions for the in-depth elucidation of the molecular mechanisms underlying the oncoprotein role of SEC61G.
HPV infection and SEC61G
There exists a potential correlation between SEC61G expression and the outcome of temozolomide treatment and radiotherapy for glioblastoma patients [8]. Due to limited clinical evidence, we could not perform a relative analysis regarding the association between SEC61G expression and the sensitivity of HNSCC to radiotherapy and chemotherapy. Human HPV infection is associated with the effect of clinical radiotherapy and chemotherapy in head and neck cancer patients [27,28,29]. Among the TCGA HNSCC samples, the HNSCC-HPV (+) group mainly consisted of 23 tonsil, 10 base of tongue, and 3 larynx tissues, while the HNSCC-HPV (−) group mainly contained 23 oral tongue and 18 larynx tissues. The expression level of SEC61G in the HNSCC-HPV (+) group was lower than that in the HNSCC-HPV (−) group. We also observed a relatively lower proportion of “arm-level gain” and “high amplification” copy-number variants in the HNSCC-HPV (+) group compared with the HNSCC-HPV (−) group. These results suggested a potential association between HPV infection and SEC61G CNA. In addition, we observed a relatively strong negative correlation between the immune infiltration of B cells and SEC61G expression in the HNSCC-HPV (+) group compared with the HNSCC-HPV (−) group. HPV infection might be involved in the potential association between SEC61G expression and the clinical therapeutic effect of treatments for HNSCC patients. CNA and the tumor microenvironment are possible mechanistic points to explore further.
Taken together, our findings first revealed the prognostic value of SEC61G for HNSCC patients. We observed a relationship between high expression of SEC61G and poor survival rates in HNSCC patients. The SEC61G gene may be implicated in the pathogenesis of HNSCC through the modulation of ribosomes, spliceosomes, proteasome degradation, cell adhesion, and the immune infiltration of B and CD8+ T cells. More in-depth molecular biological elucidations are needed.
References
Greenfield JJ, High S (1999) The Sec61 complex is located in both the ER and the ER-golgi intermediate compartment. J Cell Sci 112:1477–1486 (Pt 10)
Hartmann E, Sommer T, Prehn S, Görlich D, Jentsch S, Rapoport TA (1994) Evolutionary conservation of components of the protein translocation complex. Nature 6464(367):654–657. https://doi.org/10.1038/367654a0
Luan S, Li P, Yi T (2018) Series test of cluster and network analysis for lupus nephritis, before and after IFN-K-immunosuppressive therapy. Nephrology (Carlton) 11(23):997–1006. https://doi.org/10.1111/nep.13159
Staneva R, Rukova B, Hadjidekova S, Nesheva D, Antonova O, Dimitrov P, Simeonov V, Stamenov G, Cukuranovic R, Cukuranovic J, Stefanovic V, Polenakovic M, Dimova I, Hlushchuk R, Djonov V, Galabov A, Toncheva D (2013) Whole genome methylation array analysis reveals new aspects in Balkan endemic nephropathy etiology. BMC Nephrol 14:225. https://doi.org/10.1186/1471-2369-14-225
Stefanovic V, Toncheva D, Polenakovic M (2015) Balkan nephropathy. Clin Nephrol 1(83):64–69. https://doi.org/10.5414/cnp83s064 (7 Suppl)
Han B (2019) LncRNA LINC02418 regulates proliferation and apoptosis of non-small cell lung cancer cells by regulating miR-4677–3p/SEC61G. Eur Rev Med Pharmacol Sci 23(23):10354–10362
Servidei T, Meco D, Muto V, Bruselles A, Ciolfi A, Trivieri N, Lucchini M, Morosetti R, Mirabella M, Martini M, Caldarelli M, Lasorella A, Tartaglia M, Riccardi R (2017) Novel SEC61G-EGFR fusion gene in pediatric ependymomas discovered by clonal expansion of stem cells in absence of exogenous mitogens. Cancer Res 21(77):5860–5872. https://doi.org/10.1158/0008-5472.Can-17-0790
Liu B, Liu J, Liao Y, Jin C, Zhang Z, Zhao J, Liu K, Huang H, Cao H, Cheng Q (2019) Identification of SEC61G as a novel prognostic marker for predicting survival and response to therapies in patients with glioblastoma. Med Sci Monit 25:3624–3635
Chai YJ, Chae H, Kim K, Lee H, Choi S, Lee KE, Kim SW (2019) Comparative gene expression profiles in parathyroid adenoma and normal parathyroid tissue. J Clin Med 3(8):297. https://doi.org/10.3390/jcm8030297
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res W1(47):W556–W560. https://doi.org/10.1093/nar/gkz430
Mizuno H, Kitada K, Nakai K, Sarai A (2009) Prognoscan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2:18. https://doi.org/10.1186/1755-8794-2-18
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 269(6):1. https://doi.org/10.1126/scisignal.2004088
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 5(2):401–404. https://doi.org/10.1158/2159-8290.Cd-12-0095
Vasaikar SV, Straub P, Wang J, Zhang B (2018) LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res D1(46):D956–D963. https://doi.org/10.1093/nar/gkx1090
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res W1(W48):W509–W514
Neidert MC, Schoor O, Trautwein C, Trautwein N, Christ L, Melms A, Honegger J, Rammensee HG, Herold-Mende C, Dietrich PY, Stevanović S (2013) Natural HLA class I ligands from glioblastoma: extending the options for immunotherapy. J Neurooncol 3(111):285–294. https://doi.org/10.1007/s11060-012-1028-8
Lu Z, Zhou L, Killela P, Rasheed AB, Di C, Poe WE, McLendon RE, Bigner DD, Nicchitta C, Yan H (2009) Glioblastoma proto-oncogene SEC61gamma is required for tumor cell survival and response to endoplasmic reticulum stress. Cancer Res 23(69):9105–9111. https://doi.org/10.1158/0008-5472.Can-09-2775
Sheu JJ, Hua CH, Wan L, Lin YJ, Lai MT, Tseng HC, Jinawath N, Tsai MH, Chang NW, Lin CF, Lin CC, Hsieh LJ, Wang TL, Shih Ie M, Tsai FJ (2009) Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res 6(69):2568–2576. https://doi.org/10.1158/0008-5472.Can-08-3199
Tsukamoto Y, Uchida T, Karnan S, Noguchi T, Nguyen LT, Tanigawa M, Takeuchi I, Matsuura K, Hijiya N, Nakada C, Kishida T, Kawahara K, Ito H, Murakami K, Fujioka T, Seto M, Moriyama M (2008) Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol 4(216):471–482. https://doi.org/10.1002/path.2424
Armstrong DL, Zidovetzki R, Alarcón-Riquelme ME, Tsao BP, Criswell LA, Kimberly RP, Harley JB, Sivils KL, Vyse TJ, Gaffney PM, Langefeld CD, Jacob CO (2014) GWAS identifies novel SLE susceptibility genes and explains the association of the HLA region. Genes Immun 6(15):347–354. https://doi.org/10.1038/gene.2014.23
Qadir F, Aziz MA, Sari CP, Ma H, Dai H, Wang X, Raithatha D, Da Silva LGL, Hussain M, Poorkasreiy SP, Hutchison IL, Waseem A, Teh MT (2018) Transcriptome reprogramming by cancer exosomes: identification of novel molecular targets in matrix and immune modulation. Mol Cancer 1(17):97. https://doi.org/10.1186/s12943-018-0846-5
Navadgi-Patil VM, Burgers PM (2011) Cell-cycle-specific activators of the Mec1/ATR checkpoint kinase. Biochem Soc Trans 2(39):600–605. https://doi.org/10.1042/bst0390600
Niida H, Nakanishi M (2006) DNA damage checkpoints in mammals. Mutagenesis 1(21):3–9. https://doi.org/10.1093/mutage/gei063
Zhou ZQ, Zhao JJ, Chen CL, Liu Y, Zeng JX, Wu ZR, Tang Y, Zhu Q, Weng DS, Xia JC (2019) HUS1 checkpoint clamp component (HUS1) is a potential tumor suppressor in primary hepatocellular carcinoma. Mol Carcinog 1(58):76–87. https://doi.org/10.1002/mc.22908
Fridman WH, Galon J, Dieu-Nosjean MC, Cremer I, Fisson S, Damotte D, Pages F, Tartour E, Sautes-Fridman C (2011) Immune infiltration in human cancer: prognostic significance and disease control. Curr Top Microbiol Immunol 344:1–24. https://doi.org/10.1007/82_2010_46
Steven A, Seliger B (2018) The role of immune escape and immune cell infiltration in breast cancer. Breast Care (Basel) 1(13):16–21. https://doi.org/10.1159/000486585
Rahimi S (2020) HPV-related squamous cell carcinoma of oropharynx: a review. J Clin Pathol 10(73):624–629. https://doi.org/10.1136/jclinpath-2020-206686
Lechien JR, Descamps G, Seminerio I, Furgiuele S, Dequanter D, Mouawad F, Badoual C, Journe F, Saussez S (2020) HPV involvement in the tumor microenvironment and immune treatment in head and neck squamous cell carcinomas. Cancers (Basel) 5(12):1060. https://doi.org/10.3390/cancers12051060
De Virgilio A, Costantino A, Mercante G, Petruzzi G, Sebastiani D, Franzese C, Scorsetti M, Pellini R, Malvezzi L, Spriano G (2020) Present and future of de-intensification strategies in the treatment of oropharyngeal carcinoma. Curr Oncol Rep 9(22):91. https://doi.org/10.1007/s11912-020-00948-1
Funding
This work was supported by Key Clinical Discipline of Tianjin; National Natural Science Foundation of China (81971698); Tianjin Natural Science Foundation (19JCYBJC27200).
Author information
Authors and Affiliations
Contributions
GZ conceived and designed the project. YS, YL, WW, and GZ performed the analyses of expression, survival prognosis, genetic alteration, gene enrichment, and immune infiltration. YS and GZ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared that no conflict of interest exists.
Ethical approval
Ethics approval and patient written informed consent were not required because all analyses in our study were mainly performed based on data from the TCGA/GEO database.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
405_2021_6955_MOESM2_ESM.tif
Supplementary file2 Fig. S2 Three factors were analyzed in the study of the correlation between SEC61G expression and the survival prognosis of TCGA HNSCC patients. a Race; b histological grade; c sex (TIF 2147 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, Y., Liang, Y., Wang, W. et al. SEC61G identified as a prognostic biomarker of head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol 279, 2039–2048 (2022). https://doi.org/10.1007/s00405-021-06955-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-021-06955-7