Abstract
Inclusion body myositis (IBM) is unique across the spectrum of idiopathic inflammatory myopathies (IIM) due to its distinct clinical presentation and refractoriness to current treatment approaches. One explanation for this resistance may be the engagement of cell-autonomous mechanisms that sustain or promote disease progression of IBM independent of inflammatory activity. In this study, we focused on senescence of tissue-resident cells as potential driver of disease. For this purpose, we compared IBM patients to non-diseased controls and immune-mediated necrotizing myopathy patients. Histopathological analysis suggested that cellular senescence is a prominent feature of IBM, primarily affecting non-myogenic cells. In-depth analysis by single nuclei RNA sequencing allowed for the deconvolution and study of muscle-resident cell populations. Among these, we identified a specific cluster of fibro-adipogenic progenitors (FAPs) that demonstrated key hallmarks of senescence, including a pro-inflammatory secretome, expression of p21, increased β-galactosidase activity, and engagement of senescence pathways. FAP function is required for muscle cell health with changes to their phenotype potentially proving detrimental. In this respect, the transcriptomic landscape of IBM was also characterized by changes to the myogenic compartment demonstrating a pronounced loss of type 2A myofibers and a rarefication of acetylcholine receptor expressing myofibers. IBM muscle cells also engaged a specific pro-inflammatory phenotype defined by intracellular complement activity and the expression of immunogenic surface molecules. Skeletal muscle cell dysfunction may be linked to FAP senescence by a change in the collagen composition of the latter. Senescent FAPs lose collagen type XV expression, which is required to support myofibers’ structural integrity and neuromuscular junction formation in vitro. Taken together, this study demonstrates an altered phenotypical landscape of muscle-resident cells and that FAPs, and not myofibers, are the primary senescent cell type in IBM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spectrum of idiopathic inflammatory myopathies (IIM) is characterized by muscle inflammation as a pathogenic hallmark [1, 36]. Among IIM, inclusion body myositis (IBM) is a unique entity due to its characteristic clinical presentation, advanced age at onset, and notable refractoriness to contemporary treatment strategies [17]. As such, IBM leads to progressive muscle damage and weakness resulting in an unmet need for novel treatment approaches.
One explanation for the treatment refractoriness of IBM might be the instigation of cell-autonomous mechanisms that promote disease progression independent of ongoing inflammatory activity. Skeletal muscle is constituted of myogenic and non-myogenic cells. The complex interplay of these cells is required to maintain muscle health and for the resolution of inflammation [39, 43]. However, IBM imposes a burden of chronic inflammation on the compartment, potentially shifting the cellular homeostasis to a detrimental muscle phenotype [7, 17, 40]. Indeed, in response to chronic stress or damage, cells may assume a state of stable cell cycle arrest termed cellular senescence [44]. These cells continue to influence their environment by engaging a specific senescence-associated secretory phenotype (SASP) characterized by immunomodulatory cytokines, growth factors, and proteases. Following this line of argumentation, accumulation of senescent cells in skeletal muscle may constitute a cell-autonomous mechanism by which muscle-resident cells promote compartmentalized inflammation and fibrotic remodelling in IBM.
Succeeding the identification and characterization of highly cytotoxic, terminally differentiated CD8 T cells [16, 18], renewed attention has been focused on the development of immunomodulatory strategies for IBM, e.g. the depletion of KLRG1+ CD8 T cells [14]. However, addressing the deterioration of the skeletal muscle compartment might improve efficacy of those treatment strategies.
To understand whether IBM instigates cell-autonomous mechanisms, we studied cellular senescence in the skeletal muscles of IBM patients and compared them with those of non-diseased controls (NDC) and immune-mediated necrotizing myopathies (IMNM) as diseased control. Employing a single-nuclei transcriptomic approach, we describe a novel population of senescent fibro-adipogenic progenitors (FAPs) that may induce distinct alterations of the myogenic compartment. Given the growing interest and availability of senotherapeutics [21, 44], targeting these cells may provide an innovative strategy to ameliorate the muscle phenotype of IBM.
Methods
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of the Heinrich Heine University Duesseldorf (2016-053-f-S and 2021-1417) and the Charité Berlin (EA2/163/17). This study analysed skeletal muscle biopsies acquired from IIM patients and non-diseased controls (NDCs). All patients signed written informed consent before acquisition of the biopsy and clinical metadata.
Patient recruitment and clinical data
Patients were recruited from three tertiary centres specialized in the management of IIM (University Hospital Duesseldorf, Charité–University Medicine Berlin, and University Medical Center Göttingen). Patients treated in the outpatient clinic and patients admitted to the hospital were asked for study inclusion as well as written consent. Patients were recruited from January 2014 to January 2022. As disease control, we chose immune-mediated necrotizing myopathy (IMNM) as these patients rarely demonstrate extra-muscular manifestations, similar to IBM. IBM and IMNM patients were required to meet the European Neuromuscular Centre (ENMC) criteria for diagnosis [1, 2, 54]. NDCs served as an additional control cohort. As previously proposed by our group [53], these patients underwent muscle biopsy for diagnostic purposes, e.g. for myalgia. These patients were required not to have any objective muscle weakness or abnormal creatine kinase levels. On muscle histology, the patient's specimens were required to demonstrate no signs of inflammation or any other structural abnormalities. These patients had no myositis-specific or myositis-associated antibodies. Given the impact of age on the study readout of cellular senescence, NDC and IMNM patients were age-matched to IBM patients (maximum difference in age of 3 years). This study included 16 IBM, 16 NDC and 16 IMNM patients. The individual number of patients is given for each experiment as indicated. The disease duration was defined as the time between the first symptoms as reported by the patients to the time of muscle biopsy.
Biomaterial
All skeletal muscle specimens had been cryopreserved at − 80 °C before analysis according to the predefined standard operating procedure at the local biobank of the Heinrich Heine University Duesseldorf and the Charité Berlin.
Immunofluorescence
Immunofluorescence (IF) was performed as previously described [53]. Briefly, all stains were performed on 8 µm cryostat sections. We used irrelevant antibody stains (either mouse/rabbit monoclonal/polyclonal isotype controls) as negative controls, as well as omission of the primary antibody. The following antibodies were used for staining procedures: Laminin-β1 (Rabbit, 1:100, Novus Biologicals), p21 (Mouse, 1:50, Novus Biologicals), C3 (Rat, 1:100, Santa Cruz), PDGFRa (Mouse, 1:50, Novus Biologicals), α-bungarotoxin (Snake, 1:50, Invitrogen). The secondary antibodies for immunofluorescence were anti-rabbit (Alpaca, 1:200, Jackson ImmunoResearch), anti-rat (Alpaca, 1:200, Jackson ImmunoResearch) and anti-mouse (Alpaca, Jackson ImmunoResearch). Specimen were analysed using Zeiss Axio (for 10 and 20-fold magnification) or LSM 880, Zeiss (for 40 and 63-fold magnification) in cooperation with the Core Facility for Advanced Light Microscopy, Heinrich Heine University Düsseldorf. The biopsies were blinded for quantification with the diagnosis not possible to identify from the label. The scoring was performed in randomly distributed 10 high-power fields (HPF, based on the microscope used and the respective oculars ≙ 0.16 mm2) as previously described [53].
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from muscle specimens as previously described [53]. cDNA was synthesized using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). For reactions, 10 ng of cDNA were used on the 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) with the following running conditions: 95 °C 0:20, 95 °C 0:01, 60 °C 0:20, 45 cycles (values above 40 cycles were defined as “not expressed”). All targeted transcripts were run as triplicates. For each of these runs, the reference gene PGK1 has been included as internal control to normalize the relative expression of the targeted transcripts. The 2−ΔΔCT method was used to quantify gene expression of IBM and IMNM patients compared to NDCs.
Isolation, purification, and cultivation of primary human muscle cells
Muscle tissue was dissociated using a muscle dissociation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. For primary human skeletal muscle cell (PHMC) purification, we used a CD56 (clone N901, Beckman Coulter) antibody combined with microbead magnetic separation. For cultivation, we used Biolaminin 521 LN coated 6-well plates and skeletal muscle cell growth medium (PELOBiotech, Planegg, Germany) according to the manufacturer's instructions. For all experiments, we used differentiated PHMCs by allowing cells to grow to 100% confluence. These PHMC cultures contain different degrees of myoblast differentiation as well as myotubes.
Flow cytometry
PHMC were analysed by flow cytometry. Cells were treated and cultivated as indicated in the corresponding section. Cells were then washed with ice-cold PBS and scraped. The cells were centrifuged, and the pellet was washed before staining. PHMC were stained with the α-bungarotoxin antibody (CF555, Invitrogen). 1 × 106 PHMC per well was co-cultivated with the indicated concentrations of collagen for 24 h. Recombinant COL1A1 and COL15A1 were purchased from Abbexa, Cambridge, England.
Live/dead assay
For the assessment of viability, we used the LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells (Invitrogen, Waltham, Massachusetts) according to the manufacturer's instructions. Briefly, 1 × 106 PHMC per well were co-cultivated with the indicated concentrations of collagen for 24 h. Cell viability was assessed by fluorescence microscopy. Live cells were identified by green-fluorescent calcein-AM indicating intracellular esterase activity. Dead cells were identified by red-fluorescent ethidium homodimer-1 indicating a loss of membrane integrity. For each sample, 100 cells were counted and the frequencies of live or dead cells were recorded.
ATPase staining
ATPase enzyme histochemical preparations at pH 4.3, 4.6, and 9.4 were performed according to standard protocols to highlight type 1 and type 2A and 2X fibers [10].
Isolation of single nuclei from muscle biopsies
Single nuclei were isolated from frozen muscle biopsy specimens. Approximately 60 mg of muscle was used for each sample. All biopsies were taken from the quadriceps muscle (vastus medialis) approximately 3 cm proximal to the knee joint.
The single nuclei suspension was obtained using GEXSCOPE® Single Nucleus RNA Library Kit V2 (Singleron Biotechnologies) as previously described [34]. Briefly, on ice, the tissue was immersed in a cold nucleus separation solution and cut into small pieces. Further homogenization was achieved by performing 5 strokes with pestle A and 5 strokes with pestle B of the Kimble douncer (KIMBLE® KONTES® Dounce Tissue Grinder). The sample was then incubated on ice for 15 min, where the state of dissociation was monitored every 5 min under a light microscope. Following homogenization and digestion, the suspension was filtered using a 40 µm sterile strainer. The nuclei suspension was centrifuged at 200×g for 2 min at 4 °C, and the supernatant was centrifuged at 500×g for 5 min at 4 °C. The resulting pellet containing nuclei was resuspended in 0.25 ml cold nuclei suspension buffer. The quality of the nuclei was assessed by Trypan Blue staining (0.4% w/v, Gibco) under a light microscope. The nuclei were counted using propidium iodine with a Luna FX7 automated cell counter (Logos Biosystems, Villeneuve d’Ascq, France).
Library generation and sequencing
A total of 30 000 nuclei were loaded onto a microfluidic chip (Singleron GEXSCOPE® Single Nucleus RNA Library Kit V2) for a minimal capture of 6000 nuclei for each sample. Barcoded beads containing a unique cell barcode were loaded into the chip, and nuclei were lysed. After nuclei lysis, polyadenylated RNA was captured onto the Barcode Beads by the poly (dT) sequence. Barcode Beads with captured mRNA molecules were collected and subjected to reverse transcription reaction. The cDNA was then amplified and QC. NGS libraries generated were sequenced on an Illumina NovaSeq 6000 instrument using a paired-end 150 bp approach. The reads were demultiplexed on Illumina’s BaseCloud, and fastq files were used to initiate data analysis.
Bioinformatics workflow
Illumina reads (fastq files) were processed to gene expression matrices using CeleScope™ (v1.14.1., www.github.com/singleron-RD/CeleScope; Singleron Biotechnologies). Briefly, fastq files were demultiplexed according to their respective cell barcodes and UMIs. Adapter sequences and poly-A tails were trimmed (cutadapt; https://cutadapt.readthedocs.io/en/stable/installation.html) and the trimmed Read2 reads were aligned to the GRCh38 version of the human genome with Ensembl version 92 gene annotations (STAR v2.6.1a_08-27 [https://github.com/alexdobin/STAR] and featureCounts 2.0.1). We excluded cells with a unique feature count over 2500 or below 200 or cells with a mitochondrial ratio of more than 5%. A combined digital expression matrix was constructed, containing all sequenced experiments, for downstream analysis. Analysis was performed in Seurat (v4.3.0). Data were normalized using the NormalizeData function and 2000 features with high cell-to-cell variation were calculated using the FindVariableFeatures function. For data integration of IBM and NDC samples, we employed the integration workflow as proposed for the Seurat workspace [57]. The FindIntegrationAnchors function was used to assemble all snRNA-seq datasets into an integrated and unbatched dataset. Next, to reduce dimensionality of the datasets, the RunPCA function was conducted with default parameters on linear-transformation scaled data generated by the ScaleData function. After integration, we performed a modularity-optimized Louvain clustering with a resolution of 0.8.
Cluster annotations
The FindAllMarkers function was used to identify cluster-specific markers with a fold-change threshold of 0.25 and min.pct set to 0.25. Clusters were then classified and annotated based on expressions of canonical markers of particular cell types. The complete list of markers for all clusters is available as Supplemental File 1.
Subclustering
As indicated, clusters of interest were extracted from the integrated dataset for in-depth analysis and subclustering. Principal component analysis and clustering were repeated as described above.
Differential expression testing and functional enrichment analysis
Differential gene expression testing was performed using the FindMarkers function using Wilcox testing and a fold-change threshold of 0.25. The Bonferroni correction was used based on the total number of genes in the dataset. Differentially expressed genes (DEGs) were selected based on the adjusted p value.
DEGs were used for gene set enrichment analysis (GSEA) using the ClusterProfiler package (v4.3.1) and the Gene Ontology (GO) “Biological Process” and the “Reactome” databases.
For targeted GSEA analysis, we used Genetrail (https://genetrail.bioinf.uni-sb.de/start.html) with the SenMayo gene set (v2023.1.Hs, available from https://www.gsea-msigdb.org/gsea/index.jsp as “SAUL_SEN_MAYO”).
Cell–cell communication was analysed using the CellChat package (v1.5.0) as proposed by the developers.
Statistical analysis
Statistical analysis was performed using R 3.5.3. Data were presented as median with IQR, mean ± standard deviation (SD), as absolute (n) or relative frequencies (%). Differences between the two groups were analysed using the Mann–Whitney U test. The Kruskal–Wallis test was used for multiple groups.
p > 0.05 was classified as not significant, p < 0.05 as significant (*), p < 0.01 (**), p < 0.001 (***).
Results
p21+ senescent cells are abundant in the muscle of IBM
First, we sought to determine whether the number of senescent cells is altered in IBM compared to NDC and IMNM patients. For this purpose, we recruited a multicentric cohort of 48 patients (16 per group) in three tertiary centres specialized in managing IIMs. The clinical and epidemiological characteristics are given in Table 1. Given that age is likely to have a major impact on the development of cellular senescence, NDC and IMNM patients were matched to IBM by age to account for this confounder. For diagnosis of IBM and IMNM, patients were required to meet the current ENMC criteria for diagnosis, respectively [2, 35]. Briefly, IBM patients were 73 (45–87) years old with 7 males and 9 females. 5 out of 16 patients received intravenous immunoglobulin treatment at the time of biopsy. No other immunosuppressants were recorded for the IBM cohort.
As the first readout, we focused on the study of p21, an established marker for cellular senescence [15]. This choice was motivated by several recent studies in murine and human tissues demonstrating that p21 governs a senescent program in skeletal muscle and serves as a marker for muscle cell senescence [12, 50, 64]. We assessed the number of p21+ cells by IF (Fig. 1a). Given the nuclear expression of p21, only cells that demonstrated clear co-localization of p21 and DAPI were considered to be p21+. Indeed, the number of p21+ cells was higher in IBM as compared to NDC and IMNM patients (Fig. 1b). To further corroborate this data, we also analysed p21 at the gene level by PCR observing an increased expression of CDKN1A (coding for p21) in IBM compared to NDC or IMNM patients (Fig. 1c). Intriguingly, a substantial number of p21+ cells were detected in the perimysial area in IBM. To clarify the origin of those p21+ cells, we also counted the number of p21+ myonuclei (Fig. 1d). Myonuclei were defined as nuclei localized inside the boundaries of the corresponding myofiber (Fig. 1e). IBM and IMNM patients demonstrated similar numbers of p21+ myonuclei leading us to hypothesize that non-myogenic cells resident in skeletal muscle might primarily assume a senescent phenotype in IBM. Of note, longer disease durations were associated with an increased number of p21+ cells (Fig. 1f). Following this line of argumentation, we aimed to study senescence at the level of single cells (or nuclei) in the landscape of myogenic and non-myogenic cells in the IBM muscle.
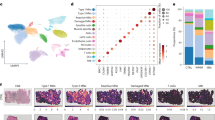
p21+ senescent cells are abundant in muscle of IBM. a Immunofluorescence staining of p21 (red), laminin-β1 (green), and DAPI (blue) in muscle specimens of NDCs (n = 16), IBM (n = 16), and IMNM (n = 16) patients. Patients were matched by age. Arrows indicate single p21+ cells or clusters of p21+ cells. b p21 + cells were counted in randomly distributed 10 HPF (≙ 0.16 mm2). The biopsies were blinded for quantification, with the diagnosis impossible to identify from the label. p21+ cells were defined as cells with a clear expression of p21 in the nucleus. c RT-qPCR analysis of CDKN1A coding for p21 in muscle specimen (n = 16 per group). The 2−ΔΔCT method was used for normalization. CDKN1A is significantly over-expressed to NDC for both IBM and IMNM. d Quantification of p21+ myonuclei in muscle in muscle specimen (n = 16 per group). p21+ myonuclei were defined as nuclei located inside a muscle fiber. e Exemplary image of a p21+ nucleus located inside a myofiber. Differences between groups were analysed by Kruskal–Wallis test followed by post hoc testing. f Simple linear regression of the number of p21+ cells and the disease duration of each IBM patient. The disease duration was defined as the time in months between the first symptoms as reported by the patient and the time of biopsy. The dotted line indicates the 95% confidence interval. Significance was tested by the likelihood test. *p < 0.05, **p < 0.01, ***p < 0.001. NDC non-diseased control; HPF high-power field; IBM inclusion body myositis; IMNM immune-mediated necrotizing myopathy; r2 coefficient of determination; RT-qPCR real-time quantitative polymerase chain reaction
Single-nuclei RNA sequencing deconvolutes distinct cell populations and allocates senescence in IBM muscle mainly to fibro-adipogenic progenitors
For this purpose, we performed snRNA-seq of three IBM patients and three NDCs. We chose snRNA-seq as an alternative to single-cell RNA-seq due to the large size of myofibers that restricts the latter’s use for studying skeletal muscle [30, 50]. For snRNA-seq, all muscle specimens were obtained from the quadriceps muscle, specifically the vastus medialis, approximately 3 cm proximal to the knee joint. IBM patients were 65, 68, and 71 years old with two male and one female subject. NDCs were age- and sex-matched. All IBM patients and all NDCs were treatment naïve.
After quality control, ~ 30,000 nuclei of which ~ 13,000 were obtained from IBM muscle and ~ 17,000 from NDC were integrated into a batch-corrected expression matrix for downstream analysis. Using graph-based clustering of uniform manifold approximation and projection (UMAP), we observed nine major cell types or subtypes based on differential marker expression (Fig. 2a, b). Cell types were assigned according to the expression of canonical markers (a full list of all marker genes is given in Supplemental File 1) based on previous transcriptomic studies of skeletal muscle [9, 30, 50, 52]. While non-myogenic cells were separated based on their transcriptomic profiles, the classification of myogenic cells was more challenging. Briefly, human skeletal muscle may be classified into slow-twitch (type 1) and fast-twitch (type 2A and type 2X) myofibers based on the presence or absence of specific myosin heavy chains (MYH) [42]. For this study, we annotated the myogenic compartment based on this classification, although it should be noted that some myofibers are considered hybrids falling between canonical subtypes [58]. First, we annotated type 1 myonuclei based on the expression of canonical markers such as MYH7 or MYH7B (Fig. 2a, b) [50, 58]. Second, a subgroup of fast-twitch (type 2X) myonuclei were identified based on the expression of MYH1 and the absence of MYH7 [50, 58]. As previously described by Murgia et al., specific markers, except MYH2, for the type 2A subgroup of fast-twitch myofibers are currently lacking [42]. Following this line of argumentation, we annotated the final myonuclei cluster as type 2A given the absence of MYH7 and MYH1 expression specific for type 1 and type 2X fibers, respectively.
Single-nuclei RNA sequencing of IBM and NDC muscle. A total of 6 frozen muscle specimens were processed for single nuclei-RNAseq (3 samples per group). A total of ~ 30,000 nuclei were included for downstream processing and clustering after quality control. a UMAP embedding demonstrating distinct clusters of cell types and subtypes. b Clustered dot plot visualization of top-regulated marker genes. The mean expression for each cluster is indicated by colour code. The dot size indicates the percent of expressing cells. Clusters were annotated based on marker genes. c Expression of CKDN1A (coding for p21) across the UMAP embedding. The mean expression for each cell is indicated by the colour code. d Frequency of CDKN1A+ cells for each cell cluster as indicated for IBM patients and NDC. Differences between groups were analysed by the Kruskal–Wallis test followed by post hoc testing. e Gene set enrichment analysis (GSEA) for the SenMayo dataset. Differentially expressed genes were determined by the FindMarkers function using Wilcox testing and a fold-change threshold of 0.25. The Bonferroni correction was used for correction for multiple testing. DEGs specific to the IBM dataset were entered into the GSEA. The Kolmogorov–Smirnov test, followed by post hoc correction, was used to determine the significance. **p < 0.01. ACTA2 actin alpha 2; CDKN1A cyclin dependent kinase inhibitor 1A; CDH5 cadherin 5; COL collagen; EMCN endomucin; FBN1 fibrillin-1; ITGAL integrin subunit alpha L; LILRB5 leukocyte immunoglobulin like receptor B5; MRC1 mannose receptor C-type 1; MYBPC2 myosin binding protein C2; MYL9 myosin light chain 9; NDC non-diseased control; IBM inclusion body myositis; SIGLEC1 sialic acid binding Ig like lectin 1; TPM3 tropomyosin 3; PECAM1 platelet and endothelial cell adhesion molecule 1; PAX7 paired box 7; PTPRC protein tyrosine phosphatase type C (CD45); UMAP uniform manifold approximation and projection
To understand the distribution of senescence in myogenic and non-myogenic cells, we determined the expression of CDKN1A across the dataset (Fig. 2c). To compare the cluster-specific frequencies, we calculated the number of CDKN1A expressing cells as percentage of all cells in a cluster for IBM and NDC (Fig. 2d). As described in the previous section, the relative frequencies of CDKN1A+ nuclei were comparable between IBM and NDC for myonuclei. Intriguingly, the frequencies of CDKN1A expressing FAPs and monocytes/macrophages were strongly increased in IBM compared to NDC. However, it should be noted that the expression of a single gene is unlikely to capture the heterogeneity of cellular senescence across tissues. To address this caveat and to further corroborate the engagement of senescence pathways, we determined the differentially expressed genes (DEGs) between IBM and NDC for all cell clusters. Next, we employed the SenMayo gene set for enrichment analysis [56]. Briefly, the SenMayo gene set is a panel composed of 125 key genes associated with senescence signalling pathways. We chose SenMayo as this gene set has been benchmarked against existing senescence or SASP gene sets and outperformed the latter in the detection of senescent cells [56]. DEG lists of all clusters were entered into GSEA for the SenMayo gene set (Fig. 2e). Only the FAPs cell cluster was enriched for SenMayo in IBM compared to NDC. SnRNA-seq demonstrates the capability of discerning distinct cellular subtypes and suggests that non-myogenic cells assume a senescent phenotype in IBM.
Single-nuclei RNA sequencing identifies a novel population of senescent FAPs that resides in IBM muscle
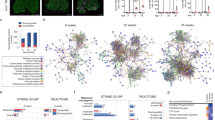
Next, we focused on the FAP population and extracted this cell cluster for in-depth analysis (Fig. 3a). A total of ~ 3200 FAP nuclei were analysed. FAPs are the lineage precursors of specialized non-myogenic cells, including activated fibroblasts, adipocytes, and osteogenic cells [8]. We performed subclustering on these cells and observed 4 subpopulations of FAPs (Fig. 3b, c). We determined marker genes for all FAP subtypes and compared these with previous studies [9, 55]. Although there currently exists no consensus regarding canonical marker genes of human FAPs, Perez et al. previously reported subtypes of FAPs associated with ageing based on the expression of the ryanodine receptor 1 (RYR1) and triadin (TRDN) [50]. In our dataset, the clusters FAPs2 and FAPs3 were also characterized by RYR1 and TRDN expression. Given the advanced age of IBM patients and NDCs in our cohort, we suspect that these FAPs are reminiscent of these previously reported FAP phenotypes. In contrast, we also determined two clusters of FAPs only found in IBM patients, but not in NDCs. The first cluster was defined by the expression of lumican (LUM) and fibrillin-1 (FBN1). LUM+/FBN1+ FAPs were previously described [55] and are thought to resemble neprilysin (MME) expressing FAPs associated with fatty infiltration of skeletal muscle. Intriguingly, we also investigated the expression of CDKN1A across FAP subtypes and found that the expression of this senescence marker is largely restricted to the LUM+/FBN+ FAP population. Consequently, we termed these FAPs CDKN1A+ to underline their senescent phenotype and association with the p21 pathway. This FAP phenotype is found in IBM but not in NDCs (Fig. 3e). The identity of the fourth FAP subtype (FAPs3) is characterized by the expression of XIAP-associated factor 1 (XAF1) and dynamin-1 (DNM1). These markers were previously described in FAPs [19], however, their role is currently unknown.
A novel population of senescent FAPs resides in IBM muscle. a UMAP embedding of the full dataset. FAPs are highlighted in green. All FAPs were extracted for downstream analysis and subclustering. b Subcluster analysis of the FAP population. Four FAP populations are identified based on their marker genes. c UMAP embedding displaying the origin for each nucleus. d Clustered dot plot visualization of top-regulated marker genes. The mean expression for each cluster is indicated by colour code. The dot size indicates the percent of expressing cells. Cluster were annotated based on marker genes. e Expression of CKDN1A (coding for p21) across the UMAP embedding split into the IBM (left) and NDC (right) datasets. The mean expression for each cell is indicated by the colour code. f Gene set enrichment analysis (GSEA) for the GO-BP dataset for the CDKN1A+ FAP cluster. Differentially expressed genes were determined by the FindMarkers function using Wilcox testing and a fold-change threshold of 0.25. The Bonferroni correction was used for correction for multiple testing. DEGs specific to the IBM dataset were entered into the GSEA. The Kolmogorov–Smirnov test, followed by a post hoc correction, was used to determine the significance. g GSEA analysis for the SenMayo dataset for the DEGs obtained from the CDKN1A+ FAP cluster. The running-sum statistic is in red, with the position in the ranked DEG list in black. Genes were sorted by fold change. The Kolmogorov–Smirnov test, followed by a post hoc correction, was used to determine the significance. h Violin plots displaying the normalized gene expression of the indicated genes for each FAP cluster. CDKN1A cyclin dependent kinase inhibitor 1A; DNM1 dynamin-1; FBN1 fibrillin-1; GO-BP gene ontology biological processes; LUM lumican; NDC non-diseased control; IBM inclusion body myositis; RYR1 ryanodine receptor 1; TRDN triadin; UMAP uniform manifold approximation and projection; XAF1 XIAP-associated factor 1
Given the scope of this study, we focused our analysis on the CDKN1A+ FAP population. To understand whether these FAPs exhibit senescence features, we determined the marker genes for this cell cluster and performed GSEA for the biological processes (BP) database (Fig. 3f). Here, the humoral immune response was the most enriched term for the CDKN1A+ FAP population. This observation is consistent with current knowledge on the senescent phenotype, as these cells are known to engage a pro-inflammatory secretome (e.g. SASP). To further corroborate a senescent phenotype for these FAPs, we performed GSEA for the SenMayo gene set. Here, marker genes of CDKN1A+ FAPs were enriched for SenMayo indicating the engagement of senescence pathways in these cells (Fig. 3g). We manually screened the marker genes of CDKN1A+ FAPs and observed that these cells express endosialin (CD248), a glycoprotein and key regulator of tissue fibrosis [48, 51, 59]. CD248 expression was only detected in CDKN1A+ FAPs but not in other FAP subtypes (Fig. 3h). Besides CD248, CDKN1A+ FAPs also engage the Jun/JunB signalling pathway [37, 56]. This pathway has been reported to govern fibroblast senescence by inhibition of insulin growth factor-1 (IGF-1) [37]. Surprisingly, CDKN1A+ FAPs also demonstrate upregulation of complement factors such as complement factor 3 (C3), complement factor H (CFH) or complement factor D (CFD). In respect to signalling, CDKN1A+ FAPs express CD44, a pro-inflammatory cell-to-cell signalling receptor, as well as receptors for transforming growth factor beta (TGF-β, TGFBR2, and TGFBR3). To corroborate that the p21+ cells in the perimysium/endomysium are indeed the FAP phenotype detected in the transcriptomic dataset, we performed IF staining of IBM biopsy specimens (Suppl. Figure 1). Here, p21+ cells located in the perimysium were identified by the canonical FAP marker platelet-derived growth factor receptor α (PDGRA).
To further cross-validate the engagement of senescence in these cells, we additionally investigated the activity of the senescence-associated β-galactosidase (SA-β-Gal) [38, 60]. To enable intracellular staining in tissue sections, we employed SPiDER-β-Gal which exerts higher cell permeability than traditional SA-β-Gal [29]. Indeed, p21 (CDKN1A)+ FAPs were detected in the perimysium of IBM patients and stained positive for SA-β-Gal as an additional biomarker for cellular senescence (Suppl. Fig. 2a).
Succinctly, CDKN1A + FAPs demonstrate key features of cellular senescence including expression of senescence markers, engagement of a pro-inflammatory secretory phenotype and pro-fibrotic surface molecules, the activity of SA-β-gal and a senescence-associated transcriptomic signature.
IBM demonstrates a pronounced loss of type 2A muscle fibers
Next, we analysed the myonuclear compartment of IBM and compared it to NDCs (Fig. 4a). Consistent with the current knowledge on IBM histopathology [17], the immune cell clusters of monocytes/macrophages and T, B, and NK cells were expanded in IBM compared to NDC (Fig. 4a, b). Further, we also observed that the number of type 2A and to a lesser extent type 2X myofibers were strongly reduced in IBM compared to NDC.
IBM demonstrates a pronounced loss of type 2A muscle fibers. a UMAP embedding of the full dataset split into IBM (left) and NDC (right). Cell populations are indicated by their colour code. b Relative frequency of each cell type or subtype in the IBM and the NDC dataset as a stacked bar plot. c UMAP embedding of the full dataset. Myonuclei are highlighted in dark red. These nuclei were extracted for downstream analysis d UMAP embedding of the myonuclei subclusters. A total of five populations were obtained from subclustering. e Clustered dot plot visualization of top-regulated marker genes. The mean expression for each cluster is indicated by colour code. The dot size indicates the percent of expressing cells. Clusters were annotated based on marker genes. f UMAP embedding for the myonuclei subcluster split into the IBM (left) and NDC (right) datasets. Subclusters are colour coded. g Exemplary ATPase staining for muscle specimens obtained from NDC, IBM, and IMNM patients. 8 patients were analysed by ATPase staining for each group. Muscle slices were incubated at a pH of 4.6, inactivating the myosin-ATPase of specific muscle fiber types. Type 1 muscle fibers are dark brown, type 2A is light brown, and type 2X are of an intermediate colour. The contrast and intensity vary between muscle specimens. A total of 10 high-power fields were counted. The corresponding statistical analysis is displayed in Suppl. Fig. 1B. ACHE acetylcholine esterase; CHRNA1 cholinergic receptor nicotinic alpha 1 subunit; FAP fibro-adipogenic progenitor; HLA human leukocyte antigen; IMNM immune-mediated necrotizing myopathy; MYH myosin heavy chain; MuSC muscle stem cell; NDC non-diseased control; IBM inclusion body myositis; TNFAIP2 TNF Alpha Induced Protein 2; UMAP uniform manifold approximation and projection
To gain further insight into the phenotype of myogenic cells in IBM, we extracted the myonuclei from the full dataset (Fig. 4c) and performed subclustering on these nuclei (Fig. 4d). ~ 20,000 myonuclei were analysed by this approach. Subclustering revealed five distinct populations of myonuclei. Briefly, we detected type 1 myonuclei defined by MYH7 and MYH7B expression as well as type 2X myonuclei expressing MYH1 (Fig. 4e). Type 2A myonuclei were defined, as above, by the lack of MYH1 and MYH7 while expressing MYH2. This subclustering revealed two additional clusters of myonuclei. First, we detected a population of myonuclei that were defined by the expression of the acetylcholine receptor (CHRNA1), the acetylcholine esterase (ACHE) and the muscle associated receptor tyrosine kinase (MUSK). Given their unique expression profile of NMJ proteins, these nuclei are likely to belong to myonuclei constituting the neuromuscular synapse (NMJ). Second, a cluster of myonuclei emerged that expressed high levels of pro-inflammatory molecules, such as HLA-A, HLA-B or HLA-C constituting the major histocompatibility complex (MHC) class 1. Further, these myonuclei displayed TNF alpha-induced protein 2 (TNFAIP2) and complement protein 3 (C3) as marker genes. Given their unique expression profile of pro-inflammatory genes, we termed these myonuclei as inflammatory. Phenotypically, these myonuclei express MYH7 and MYH7B suggesting that they resemble type 1 myofibers. Comparing IBM and NDC, there was a strong reduction in the number of type 2A myonuclei (Fig. 4f). Concurrently, CHRNA1+ myonuclei were also strongly reduced, while type 1 and type 2X myonuclei appeared largely unaltered.
For cross-validation of the specific loss of type 2A myofibers, we determined the frequency of myofiber types based on their ATPase staining after incubation at pH 4.6 [58]. We chose this approach to demonstrate the reduction in type 2A myofibers on the protein level and in a method independent of the MYH classification. Here, type 2A myofibers were markedly reduced in IBM compared to NDCs or IMNM patients (Fig. 4g, Suppl. Fig. 2b). In line, the relative frequency of type 1 myofibers was higher in IBM compared to the other groups. As such, a selective loss of type 2A myofibers appears to characterize the pathomorphology of IBM.
Skeletal muscle cells assume an inflammatory reprogramming in IBM
Next, we focused on the subpopulation of inflammatory myonuclei. Intriguingly, while this population was detected in IBM and NDC, their phenotype was altered depending on their origin (Fig. 5a). These myonuclei demonstrated high expression of marker genes such as C3 or HLA-A only in IBM, but not in NDC. Notably, these inflammatory myonuclei were also the primary source of transforming growth factor beta (TGFB1). Further, we sought to cross-validate the presence of these myonuclei in muscle. For this purpose, we performed IF staining for intracellular C3 as expression for this gene was only detected in inflammatory myonuclei from IBM patients, but not in other subtypes of myonuclei in either NDC or IBM suggesting sufficient specificity to serve as a marker (Fig. 5b). Indeed, myofibers with intracellular C3 staining were abundant in IBM compared to NDC or IMNM muscle (Fig. 5c, d). To further characterize the phenotype of these myofibers, we computed the DEGs comparing inflammatory myonuclei between IBM and NDC. GSEA analysis of these DEGs for the GO-BP database indicated engagement of pro-inflammatory pathways, including the immunoglobulin mediated immune response, adaptive immune response, innate immune response and, interestingly, the B cell mediated immune response (Fig. 5e).
Skeletal muscle cells assume an inflammatory reprogramming in IBM. a UMAP embedding of the myonuclei split into IBM (left) and NDC (right). The expression of each gene indicated on the right is colour coded. b Violin plot displaying the expression of complement factor 3 (C3) for each myonuclei subset for IBM and NDC. Only inflammatory myonuclei obtained from IBM patients express C3. c Exemplary staining for C3 (red) laminin-β1 (green) and DAPI (blue) for NDC, IBM, and IMNM patients. 16 patients were analysed for each group by immunofluorescence. d C3+ myofibers were counted in randomly distributed 10 HPF (≙ 0.16 mm2). The biopsies were blinded for quantification, making the diagnosis impossible to identify from the label. e Visualization of the gene set enrichment analysis (GSEA) for the GO-BP dataset as a gene concept network. GSEA was performed from the DEGs comparing inflammatory myonuclei from IBM patients and NDC. The Kolmogorov–Smirnov test followed by post hoc correction was used to determine the significance. The top five GO terms for the BP dataset are colour coded. A line connects the corresponding genes constituting each term. The fold change for each gene comprising the GO term comparing IBM and NDC is indicated in red. The number of genes for each GO term is indicated as a dot size. CHRNA1 cholinergic receptor nicotinic alpha 1 subunit; HLA human leukocyte antigen; HPF high-power field; MYH myosin heavy chain; NDC non-diseased control; IBM inclusion body myositis; TGFB1 transforming growth factor beta 1; UMAP uniform manifold approximation and projection
IBM myofibers lose their potential for endplate formation
To further characterize the changes to the myogenic compartment in IBM, we aimed to validate the reduction in AChR (CHRNA1) expressing muscle fibers (Fig. 6a). On the transcriptomic level, cells concurrently expressing the AChR as well as the acetylcholine esterase (ACHE) were reduced in IBM (Fig. 6b). Similarly, the expression of CHRNA1 in myonuclei was detected in NDC, but not in IBM (Fig. 6c). Formation of the NMJ is required for nerve-muscle communication. To visualize the neuromuscular endplate, we performed IF for α-bungarotoxin binding the AChR and counted the number of NMJs. Here, the number of NMJs was strongly reduced in IBM compared to NDCs or IMNM patients (Fig. 6d, e). NMJs were defined by the concurrent binding of α-bungarotoxin and their characteristic topography. These data suggest that endplate formation is impaired in IBM muscle potentially affecting nerve-muscle communication.
IBM myofibers lose their potential for endplate formation. a UMAP embedding of the myonuclei split into IBM (left) and NDC (right). The myonuclei cluster expressing the acetylcholine receptor (CHRNA1) is marked in orange. b Density plot generated with the Nebulosa package of cells expressing acetylcholine receptor and acetylcholine esterase (ACHE). Areas of high cellular density are indicated in orange. c Violin plot for the CHRNA1 expression across the dataset. CHRNA1-expressing nuclei are largely absent in IBM muscle. d Exemplary staining of a NMJ in NDC, IBM, and IMNM patients, respectively. α-bungarotoxin was used to label the acetylcholine receptors of the NMJ in green. 12 patients were analysed for each group. e While NMJ was detected in all muscle samples, their frequency was strongly reduced in IBM. NMJ was defined by its characteristic topography and expression of the acetylcholine receptor. NMJ was counted in randomly distributed 10 HPF (≙ 0.16 mm2). The biopsies were blinded for quantification, with the diagnosis impossible to identify from the label. 12 patients were analysed by immunofluorescence for each group. f Frequencies of NMJ in NDC, IBM, and IMNM patients. Differences between groups were analysed by Kruskal–Wallis test followed by post hoc testing. ***p < 0.001. ACHE acetylcholine esterase; CHRNA1 cholinergic receptor nicotinic alpha 1 subunit; HPF high-power field; NDC non-diseased control; IBM inclusion body myositis; IMNM immune-mediated necrotizing myopathy; UMAP uniform manifold approximation and projection
Conclusively, single nuclei analysis suggests that the myogenic compartment of IBM is characterized by the loss of type 2A myonuclei and of myonuclei that constitute the NMJ. Conversely, C3 expression identifies a distinct pro-inflammatory subpopulation of myonuclei residing in IBM muscle.
FAPs demonstrate a shifted collagen homeostasis with potential consequences for muscle health in IBM
Finally, we aimed to determine whether senescence of FAPs may be linked to changes to the myogenic compartment. FAPs and their fibroblast lineage are the primary sources of extracellular matrix proteins required for skeletal muscle homeostasis [6]. Manual screening of the top dysregulated genes in CDKN1A+ FAPs revealed a shift in the expression of collagens, specifically of COL15A1, an isoform coding for collagen type XV, as well as COL1A1 and COL1A2, coding for collagen type I. Expression of all three collagen coding genes is allocated to the FAP cluster in both NDC and IBM (Fig. 7a). However, further analysis of the FAP subcluster indicates a shift of collagen expression between IBM and NDCs with CDKN1A+ FAPs demonstrating a sharp downregulation of COL15A1 (p = 5 × 10–32), while engaging COL1A1 (p = 1 × 10–87) as well as COL1A2 (p = 1 × 10–70) expression (Fig. 7b, c). Both types of collagens are required to stabilize skeletal muscle cells and their differentiation. However, they vary in their biological functions [6, 24].
FAPs demonstrate a shifted collagen homeostasis with potential consequences for muscle health in IBM. a UMAP embedding of the full dataset with the corresponding gene expression indicated by the colour code. The FAP cluster is in the upper left. b UMAP of the FAP subclusters split into their origin of IBM patients or NDC. The annotation of the corresponding subcluster is given beside the plot. CDKN1A+ FAPs are in the lower left. c UMAPs demonstrating COL1A1, COL1A2, and COL15A1 expression across the FAP cluster and its subclusters. COL15A1 is largely absent in the CDKN1A+ FAP subcluster. d Analysis of the cell–cell communication across the IBM and NDC datasets using the CellChat package. The number of the inferred ligand/receptor interactions is indicated by the size of an arrow connecting two cell populations. The larger the arrow, the higher the number of interactions between the cell populations. The ligand/receptor pairings were tested against the CellChat library for significance. e Representative flow cytometry scatter plots displaying SSC vs α-bungarotoxin for primary human muscle cells (PHMC) treated with COL15A1 or COL1A1. f Percentage of α-bungarotoxin + PHMC treated with COL15A1 or COL1A1 or vehicle. N = 8 per group. g Representative live/dead staining for PHMCs incubated with COL15A1 or COL1A1 in addition to 10 ng/ml interferon-γ (INF-γ). Live cells were identified by green-fluorescent calcein-AM, indicating intracellular esterase activity. Dead cells were identified by red-fluorescent ethidium homodimer-1, indicating a loss of membrane integrity. h 100 cells were counted for each sample, and the frequencies of live or dead cells were recorded. N = 8 per group. Differences between groups were analysed by Kruskal–Wallis test followed by post hoc testing. *p < 0.05, ***p < 0.001. CDKN1A cyclin dependent kinase inhibitor 1A; COL collagen; FAP fibro-adipogenic progenitor; NDC non-diseased control; IBM inclusion body myositis; IMNM immune-mediated necrotizing myopathy; UMAP uniform manifold approximation and projection
Following this line of argumentation, we suspected that differences in collagen might affect skeletal muscle cell homeostasis. To understand the communication between FAPs and myofibers better, we analyzed cell–cell communication across the transcriptomic dataset. We used the CellChat package to calculate the inferred intercellular communication based on an established receptor/ligand database [27]. We extracted all nuclei acquired from IBM patients and NDCs for individual analysis. Here, quantification of significantly over-expressed ligand/receptor pairings revealed that cell–cell communication is amplified across cell types and subtypes in IBM compared to NDCs (Fig. 7d). Intriguingly, FAPs appear to be a major communication hub interacting with myonuclei, immune cells and MuSCs. Indeed, the collagen and laminin signalling pathways constituted most outgoing ligands in IBM FAPs. Both signalling pathways belong to the KEGG pathway of extracellular matrix-receptor interaction (hsa04512). We suspect that a shift of the extracellular matrix composition might have functional implications for myofiber health in IBM.
To test this assumption in vitro, we cultivated primary human muscle cells (PHMC) and analysed them by flow cytometry. After reaching full confluence, PHMCs were treated with either 20 μg/ml of COL15A1 or COL1A1 [26, 33]. As specific collagen compositions are required for the successful maturation of the NMJ [33, 47], we analysed the amount of NMJ forming myofibers by quantification of α-bungarotoxin (Fig. 7e). In this setup, treatment with recombinant COL15A1 or with vehicle improved NMJ formation as compared to COL1A1. Next, we also aimed to understand if differences in collagen composition might affect PHMC survival. For this purpose, differentiated PHMCs were treated with 20 μg/ml of COL15A1 or 20 μg/ml COL1A1. To reflect inflammatory conditions in IBM, PHMCs were additionally treated with 10 ng/ml interferon-γ (INF-γ). After 48 h of incubation, we determined the number of dead PHMCs by live/dead staining (Fig. 7g, h). Interestingly, COL15A1 improved PHMC survival in response to INF-γ compared to COL1A1 or vehicle. These data suggest that senescent FAPs demonstrate an altered collagen homeostasis shifted towards collagen type I.
Discussion
FAPs are a mesenchymal cell population with high phenotypical plasticity that is crucially involved in skeletal muscle homeostasis and regeneration [5, 23]. Here, we report that tissue-resident FAPs, not myofibers, are the main cell type assuming a senescent phenotype in IBM. Depending on environmental cues, FAPs may differentiate into fibroblasts or adipocytes. In response to muscle damage, FAPs proliferate, expand and accumulate, constituting the main source of extracellular matrix proteins [23, 41]. Depletion of FAPs hinders muscle repair underscoring their functional importance [23, 41]. Conversely, in conditions of chronic muscle damage, FAPs may prove detrimental to muscle health. Their persistent activity cumulates in progressive tissue fibrosis and loss of normal tissue architecture [22, 23]. In line, we describe a novel population of senescent FAPs that reside in IBM muscle. These FAPs exhibit key hallmarks of cellular senescence including a pro-inflammatory secretome, engagement of the Jun/JunB-pathway and expression of senescence biomarkers (p21 and SA-β-Gal).
Besides their inflammatory properties, these FAPs also exhibit a shift in their collagen balance losing collagen type XV and increasing collagen type I expression. Interestingly, Col15a1 knockout mice lacking collagen type XV demonstrate a myopathy vulnerable to exercise-induced muscle injury, suggesting that this collagen type is required for muscle homeostasis [11]. Histopathologically, this myopathy is characterized by areas of degeneration and regeneration as well as variations in myofiber size [11]. Unlike other collagen types, collagen type XV is only encoded by a single gene (COL15A1) [25]. As such, a loss of COL15A1 expression, as seen in CDKN1A+ FAPs, will likely result in impaired collagen type XV production. Collagen interacts with skeletal muscle via a number of receptors, such as integrins, mediating a plethora of downstream effects [63]. Besides differentiation and regeneration, collagen-muscle cell interaction is also required for the maturation and formation of the NMJ. Animal studies demonstrated that collagen type XIII and type XIX are needed for successful NMJ genesis [33, 47]. While the exact molecular pathways remain to be studied, we hypothesize that collagen dysregulation and persistent pro-inflammatory activity could link FAP senescence and the loss of structural integrity of the muscle compartment in IBM. Current immunosuppressive approaches are unlikely to inhibit or even affect the persistent activity of FAPs. Following this line of argumentation, senescent FAPs may represent a cell-autonomous mechanism that sustains inflammation and fibrotic remodelling in IBM despite therapeutically addressing immune cell pathology warranting further research.
Expanding on previous proteomics data that demonstrated a loss of type 2 myofibers linked to impaired glycolysis in IBM [49], snRNA-seq provides further insight into changes to the myogenic compartment in IBM at a high resolution. Here, the loss of type 2 fibers allocates specifically to the subtype of type 2A myofibers. Further, the transcriptomic landscape of IBM displays a rarefication of neuromuscular endplates and the engagement of an inflammatory muscle phenotype. Differences in composition and metabolic demands render individual myofibers vulnerable to distinct conditions. As such, type 2A myofibers rely primarily on oxidative phosphorylation for their ATP supply, while type 2X myofibers have an effective glycolytic ATP production [58]. Mitochondrial dysfunction is a primary feature of IBM [46], thus providing a potential explanation for the pattern of myofiber loss observed. We hypothesize that type 2A myofibers are unable to meet their metabolic demand due to mitochondrial dysfunction in IBM, while type 2X myofibers may rely on glycolytic ATP. Further research on muscle metabolism and immunogenicity might prove valuable to better understand this understudied aspect of IBM pathophysiology. The importance of the myogenic compartment is further stressed by the possibility of denervation contributing to pathology. Clinical, histomorphological and electrophysiological data suggest a neurogenic component and a failure of NMJ transmission in IBM [17, 45]. Here, we provide molecular evidence in favour of this hypothesis as myofibers constituting the NMJ appear to decline in IBM. Further research is warranted to understand this aspect of pathomorphology and whether treatment strategies addressing the NMJ, such as pyridostigmine, could prove valuable for the clinical management of IBM.
A recurrent observation across the transcriptomic dataset was the dysregulation of intracellular complement components. Historically, complement has been viewed as a serum-effective system that mediates the detection and removal of invading pathogens [62]. Intriguingly, recent studies challenged this classical view by demonstrating that complement activation not only occurs on the cell surface, but also in intracellular compartments, thereby regulating cell physiology of immune and non-immune cells [3, 31, 62]. In this study, we observed that inflammatory states of both myogenic and non-myogenic cells were accompanied by the upregulation of complement proteins, particularly of C3. Indeed, intracellular complement proteins are coded by the same set of genes that give rise to the circulatory complement system [62]. Moreover, intracellular complement regulates tissue inflammation and the transcriptomic programming of fibroblasts [13]. In response to repeated inflammatory challenges, fibroblasts in humans and mice assume a ‘primed’ phenotype that renders tissue susceptible to sustained inflammation. This transcriptomic program is controlled by C3 and the C3a-receptor as well as the mammalian target of rapamycin (mTOR). Following this line of argumentation, we suspect that the engagement of complement factors in this transcriptomic dataset suggests altered intracellular complement signalling as a potential contributor to IBM pathophysiology warranting further research.
Concurrently, an interesting argument for cellular senescence might arise from the effect of sirolimus in IBM. In a recent randomized, placebo-controlled trial for IBM, the mTOR inhibitor sirolimus did not meet the primary outcome of maximal voluntary isometric knee extension strength; however, secondary outcome measures such as 6-min walking distance, forced vital capacity and thigh fat fraction were ameliorated [4]. Aside from immunoregulatory effects, mTOR signalling has been linked to cellular senescence. In vivo and in vitro evidence demonstrated that the mTOR pathway suppresses the SASP, e.g. via Mitogen-activated protein kinase 2 (MAPK) signalling, of senescent cells [20, 32]. Hence, sirolimus is likely to inhibit pro-inflammatory fibroblasts requiring mTOR signalling, as described above [13]. Thus, it is tempting to speculate that the therapeutic effect of sirolimus is—at least in part—mediated by modulation of the senescent fibroblasts.
This study employs snRNA-seq as opposed to scRNA-seq. Both methodological approaches offer specific advantages and disadvantages. In respect to myofibers, the large size of individual cells limits the use of scRNA-seq rendering snRNA-seq a preferential choice [9, 50]. Concurrently, snRNA-seq also allows for the analysis of frozen tissues, thereby enabling the collection and analysis in a single run reducing the risk for batch effects [30]. A limitation of this approach is the complexity of multinucleated cells, as we cannot determine if certain nuclei originated from the same myofiber. Further, depending on the chosen method [59], specific cell populations might be underrepresented. In the study of liver tissue, snRNA-seq detected a lower number of immune cells than scRNA-seq [61]. Combining snRNA-seq with scRNA-seq might provide a more comprehensive picture of the muscle compartment than an individual method alone. Finally, a potential limitation to this study is introduced by the focus on the distal segment of the quadriceps muscle. The highest concentration of NMJs is located within the equatorial region of the muscle [28]. This topographic disparity should be considered when interpreting these findings as further research is warranted to understand whether the reduction of NMJs in IBM patients is also present in the high-density equatorial segment of the muscle.
Taken together, this study highlights the contribution of dysfunctional tissue-resident cells to IBM pathophysiology. Treatment strategies for IBM may benefit from exploring an integrative approach that targets invading immune cells while also addressing detrimental tissue-resident cells such as FAPs and their senescent phenotype.
Data availability
The raw transcriptomic data are publicly available under the accession number: SCP2253 at https://singlecell.broadinstitute.org/single_cell. All further data are available from the corresponding author upon reasonable request.
References
Allenbach Y, Benveniste O, Goebel H-H, Stenzel W (2017) Integrated classification of inflammatory myopathies. Neuropathol Appl Neurobiol 43:62–81. https://doi.org/10.1111/nan.12380
Allenbach Y, Mammen AL, Benveniste O, Stenzel W, Allenbach Y, Amato A et al (2018) 224th ENMC international workshop: clinico-sero-pathological classification of immune-mediated necrotizing myopathies Zandvoort, the Netherlands, 14–16 October 2016. Neuromuscul Disord 28:87–99. https://doi.org/10.1016/j.nmd.2017.09.016
Arbore G, Kemper C, Kolev M (2017) Intracellular complement—the complosome—in immune cell regulation. Mol Immunol 89:2–9. https://doi.org/10.1016/j.molimm.2017.05.012
Benveniste O, Hogrel J-Y, Belin L, Annoussamy M, Bachasson D, Rigolet A et al (2021) Sirolimus for treatment of patients with inclusion body myositis: a randomised, double-blind, placebo-controlled, proof-of-concept, phase 2b trial. Lancet Rheumatol 3:e40–e48. https://doi.org/10.1016/S2665-9913(20)30280-0
Biferali B, Proietti D, Mozzetta C, Madaro L (2019) Fibro–adipogenic progenitors cross-talk in skeletal muscle: the social network. Front Physiol 10:1074. https://doi.org/10.3389/fphys.2019.01074
Chapman MA, Meza R, Lieber RL (2016) Skeletal muscle fibroblasts in health and disease. Differentiation 92:108–115. https://doi.org/10.1016/j.diff.2016.05.007
Chen Y-W, Shi R, Geraci N, Shrestha S, Gordish-Dressman H, Pachman LM (2008) Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol 9:43. https://doi.org/10.1186/1471-2172-9-43
Contreras O, Rossi FMV, Theret M (2021) Origins, potency, and heterogeneity of skeletal muscle fibro-adipogenic progenitors—time for new definitions. Skeletal Muscle 11:16. https://doi.org/10.1186/s13395-021-00265-6
De Micheli AJ, Spector JA, Elemento O, Cosgrove BD (2020) A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skeletal Muscle 10:19. https://doi.org/10.1186/s13395-020-00236-3
Dubowitz, Sewry, Oldfors muscle biopsy: a practical approach. 5th edn. ISBN: 978-7020-7471-4. Pages 21-22
Eklund L, Piuhola J, Komulainen J, Sormunen R, Ongvarrasopone C, Fássler R et al (2001) Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc Natl Acad Sci USA 98:1194–1199. https://doi.org/10.1073/pnas.98.3.1194
Englund DA, Jolliffe A, Aversa Z, Zhang X, Sturmlechner I, Sakamoto AE et al (2023) p21 induces a senescence program and skeletal muscle dysfunction. Molecular Metabolism 67:101652. https://doi.org/10.1016/j.molmet.2022.101652
Friščić J, Böttcher M, Reinwald C, Bruns H, Wirth B, Popp S-J et al (2021) The complement system drives local inflammatory tissue priming by metabolic reprogramming of synovial fibroblasts. Immunity 54:1002-1021.e10. https://doi.org/10.1016/j.immuni.2021.03.003
Goel N, Needham M, Soler-Ferran D, Cotreau MM, Escobar J, Greenberg S (2022) Pos1342 depletion of Klrg1+ T cells in a first-in-human clinical trial of Abc008 in inclusion body myositis (ibm). Ann Rheum Dis 81:1008–1009. https://doi.org/10.1136/annrheumdis-2022-eular.2141
Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C et al (2019) Cellular senescence: defining a path forward. Cell 179:813–827. https://doi.org/10.1016/j.cell.2019.10.005
Goyal NA, Coulis G, Duarte J, Farahat PK, Mannaa AH, Cauchii J et al (2022) Immunophenotyping of inclusion body myositis blood T and NK cells. Neurology 98:e1374–e1383. https://doi.org/10.1212/WNL.0000000000200013
Greenberg SA (2019) Inclusion body myositis: clinical features and pathogenesis. Nat Rev Rheumatol 15:257–272. https://doi.org/10.1038/s41584-019-0186-x
Greenberg SA, Pinkus JL, Kong SW, Baecher-Allan C, Amato AA, Dorfman DM (2019) Highly differentiated cytotoxic T cells in inclusion body myositis. Brain 142:2590–2604. https://doi.org/10.1093/brain/awz207
Grimaldi A, Comai G, Mella S, Tajbakhsh S (2022) Identification of bipotent progenitors that give rise to myogenic and connective tissues in mouse. Elife 11:e70235. https://doi.org/10.7554/eLife.70235
Herranz N, Gallage S, Mellone M, Wuestefeld T, Klotz S, Hanley CJ et al (2015) mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol 17:1205–1217. https://doi.org/10.1038/ncb3225
Hickson LJ, Langhi Prata LGP, Bobart SA, Evans TK, Giorgadze N, Hashmi SK et al (2019) Senolytics decrease senescent cells in humans: preliminary report from a clinical trial of dasatinib plus quercetin in individuals with diabetic kidney disease. EBioMedicine 47:446–456. https://doi.org/10.1016/j.ebiom.2019.08.069
Hinz B, Lagares D (2020) Evasion of apoptosis by myofibroblasts: a hallmark of fibrotic diseases. Nat Rev Rheumatol 16:11–31. https://doi.org/10.1038/s41584-019-0324-5
Hogarth MW, Uapinyoying P, Mázala DAG, Jaiswal JK (2022) Pathogenic role and therapeutic potential of fibro-adipogenic progenitors in muscle disease. Trends Mol Med 28:8–11. https://doi.org/10.1016/j.molmed.2021.10.003
Hu L-Y, Mileti CJ, Loomis T, Brashear SE, Ahmad S, Chellakudam RR et al (2021) Skeletal muscle progenitors are sensitive to collagen architectural features of fibril size and cross linking. Am J Physiol Cell Physiol 321:C330–C342. https://doi.org/10.1152/ajpcell.00065.2021
Huebner K, Cannizzaro LA, Jabs EW, Kiyirikko S, Manzone H, Pihlajaniemi T et al (1992) Chromosomal assignment of a gene encoding a new collagen type (COL15A1) to 9q21 → q22. Genomics 14:220–224. https://doi.org/10.1016/S0888-7543(05)80209-5
Hurskainen M, Ruggiero F, Hägg P, Pihlajaniemi T, Huhtala P (2010) Recombinant human collagen XV regulates cell adhesion and migration. J Biol Chem 285:5258–5265. https://doi.org/10.1074/jbc.M109.033787
Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H et al (2021) Inference and analysis of cell-cell communication using cell chat. Nat Commun 12:1088. https://doi.org/10.1038/s41467-021-21246-9
Jones RA, Harrison C, Eaton SL, Llavero Hurtado M, Graham LC, Alkhammash L et al (2017) Cellular and molecular anatomy of the human neuromuscular junction. Cell Rep 21:2348–2356. https://doi.org/10.1016/j.celrep.2017.11.008
Kim SR, Eirin A, Zhang X, Lerman A, Lerman LO (2019) Mitochondrial protection partly mitigates kidney cellular senescence in swine atherosclerotic renal artery stenosis. Cell Physiol Biochem 52:617–632. https://doi.org/10.33594/000000044
Kim M, Franke V, Brandt B, Lowenstein ED, Schöwel V, Spuler S et al (2020) Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat Commun 11:6375. https://doi.org/10.1038/s41467-020-20064-9
Kolev M, Friec GL, Kemper C (2014) Complement — tapping into new sites and effector systems. Nat Rev Immunol 14:811–820. https://doi.org/10.1038/nri3761
Laberge R-M, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L et al (2015) MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol 17:1049–1061. https://doi.org/10.1038/ncb3195
Latvanlehto A, Fox MA, Sormunen R, Tu H, Oikarainen T, Koski A et al (2010) Muscle-derived collagen XIII regulates maturation of the skeletal neuromuscular junction. J Neurosci 30:12230–12241. https://doi.org/10.1523/JNEUROSCI.5518-09.2010
Li M-L, Wu S-H, Song B, Yang J, Fan L-Y, Yang Y et al (2022) Single-cell analysis reveals transcriptomic reprogramming in aging primate entorhinal cortex and the relevance with Alzheimer’s disease. Aging Cell 21:e13723. https://doi.org/10.1111/acel.13723
Lloyd TE, Mammen AL, Amato AA, Weiss MD, Needham M, Greenberg SA (2014) Evaluation and construction of diagnostic criteria for inclusion body myositis. Neurology 83:426–433. https://doi.org/10.1212/WNL.0000000000000642
Lundberg IE, de Visser M, Werth VP (2018) Classification of myositis. Nat Rev Rheumatol 14:269–278. https://doi.org/10.1038/nrrheum.2018.41
Maity P, Singh K, Krug L, Koroma A, Hainzl A, Bloch W et al (2021) Persistent JunB activation in fibroblasts disrupts stem cell niche interactions enforcing skin aging. Cell Rep 36:109634. https://doi.org/10.1016/j.celrep.2021.109634
Martínez-Zamudio RI, Dewald HK, Vasilopoulos T, Gittens-Williams L, Fitzgerald-Bocarsly P, Herbig U (2021) Senescence-associated β-galactosidase reveals the abundance of senescent CD8+ T cells in aging humans. Aging Cell 20:e13344. https://doi.org/10.1111/acel.13344
McLeish E, Slater N, Sooda A, Wilson A, Coudert JD, Lloyd TE et al (2022) Inclusion body myositis: the interplay between ageing, muscle degeneration and autoimmunity. Best Pract Res Clin Rheumatol 36:101761. https://doi.org/10.1016/j.berh.2022.101761
Morosetti R, Broccolini A, Sancricca C, Gliubizzi C, Gidaro T, Tonali PA et al (2010) Increased aging in primary muscle cultures of sporadic inclusion-body myositis. Neurobiol Aging 31:1205–1214. https://doi.org/10.1016/j.neurobiolaging.2008.08.011
Mozzetta C, Consalvi S, Saccone V, Tierney M, Diamantini A, Mitchell KJ et al (2013) Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Mol Med 5:626–639. https://doi.org/10.1002/emmm.201202096
Murgia M, Nogara L, Baraldo M, Reggiani C, Mann M, Schiaffino S (2021) Protein profile of fiber types in human skeletal muscle: a single-fiber proteomics study. Skeletal Muscle 11:24. https://doi.org/10.1186/s13395-021-00279-0
Nelke C, Dziewas R, Minnerup J, Meuth SG, Ruck T (2019) Skeletal muscle as potential central link between sarcopenia and immune senescence. EBioMedicine 49:381–388. https://doi.org/10.1016/j.ebiom.2019.10.034
Nelke C, Schroeter CB, Pawlitzki M, Meuth SG, Ruck T (2022) Cellular senescence in neuroinflammatory disease: new therapies for old cells? Trends Mol Med 28:850–863. https://doi.org/10.1016/j.molmed.2022.07.003
Noda S, Murakami A, Kazuta T, Hirano S, Kimura S, Nakanishi H et al (2022) Clinical implication of denervation in sporadic inclusion body myositis. J Neurol Sci 439:120317. https://doi.org/10.1016/j.jns.2022.120317
Oldfors A, Moslemi AR, Fyhr IM, Holme E, Larsson NG, Lindberg C (1995) Mitochondrial DNA deletions in muscle fibers in inclusion body myositis. J Neuropathol Exp Neurol 54:581–587. https://doi.org/10.1097/00005072-199507000-00012
Oprişoreanu A-M, Smith HL, Arya S, Webster R, Zhong Z, Eaton-Hart C et al (2019) Interaction of axonal chondrolectin with collagen XIXa1 is necessary for precise neuromuscular junction formation. Cell Rep 29:1082-1098.e10. https://doi.org/10.1016/j.celrep.2019.09.033
Pai C-H, Lin S-R, Liu C-H, Pan S-Y, Hsu H, Chen Y-T et al (2020) Targeting fibroblast CD248 attenuates CCL17-expressing macrophages and tissue fibrosis. Sci Rep 10:16772. https://doi.org/10.1038/s41598-020-73194-x
Parker KC, Kong SW, Walsh RJ, Salajegheh M, Moghadaszadeh B, Amato AA et al (2009) Fast-twitch sarcomeric and glycolytic enzyme protein loss in inclusion body myositis. Muscle Nerve 39:739–753. https://doi.org/10.1002/mus.21230
Perez K, Ciotlos S, McGirr J, Limbad C, Doi R, Nederveen JP et al (2022) Single nuclei profiling identifies cell specific markers of skeletal muscle aging, frailty, and senescence. Aging (Albany NY) 14:9393–9422. https://doi.org/10.18632/aging.204435
Petrus P, Fernandez TL, Kwon MM, Huang JL, Lei V, Safikhan NS et al (2019) Specific loss of adipocyte CD248 improves metabolic health via reduced white adipose tissue hypoxia, fibrosis and inflammation. eBioMedicine 44:489–501. https://doi.org/10.1016/j.ebiom.2019.05.057
Pinal-Fernandez I, Quintana A, Milisenda JC, Casal-Dominguez M, Muñoz-Braceras S, Derfoul A et al (2023) Transcriptomic profiling reveals distinct subsets of immune checkpoint inhibitor induced myositis. Ann Rheum Dis 82:829–836. https://doi.org/10.1136/ard-2022-223792
Preuße C, Paesler B, Nelke C, Cengiz D, Müntefering T, Roos A et al (2022) Skeletal muscle provides the immunological micro-milieu for specific plasma cells in anti-synthetase syndrome-associated myositis. Acta Neuropathol. https://doi.org/10.1007/s00401-022-02438-z
Rose MR, ENMC IBM Working Group (2013) 188th ENMC International Workshop: Inclusion Body Myositis, 2–4 December 2011, Naarden, The Netherlands. Neuromuscul Disord 23:1044–1055. https://doi.org/10.1016/j.nmd.2013.08.007
Rubenstein AB, Smith GR, Raue U, Begue G, Minchev K, Ruf-Zamojski F et al (2020) Single-cell transcriptional profiles in human skeletal muscle. Sci Rep 10:229. https://doi.org/10.1038/s41598-019-57110-6
Saul D, Kosinsky RL, Atkinson EJ, Doolittle ML, Zhang X, LeBrasseur NK et al (2022) A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun 13:4827. https://doi.org/10.1038/s41467-022-32552-1
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM et al (2019) Comprehensive integration of single-cell data. Cell 177:1888-1902.e21. https://doi.org/10.1016/j.cell.2019.05.031
Talbot J, Maves L (2016) Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip Rev Dev Biol 5:518–534. https://doi.org/10.1002/wdev.230
Teicher BA (2019) CD248: a therapeutic target in cancer and fibrotic diseases. Oncotarget 10:993–1009. https://doi.org/10.18632/oncotarget.26590
Valieva Y, Ivanova E, Fayzullin A, Kurkov A, Igrunkova A (2022) Senescence-associated β-galactosidase detection in pathology. Diagnostics (Basel) 12:2309. https://doi.org/10.3390/diagnostics12102309
Wen F, Tang X, Xu L, Qu H (2022) Comparison of single-nucleus and single-cell transcriptomes in hepatocellular carcinoma tissue. Mol Med Rep 26:339. https://doi.org/10.3892/mmr.2022.12855
West EE, Kemper C (2023) Complosome—the intracellular complement system. Nat Rev Nephrol. https://doi.org/10.1038/s41581-023-00704-1
Zhang W, Liu Y, Zhang H (2021) Extracellular matrix: an important regulator of cell functions and skeletal muscle development. Cell Biosci 11:65. https://doi.org/10.1186/s13578-021-00579-4
Zhang X, Habiballa L, Aversa Z, Ng YE, Sakamoto AE, Englund DA et al (2022) Characterization of cellular senescence in aging skeletal muscle. Nat Aging 2:601–615. https://doi.org/10.1038/s43587-022-00250-8
Acknowledgements
We thank the patients and their families for their support. We acknowledge the support from the Core Facility for Advanced Light Microscopy and the support from the Friedrich-Baur-Institut.
Funding
Open Access funding enabled and organized by Projekt DEAL. The funding source had no influence on the study design, on the collection, analysis, and interpretation of data, on the writing of the manuscript, and the decision to submit the paper for publication. This work was directly funded by the Else Kröner-Fresenius-Stiftung to CN (2023_EKEA.38). Further, this work was supported by the internal research funding program of the Heinrich Heine University to CN, CBS and TR. This research was supported by the Myositis Netz e.V. for CN. Further study funding from the BMBF (Bundesministeriums für Bildung und Forschung, 01EC1901A) to TR, by the EKFS (Else Kröner-Fresenius-Stiftung, 2018_A03) to TR, the DGM (Deutsche Gesellschaft für Muskelkranke, Ru2/1 and Sc22/4) to TR, and to CN (Deutsche Gesellschaft für Muskelkranke, Ne4/1) and by the DFG (RU 2169/2-1, Deutsche Forschungsgemeinschaft) to TR.
Author information
Authors and Affiliations
Contributions
CN and TR designed the study and methods. CN, LT, CP, and VD performed the experiments. CP and WS provided clinical data. BS, AB, ENJ, JZ, and SGM provided resources. CN and TR wrote the manuscript. SGM, WS, and TR supervised the study. The manuscript was revised by CBS, LT, CP, MP, SR, VD, FK, AR, BS, AB, ENJ, and JZ.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
401_2023_2637_MOESM2_ESM.tiff
Supplementary file2 a Representative immunofluorescence staining of IBM muscle specimen. Muscle slices were incubated with SPiDER-βGAL at a pH of 6. SPiDER-βGAL indicates the activity of the senescence-associated β-galactosidase. SPiDER-βGAL stains green. Senescent FAPs were identified in the perimysium by p21 staining in red. b Quantification of myofiber types by ATPase 4.6 staining. An exemplary set of staining is given in Fig. 4. A total of 100 myofibers were counted for each sample. N = 8 per group. Differences between groups were analysed by Kruskal–Wallis test followed by post hoc testing. *p < 0.05, ***p < 0.001. NDC non-diseased control; IBM inclusion body myositis; IMNM immune-mediated necrotizing myopathy (TIFF 2509 KB)
401_2023_2637_MOESM3_ESM.tiff
Supplementary file3 a Representative immunofluorescence staining of IBM muscle specimen. FAPs were detected in the perimysium, and their phenotype was confirmed by staining for p21 (red), PDGFRA (green), CD248 (brown), and DAPI (blue). Here, two senescent FAPs are seen between two muscle fibers. IBM inclusion body myositis; PDGFRA platelet-derived growth factor receptor alpha (TIFF 2494 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nelke, C., Schroeter, C.B., Theissen, L. et al. Senescent fibro-adipogenic progenitors are potential drivers of pathology in inclusion body myositis. Acta Neuropathol 146, 725–745 (2023). https://doi.org/10.1007/s00401-023-02637-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-023-02637-2