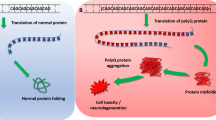
Abstract
GEMIN5 is essential for core assembly of small nuclear Ribonucleoproteins (snRNPs), the building blocks of spliceosome formation. Loss-of-function mutations in GEMIN5 lead to a neurodevelopmental syndrome among patients presenting with developmental delay, motor dysfunction, and cerebellar atrophy by perturbing SMN complex protein expression and assembly. Currently, molecular determinants of GEMIN5-mediated disease have yet to be explored. Here, we identified SMN as a genetic suppressor of GEMIN5-mediated neurodegeneration in vivo. We discovered that an increase in SMN expression by either SMN gene therapy replacement or the antisense oligonucleotide (ASO), Nusinersen, significantly upregulated the endogenous levels of GEMIN5 in mammalian cells and mutant GEMIN5-derived iPSC neurons. Further, we identified a strong functional association between the expression patterns of SMN and GEMIN5 in patient Spinal Muscular Atrophy (SMA)-derived motor neurons harboring loss-of-function mutations in the SMN gene. Interestingly, SMN binds to the C-terminus of GEMIN5 and requires the Tudor domain for GEMIN5 binding and expression regulation. Finally, we show that SMN upregulation ameliorates defective snRNP biogenesis and alternative splicing defects caused by loss of GEMIN5 in iPSC neurons and in vivo. Collectively, these studies indicate that SMN acts as a regulator of GEMIN5 expression and neuropathologies.







Similar content being viewed by others
Data availability
The data that support the findings of this study are all included throughout the manuscript and available on request from the corresponding author.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- ASO:
-
Antisense oligonucleotide
- CB:
-
Cajal body
- iPSC:
-
Induced pluripotent stem cell
- SMA:
-
Spinal muscular atrophy
- SMN:
-
Survival motor neuron
References
Anderson EN, Gochenaur L, Singh A, Grant R, Patel K, Watkins S et al (2018) Traumatic injury induces stress granule formation and enhances motor dysfunctions in ALS/FTD models. Hum Mol Genet 27:1366–1381. https://doi.org/10.1093/hmg/ddy047
Anhuf D, Eggermann T, Rudnik-Schoneborn S, Zerres K (2003) Determination of SMN1 and SMN2 copy number using TaqMan technology. Hum Mutat 22:74–78. https://doi.org/10.1002/humu.10221
Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J et al (2006) The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71:313–320. https://doi.org/10.1101/sqb.2006.71.001
Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G (2006) The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell 23:273–279. https://doi.org/10.1016/j.molcel.2006.05.036
Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonne R (2011) Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet 20:641–648. https://doi.org/10.1093/hmg/ddq508
Burghes AH, Beattie CE (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10:597–609. https://doi.org/10.1038/nrn2670
Calucho M, Bernal S, Alias L, March F, Vencesla A, Rodriguez-Alvarez FJ et al (2018) Correlation between SMA type and SMN2 copy number revisited: An analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul Disord 28:208–215. https://doi.org/10.1016/j.nmd.2018.01.003
Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M (1999) The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol 147:715–728. https://doi.org/10.1083/jcb.147.4.715
Castello A, Fischer B, Hentze MW, Preiss T (2013) RNA-binding proteins in Mendelian disease. Trends Genet 29:318–327. https://doi.org/10.1016/j.tig.2013.01.004
Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M et al (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156. https://doi.org/10.1038/nature05954
Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B (2002) Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 70:358–368. https://doi.org/10.1086/338627
Francisco-Velilla R, Azman EB, Martinez-Salas E (2019) Impact of RNA-Protein Interaction Modes on Translation Control: The Versatile Multidomain Protein Gemin5. Bioessays 41:e1800241. doi: https://doi.org/10.1002/bies.201800241
Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L (2007) Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2:e921. doi: https://doi.org/10.1371/journal.pone.0000921
Garbes L, Heesen L, Holker I, Bauer T, Schreml J, Zimmermann K et al (2013) VPA response in SMA is suppressed by the fatty acid translocase CD36. Hum Mol Genet 22:398–407. https://doi.org/10.1093/hmg/dds437
Genabai NK, Kannan A, Ahmad S, Jiang X, Bhatia K, Gangwani L (2017) Deregulation of ZPR1 causes respiratory failure in spinal muscular atrophy. Sci Rep 7:8295. https://doi.org/10.1038/s41598-017-07603-z
Gleixner AM, Verdone BM, Otte CG, Anderson EN, Ramesh N, Shapiro OR et al (2022) NUP62 localizes to ALS/FTLD pathological assemblies and contributes to TDP-43 insolubility. Nat Commun 13:3380. https://doi.org/10.1038/s41467-022-31098-6
Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G (2002) Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem 277:5631–5636. https://doi.org/10.1074/jbc.M109448200
Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R et al (2017) HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun 8:861. https://doi.org/10.1038/s41467-017-00911-y
le Hao T, Fuller HR, le Lam T, Le TT, Burghes AH, Morris GE (2007) Absence of gemin5 from SMN complexes in nuclear Cajal bodies. BMC Cell Biol 8:28. https://doi.org/10.1186/1471-2121-8-28
Heesen L, Peitz M, Torres-Benito L, Holker I, Hupperich K, Dobrindt K et al (2016) Plastin 3 is upregulated in iPSC-derived motoneurons from asymptomatic SMN1-deleted individuals. Cell Mol Life Sci 73:2089–2104. https://doi.org/10.1007/s00018-015-2084-y
Irwin M, Tare M, Singh A, Puli OR, Gogia N, Riccetti M et al (2020) A Positive Feedback Loop of Hippo- and c-Jun-Amino-Terminal Kinase Signaling Pathways Regulates Amyloid-Beta-Mediated Neurodegeneration. Front Cell Dev Biol 8:117. https://doi.org/10.3389/fcell.2020.00117
Jablonka S, Holtmann B, Meister G, Bandilla M, Rossoll W, Fischer U et al (2002) Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc Natl Acad Sci U S A 99:10126–10131. https://doi.org/10.1073/pnas.152318699
Jin W, Wang Y, Liu CP, Yang N, Jin M, Cong Y et al (2016) Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev 30:2391–2403. https://doi.org/10.1101/gad.291377.116
Kannan A, Bhatia K, Branzei D, Gangwani L (2018) Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res 46:8326–8346. https://doi.org/10.1093/nar/gky641
Kannan A, Cuartas J, Gangwani P, Branzei D, Gangwani L (2022) Mutation in senataxin alters the mechanism of R-loop resolution in amyotrophic lateral sclerosis 4. Brain 145:3072–3094. https://doi.org/10.1093/brain/awab464
Kannan A, Jiang X, He L, Ahmad S, Gangwani L (2020) ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy. Brain 143:69–93. https://doi.org/10.1093/brain/awz373
Kapeli K, Martinez FJ, Yeo GW (2017) Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet 136:1193–1214. https://doi.org/10.1007/s00439-017-1830-7
Kour S, Rajan DS, Fortuna TR, Anderson EN, Ward C, Lee Y et al (2021) Loss of function mutations in GEMIN5 cause a neurodevelopmental disorder. Nat Commun 12:2558. https://doi.org/10.1038/s41467-021-22627-w
Lanfranco M, Cacciottolo R, Borg RM, Vassallo N, Juge F, Bordonne R et al (2017) Novel interactors of the Drosophila Survival Motor Neuron (SMN) Complex suggest its full conservation. FEBS Lett 591:3600–3614. https://doi.org/10.1002/1873-3468.12853
Lanfranco M, Vassallo N, Cauchi RJ (2017) Spinal Muscular Atrophy: From Defective Chaperoning of snRNP Assembly to Neuromuscular Dysfunction. Front Mol Biosci 4:41. https://doi.org/10.3389/fmolb.2017.00041
Lanson NA Jr, Maltare A, King H, Smith R, Kim JH, Taylor JP et al (2011) A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet 20:2510–2523. https://doi.org/10.1093/hmg/ddr150
Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L et al (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165. https://doi.org/10.1016/0092-8674(95)90460-3
Liu Q, Fischer U, Wang F, Dreyfuss G (1997) The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90:1013–1021. https://doi.org/10.1016/s0092-8674(00)80367-0
Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M (2010) WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 8:e1000521. doi: https://doi.org/10.1371/journal.pbio.1000521
Martinez-Salas E, Embarc-Buh A, Francisco-Velilla R (2020) Emerging Roles of Gemin5: From snRNPs Assembly to Translation Control. Int J Mol Sci 21. doi: https://doi.org/10.3390/ijms21113868
Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK (2009) Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 17:639–647. https://doi.org/10.1016/j.devcel.2009.10.017
Mercuri E, Pera MC, Scoto M, Finkel R, Muntoni F (2020) Spinal muscular atrophy - insights and challenges in the treatment era. Nat Rev Neurol 16:706–715. https://doi.org/10.1038/s41582-020-00413-4
Morris GE (2008) The Cajal body. Biochim Biophys Acta 1783:2108–2115. https://doi.org/10.1016/j.bbamcr.2008.07.016
Moses K, Rubin GM (1991) Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev 5:583–593. https://doi.org/10.1101/gad.5.4.583
Nakamura R, Misawa K, Tohnai G, Nakatochi M, Furuhashi S, Atsuta N et al (2020) A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun Biol 3:526. https://doi.org/10.1038/s42003-020-01251-2
Nussbacher JK, Tabet R, Yeo GW, Lagier-Tourenne C (2019) Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron 102:294–320. https://doi.org/10.1016/j.neuron.2019.03.014
Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, Fischer U (2007) A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J Biol Chem 282:5825–5833. https://doi.org/10.1074/jbc.M608528200
Panopoulos AD, Ruiz S, Yi F, Herrerias A, Batchelder EM, Izpisua Belmonte JC (2011) Rapid and highly efficient generation of induced pluripotent stem cells from human umbilical vein endothelial cells. PLoS One 6:e19743. doi: https://doi.org/10.1371/journal.pone.0019743
Passon N, Pozzo F, Molinis C, Bregant E, Gellera C, Damante G et al (2009) A simple multiplex real-time PCR methodology for the SMN1 gene copy number quantification. Genet Test Mol Biomarkers 13:37–42. https://doi.org/10.1089/gtmb.2008.0084
Pineiro D, Fernandez-Chamorro J, Francisco-Velilla R, Martinez-Salas E (2015) Gemin5: A Multitasking RNA-Binding Protein Involved in Translation Control. Biomolecules 5:528–544. https://doi.org/10.3390/biom5020528
Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ et al (2009) A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet 85:408–413. https://doi.org/10.1016/j.ajhg.2009.08.002
Raj A, Chimata AV, Singh A (2020) Motif 1 Binding Protein suppresses wingless to promote eye fate in Drosophila. Sci Rep 10:17221. https://doi.org/10.1038/s41598-020-73891-7
Rajan DS, Kour S, Fortuna TR, Cousin MA, Barnett SS, Niu Z, Babovic-Vuksanovic D, Klee EW, Kirmse B, Innes M, Rydning SL, Selmer KK, Vigeland MD, Erichsen AK, Nemeth AH, Millan F, DeVile C, Fawcett K, Legendre A, Sims D, Schnekenberg RP, Burglen L, Mercier S, Bakhtiari S, Martinez-Salas E, Wigby K, Lenberg J, Friedman JR, Kruer MC, Pandey UB (2022) Autosomal Recessive Cerebellar Atrophy and Spastic Ataxia in Patients With Pathogenic Biallelic Variants in GEMIN5. Front Cell Dev Biol 10:783762. doi: https://doi.org/10.3389/fcell.2022.783762
Ramesh N, Daley EL, Gleixner AM, Mann JR, Kour S, Mawrie D et al (2020) RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3. Acta Neuropathol Commun 8:177. https://doi.org/10.1186/s40478-020-01060-y
Saida K, Tamaoki J, Sasaki M, Haniffa M, Koshimizu E, Sengoku T et al (2021) Pathogenic variants in the survival of motor neurons complex gene GEMIN5 cause cerebellar atrophy. Clin Genet 100:722–730. https://doi.org/10.1111/cge.14066
Sapaly D, Delers P, Coridon J, Salman B, Letourneur F, Dumont F et al (2020) The Small-Molecule Flunarizine in Spinal Muscular Atrophy Patient Fibroblasts Impacts on the Gemin Components of the SMN Complex and TDP43, an RNA-Binding Protein Relevant to Motor Neuron Diseases. Front Mol Biosci 7:55. https://doi.org/10.3389/fmolb.2020.00055
Sawyer IA, Dundr M (2016) Nuclear bodies: Built to boost. J Cell Biol 213:509–511. https://doi.org/10.1083/jcb.201605049
Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN et al (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 111:E5593–E5601. https://doi.org/10.1073/pnas.1419161111
Shpargel KB, Matera AG (2005) Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A 102:17372–17377. https://doi.org/10.1073/pnas.0508947102
Singh A, Chan J, Chern JJ, Choi KW (2005) Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics 171:169–183. https://doi.org/10.1534/genetics.105.044180
Singh NN, Howell MD, Androphy EJ, Singh RN (2017) How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther 24:520–526. https://doi.org/10.1038/gt.2017.34
Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN (2013) An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res 41:8144–8165. https://doi.org/10.1093/nar/gkt609
Singh RN, Seo J, Singh NN (2020) RNA in spinal muscular atrophy: therapeutic implications of targeting. Expert Opin Ther Targets 24:731–743. https://doi.org/10.1080/14728222.2020.1783241
Sleeman JE, Ajuh P, Lamond AI (2001) snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 114:4407–4419
Smolinski DJ, Wrobel B, Noble A, Zienkiewicz A, Gorska-Brylass A (2011) Periodic expression of Sm proteins parallels formation of nuclear Cajal bodies and cytoplasmic snRNP-rich bodies. Histochem Cell Biol 136:527–541. https://doi.org/10.1007/s00418-011-0861-8
Stabley DL, Harris AW, Holbrook J, Chubbs NJ, Lozo KW, Crawford TO et al (2015) SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol Genet Genomic Med 3:248–257. https://doi.org/10.1002/mgg3.141
Tapia O, Bengoechea R, Palanca A, Arteaga R, Val-Bernal JF, Tizzano EF et al (2012) Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem Cell Biol 137:657–667. https://doi.org/10.1007/s00418-012-0921-8
Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ et al (2001) Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 154:293–307. https://doi.org/10.1083/jcb.200104083
Waegaert R, Dirrig-Grosch S, Parisot F, Keime C, Henriques A, Loeffler JP, Rene F (2020) Longitudinal transcriptomic analysis of altered pathways in a CHMP2B(intron5)-based model of ALS-FTD. Neurobiol Dis 136:104710. doi: https://doi.org/10.1016/j.nbd.2019.104710
Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G et al (2016) Cajal bodies are linked to genome conformation. Nat Commun 7:10966. https://doi.org/10.1038/ncomms10966
Winkelsas AM, Grunseich C, Harmison GG, Chwalenia K, Rinaldi C, Hammond SM et al (2021) Targeting the 5’ untranslated region of SMN2 as a therapeutic strategy for spinal muscular atrophy. Mol Ther Nucleic Acids 23:731–742. https://doi.org/10.1016/j.omtn.2020.12.027
Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D et al (2005) Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev 19:2320–2330. https://doi.org/10.1101/gad.342005
Wirth B (2021) Spinal Muscular Atrophy: In the Challenge Lies a Solution. Trends Neurosci 44:306–322. https://doi.org/10.1016/j.tins.2020.11.009
Wirth B, Brichta L, Schrank B, Lochmuller H, Blick S, Baasner A et al (2006) Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet 119:422–428. https://doi.org/10.1007/s00439-006-0156-7
Wirth B, Karakaya M, Kye MJ, Mendoza-Ferreira N (2020) Twenty-Five Years of Spinal Muscular Atrophy Research: From Phenotype to Genotype to Therapy, and What Comes Next. Annu Rev Genomics Hum Genet 21:231–261. https://doi.org/10.1146/annurev-genom-102319-103602
Workman E, Kalda C, Patel A, Battle DJ (2015) Gemin5 Binds to the Survival Motor Neuron mRNA to Regulate SMN Expression. J Biol Chem 290:15662–15669. https://doi.org/10.1074/jbc.M115.646257
Xu C, Ishikawa H, Izumikawa K, Li L, He H, Nobe Y et al (2016) Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev 30:2376–2390. https://doi.org/10.1101/gad.288340.116
Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G (2004) snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol Cell Biol 24:2747–2756. https://doi.org/10.1128/MCB.24.7.2747-2756.2004
Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G (2010) Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell 38:551–562. https://doi.org/10.1016/j.molcel.2010.03.014
Acknowledgements
The authors would like to thank Dr. Joseph Gall for gifting the Drosophila anti-coilin antibody. A.S. is supported by 1RO1EY032959-01 from NIH, Schuellein Chair Endowment Fund, and STEM Catalyst Grant from the University of Dayton.
Funding
This work was supported by the Children’s Neuroscience Institute Research grant (D.S.R. and U.B.P.), Children’s Hospital of Pittsburgh of the UPMC Health system (T.R.F.), and by the German Research Foundation [Wi 945/17–1 (ID 398410809), Wi 945/19–1 (ID 417989143), SFB1451 (ID 431549029), and GRK1960 (ID 233886668)], and the European Union’s Horizon 2020 Marie Skłodowska-Curie [ID 956185 (SMABEYOND] (B.W.).
Author information
Authors and Affiliations
Contributions
Design of this study: T.R.F, S.K., A.V.C., A.M.B., E.N.A., A.S., and U.B.P.; bioinformatics and statistical analysis: T.R.F, S.K., O.C., C.O’B., and D.R.; wrote the manuscript: T.R.F., S.K., and U.B.P.; critically read and edited the manuscript: T.R.F., S.K., A.M.B., C.W., B.W., A.S., and U.B.P. All authors reviewed and approved the manuscript prior to submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fortuna, T.R., Kour, S., Chimata, A.V. et al. SMN regulates GEMIN5 expression and acts as a modifier of GEMIN5-mediated neurodegeneration. Acta Neuropathol 146, 477–498 (2023). https://doi.org/10.1007/s00401-023-02607-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-023-02607-8