Abstract
For a long time, gene editing had been a scientific concept, which was limited to a few applications. With recent developments, following the discovery of TALEN zinc-finger endonucleases and in particular the CRISPR/Cas system, gene editing has become a technique applicable in most laboratories. The current gain- and loss-of function models in basic science are revolutionary as they allow unbiased screens of unprecedented depth and complexity and rapid development of transgenic animals. Modifications of CRISPR/Cas have been developed to precisely interrogate epigenetic regulation or to visualize DNA complexes. Moreover, gene editing as a clinical treatment option is rapidly developing with first trials on the way. This article reviews the most recent progress in the field, covering expert opinions gathered during joint conferences on genome editing of the German Cardiac Society (DGK) and the German Center for Cardiovascular Research (DZHK). Particularly focusing on the translational aspect and the combination of cellular and animal applications, the authors aim to provide direction for the development of the field and the most frequent applications with their problems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is estimated that approximately 5% of newborns will suffer from a genetic disorder. This suggests that almost 400 million people worldwide are affected by genetic diseases [157], and tailored drugs and/or genetic approaches are lacking for most of them. Gene therapy and especially gene editing are the most rational and mechanistic approaches to treat genetic diseases. Gene editing therefore aims on correcting mutations in a gene, to delete or replace parts of a gene or to alter gene expression. Since the discovery of the genetic code, gene editing has, however, been more a theoretical possibility than practical reality. Limited to a few applications and utilizing imperfect tools, like retrovirus, gene editing, for a long time, did not keep its promises, but this has changed within recent years [78].
In addition to the clinical benefit of gene editing, the technology is also important for the production of transgenic crops and animals. In basic and translational sciences, gene editing also allows specific modifications of genes and manipulation of gene expression in practically all life forms, ranging from cells and organs to living animals. Contrary to transient gain or loss-of-function approaches, like RNAi or plasmid-based overexpression, the effects of gene editing can be permanent, and are therefore less prone to artefacts. The technology, however, also has limitations, like the need of shuttle systems and potential off-target effects.
Within recent years, a series of discoveries has revolutionized the toolset to edit the cellular genome. Programmable, sequence-specific DNA endonucleases now allow for precise genome editing in cultured cells and in vivo, and improvements and modifications in particular in the “clustered regularly interspaced short palindromic repeats” (CRISPR)/Cas9 technology occur at breathtaking speed. In vivo genome editing now becomes possible, and simultaneously, the novel CRISPR/Cas9 applications for basic science allow experiments, simply unimaginable just 10 years ago [154]. This development coincides with profound improvements in bioinformatics and vector development, so that cell targeting becomes more efficient and side-effects can be better predicted, minimized, and controlled.
This position paper, writing on behalf of the commission for experimental cardiology of the German Cardiac Society (DGK) and the German Center for Cardiovascular Research (DZHK), will summarize the different available options of gene editing approaches. It aims at basic and translational scientists, and covers the recent advances in gene editing technologies and their application in unbiased screens and animal studies. Moreover, a perspective for the application of gene editing in cardiovascular medicine will be provided. Finally, the limitations and ethical considerations of gene editing will be addressed and an outlook provided on the considerations for human genome editing and future developments.
The general approach of gene editing
The traditional approaches of gene editing were restricted to cells and usually based on the integration of linear DNA stretches into the genome, which were provided through transfection or microinjection into the cell. This integration was either targeted, like the technique used for stem cell modification during knockout mouse generation or random [78]. Both techniques heavily relied on antibiotic-based selection, as integration was a rare event, and thus, an antibiotic resistance gene was integrated into the genome, too. Higher integration rates were achieved with retro-viral approaches, but site-specific targeting was limited by these approaches.
The discovery of programmable, sequence-specific nucleases was a paradigm shift for gene editing. Site-specific modifications of the genetic code at fairly high efficiency became possible, which facilitated the generation of designed cellular and animal models. This new era started with the development of sequence-specific nucleases such as meganucleases, zinc-finger nucleases, and TALENs. Just a few years ago, the bacterial CRISPR/Cas9 system was the most important addition to this toolbox and rapidly became the preferred gene editing method based on its simplicity, efficiency, and universality [35]. With these new genetic tools, double-strand breaks can be precisely introduced into the genome. Depending on the subsequent cellular repair mechanism, these double-strand breaks can be utilized to create different gene editing varieties [78].
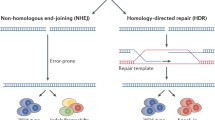
One of the most widely used CRISPR-associated enzymes is Cas9, which originates from Streptococcus pyogenes (SpCas9). SpCas9 forms a protein–RNA complex, resulting in cleavage of double-stranded DNA at target sites. SpCas9 is guided to this site by a single guide RNA (sgRNA), which is formed by the mature CRISPR-RNA (crRNA) and a trans-activating tracrRNA, and requires the simple protospacer-adjacent motif (PAM) NGG, to which the dsDNA cleavage occurs ~ 3 base pairs 5′ of the PAM (reviewed in [7]). DNA cleavage is followed by reparation of the DNA strand breaks by the cell, either through non-homologous end-joining (NHEJ) or homology-directed repair (HDR) mechanisms (Fig. 1a).
Principle CRISPR/Cas systems. NHEJ non-homologous end joining, HDR homology-directed repair, PAM protospacer-adjacent motif, Cas9 CRISPR-associated protein carrying two nuclease domains, associated genes, dCas9 catalytically dead Cas9, Cas9 nickase a Cas9 carrying only one nuclease domain to induce single-strand breaks, sgRNA single-guide RNA, pegRNA prime editing guide RNA, VP64 gene inducer protein domain, KRAB gene suppressor protein domain
CRISPR/Cas applications
For gene editing of cultured cells by the CRISPR/Cas system, most commonly plasmid-based or lentivirally guided expression methods are used [118, 125]. These systems are continuously and rapidly improved in specificity, efficacy, and applicability. For example, the Cas9 nickase mutants (D10A or H840A) can produce single-strand breaks [65, 94, 118]; other additions are high-fidelity Cas9 enzymes [72], an expanded PAM Cas9 variant called xCas9 [59] and Cas9 variants with improved proof-reading enhancing homology-directed repair (HDR) [68]. Systems for temporal and inducible control like photo-activatable CRISPR/Cas9 systems [106, 114] or doxycycline-inducible genome editing [41] have been developed and user-friendly bioinformatic tools have become available to design specific guide RNAs [34].
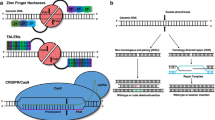
Addgene, a nonprofit plasmid repository (www.addgene.com), is of great help in the orientation phase of researchers as it provides an overview for what other purposes than knockout, knockin, and base editing CRISPR/Cas can be used. Especially the invention of the catalytically inactive dead Cas9 (dCas9), which lacks endonuclease activity and therefore does not cleave, but still can be guided to a target DNA sequence [115], opened a new field of alternative applications. One of such applications is CRISPR-mediated control of gene expression by, e.g., CRISPR interference (CRISPRi). Here, expression of targeted genes is efficiently repressed by either blocking or repressing transcription with a dCas9–KRAB fusion protein [48, 115]. In CRISPR activation (CRISPRa), dCas9 is fused to transcriptional activators (like VP64) to activate gene expression of the targeted gene [40] (Fig. 1b).
dCas9 fusions with epigenetic modifiers can be used to alter locus-specific epigenetic modifications like DNA methylation, histone methylations, and acetylations [40]. Dynamic live cell chromatin imaging profits from fluorescently labeled dCas9 to visualize genomic loci in a single-, dual-, or multicolor way [29, 91, 92, 116]. Interestingly, recent similar developments were initiated focusing on Cas13a/C2C2, Cas13b or CasRx/Cas13d, or their catalytically inactive variants. Importantly, Cas13 enzymes bind RNA and not DNA, which opens up new ways to study RNA biology [5, 32, 75, 119].
Specifically, for gene editing purposes, fusion proteins of dCas9 with other enzymes have been developed: for base editing without double-strand breaks, dCas9 was fused to cytidine deaminase, which mediates the direct conversion of C → T (or G → A) [12, 74]. This technique is more efficient (between 10 and 30%) than the knock-in approach and generates fewer unspecific indels [171]. Fusion proteins of the Cas9 nickase domain with reverse transcriptase allow a specific insertion of a sequence contained in the provided RNA into the genome and is called Prime editor approach [11, 74] (Fig. 1c).
Clonal expansion and mixed culture
The CRISPR toolbox is currently growing rapidly [154] increasing its versatility, usability, and specificity [12, 49]. CRISPR techniques in general can be established in most labs and are fairly cheap, as many of the required plasmids are provided to the scientific community at no cost. An important hurdle, yet, is the introduction of the Cas and the gRNAs into the target cell. For this, direct injection, plasmid-based transfection or virus-based transduction is required. Cell lines usually have to be subjected to antibiotic-based selection to enrich the transfected cells, a step often followed by clonal expansion [169]. This workflow, however, is not applicable to primary cells. Such cells have a limited growth capacity before entering a senescent state and de-differentiate in culture. Moreover, when primary cells become too sparse, they often stop growing, so that the culture does not recover from the antibiotic-based selection. Thus, although clonal expansion might be a possibility for some cell types, it comes with the price of substantial phenotype alterations during the process. Obviously, for non-dividing cells, selection and expansion are not an option at all, and thus, for primary or non-dividing cells, systems with high transfection efficiency are needed. Viral delivery systems such as vectors based on adeno-associated virus (AAV), adenovirus, or lentivirus are able to transduce non-dividing cells [84]. For cardiomyocytes, AAV vector delivery enables HDR in murine adult heart tissues and human cardiomyocytes differentiated from induced pluripotent stem cells (hiPSCs) independently of the cell cycle stage [73]. In contrast to AAV, for adenovirus (AdV) vectors, genomic integration is rather an exception. In cultured neonate cardiomyocytes, however, some limited integration was observed for cells which had entered S-phase [73]. Viral vector systems vary in their packaging capacity, the genetic material (DNA/RNA), and the vector genome form. Adenoviral vectors (HCAdV) are rather effective in transduction and have a large packaging capacity. They can carry within a single vector a whole CRISPR/Cas9 system with gRNAs. The system has been successfully used for targeting the human papillomavirus (HPV) 18 oncogene E6, the dystrophin gene causing Duchenne muscular dystrophy (DMD) and the HIV co-receptor C–C chemokine receptor type 5 (CCR5) [42]. Very popular vectors are adeno-associated viruses (AAVs). They have already been approved for a number of human clinical trials, showed only mild toxicity at high doses in animal experiments, and are less immunogenic than other viruses. Most importantly, AAV displays a safe integration pattern and long-term persistence in non-dividing cells mediating stable gene expression [167].
Regarding the turnout of the gene editing events, the impact on the cell population will be diverse. Non-edited cells, successful edits, failed edits with chromosomal aberrations, and off-target edits will all be present in the small culture dish. As a result, in the mixed culture, the functional consequences of gene editing on the cellular phenotype will not be as pronounced as with clonal expansion. It should, however, be mentioned that clonal expansion is only superior to the mixed culture approach if several clones are being characterized and if the clones are carefully studied for the above-mentioned limitations. Whereas demonstration of the anticipated gene editing event is usually a relatively easy task with PCR, the demonstration of the absence of off-target editing events is laborious and expensive. Clonal expansion is also more prone to artefacts than a mixed culture approach. A recent study by the Odom lab, comparing different loss-of-function methods (siRNA, LNA, CRISPRi) showed that the introduction of the dCas9-KRAB protein alone had already strong effects on the transcriptomic landscape on a clonal level, whereas the non-clonal cell line showed almost no differentially de-regulated genes compared to the parental cell line [144].
The decision on the most appropriate technique for the individual purpose should therefore consider the type of cell, the transfection efficiency, and the quality of the gene editing approach. If the particular cell model under investigation permits the expansion of single cells and if efficacy of the sgRNAs is an issue, then clonal expansion may be considered. If the cell system does not permit clonal expansion or cannot refrain from natural heterogeneity of, e.g., a primary cell population, a polyclonal design may be chosen [128].
CRISPR/Cas screening
Screening technologies are used to uncover dependencies and relations targeting a number of interactors followed by the acquisition of a specific readout. In unbiased genetic screens, expression of a large array of genes is altered randomly, and the subsequent consequences on expression of other genes or functional parameters like proliferation or differentiation are determined. The CRISPR/Cas system has revolutionized the technology of genetic screening as unbiased forward and reverse genetic screens with large sgRNA libraries targeting thousands of genes in parallel are now possible. This technique currently represents the most powerful approach to identify genotype-to-phenotype relationships. To call these interactions, current screens take advantage of pioneering RNA interference work performed in drosophila [100]. Generally, there are two types of genetic screens—positive and negative—in which phenotypes and genotypes are either enriched or depleted, respectively [39]. These screens can be performed in arrayed and pooled conditions (Fig. 2). Arrayed screens have the advantage of physically separating genotypes, enabling subcellular and morphologic read-outs, but have the disadvantage of becoming quickly unfeasible when automated plate handling and phenotype recording is not available [8]. In contrast, pooled screens have the advantage that hundreds of thousands of genotypes can be tested simultaneously in adherent, suspension, or three-dimensional culture as well as living animals. However, read-outs of pooled screens are currently limited to cell fitness effects or depend on phenotypic reporters coupled to cell enrichment steps [98, 124].
CRISPR/Cas screening possibilities. Three strategies have been developed for CRISPR/Cas screens: in array screens, single wells/dishes of cells are infected with one individual sgRNA each, the readout is typically on cellular signature signals, such as comparing bulk transcriptomes or performing surface protein expression profiles. Pooled screens involve transducing several to many sgRNAs and applying a positive or negative selection on transduced cells. Target genes that generate the desired phenotype are uncovered by deep sequencing and subsequent ranking of measured sgRNAs. Third, cells expressing any kind of CRISPR/Cas machinery are transduced with a pooled sgRNA library. Cells will express a functional sgRNA copy together with a second copy of the sgRNA that allows identification by sequencing [barcoding, poly(A)-tailing, e.g., used in, e.g., Perturb-Seq, CROP-Seq etc.]. After droplet sequencing, sgRNA-mediated perturbation can be analyzed in single cells. Multiple sgRNAs against a target gene are used to validate the phenotype
The groups of Feng Zhang, David Sabatini, and Eric Lander were the first to report the use of CRISPR technology for unbiased genetic screening. In these pioneering studies, the identification of essential human genes and 6-Thioguanin and vemurafenib resistance mechanisms served as a proof of concept and represented the beginning of the CRISPR screening revolution [131, 159]. Since these early reports, technical parameters defining library composition (gRNA design rules and number of gRNAs per target) and optimal screening conditions [multiplicity of infection (MOI) and library representation (coverage)] have been identified [108, 127]. Currently accepted conditions include the use of 4–6 highly active gRNAs per target gene, delivered to cells with an MOI of 0.2–0.5 and represented in the experiment with 500–1,000-fold coverage. Standardizing these conditions enables the comparison of different gRNA libraries and their in-screen performance in different cellular model systems. This has vastly contributed to the increase in screening reproducibility [16, 67, 108, 153]. The majority of reported CRISPR screens were designed to identify novel gene to phenotype relationships, while only few studies investigated functional aspects of non-coding sequences [23, 134, 160]. Strategies to efficiently interrogate and functionally annotate non-coding regions apply either DNA tiling or excision approaches. For both approaches, genomic regions are targeted by all possible gRNAs within a given region or the DNA connecting two juxtaposed gRNA-target sites is excised and lost, respectively [23, 76, 126]. Tiling approaches were initially used to identify functional domains in proteins, but have since been used successfully to find essential nucleotide sequences in predicted promoters, enhancers, and long non-coding RNAs [87, 126, 132]. Excision screens are currently limited to a few target sites, mostly because no technological solution exists to generate pooled gRNA libraries with predetermined gRNA combinations [130]. Noncoding sequences make up the vast majority of the human genome; hence, it will be important to solve this technical issue to enable broader unbiased investigations into this “dark” region of the genome.
For CRISPR screens, data recording is robustly and routinely performed by next-generation-sequencing (NGS), while data analysis lacks a broadly accepted approach. Several algorithms (e.g., MAGeCK, HitSelect, PinAPL.py, and ScreenBeam) with differing statistical assumptions have been developed, though it is currently unknown which pipeline performs most accurately as data on comparing hit validation are still missing [37, 82, 142]. Also, most of these tools have been repurposed from the analysis of RNAseq data and only few algorithms are designed for the needs of CRISPR screens. With the increasing number of CRISPR screens that aim at identifying gene–gene interactions, CRISPR-tailored data analysis algorithms are needed. The search recently culminated in the GEMINI algorithm to identify genetic interactions in multiplexed or combinatorial CRISPR screens [170]. As the CRISPR screening field is moving towards multiplexing, it will be crucial to establish, similar to single screens, experimental parameters and statistical models that enable high reproducibility and hit penetrance [38].
CRISPR/Cas screening in combination with scRNA-Seq
The recent rise of single-cell RNA sequencing (scRNA-Seq) permitted for further evolution of CRISPR-based genetic screens [39, 112]. While pooled screens produce rather simple, low-dimensional read-outs such as “growth” or “no growth” and typically require validation by further experimentation, scRNA-Seq of CRISPR-manipulated cell populations employ the transcriptomic signatures of individual CRISPR-manipulated cells as high-dimensional, complex phenotypes (Fig. 2). The assay principle relies on the introduction of “barcode” sequences (or a polyadenylated copy of the sgRNA itself) together with individual sgRNA sequences, thus permitting the assignment of gene expression profiles to the manipulation of defined genetic loci in one single experiment. A treatment or selection is not necessarily needed to perform such screens (although the frequency of the sgRNA still can yield some insight on, e.g., lethality of a knockout/knockdown) (Fig. 2).
Editing the genome of hiPSC for advancing disease modeling
The advent of human-induced pluripotent stem cell (hiPSCs) technology has provided a huge opportunity to establish cellular models of disease from individual patients, and to study the effects underlying genetic aberrations of inaccessible cell types [93, 141, 147]. Particularly, the investigation of molecular mechanisms and cellular phenotypes resulting from a specific mutation (rather than the individual’s genetic background) is possible in iPSC models of disease through the correction of mutated genes in diseased iPSCs (isogenic controls), or by the introduction of causative mutations in healthy iPSCs [17, 140]. It should be emphasized that, contrary to monogenic diseases, the dissection of polygenic disorders is more complicated as a large number of genetic variants are acting in a complex network, and a single variant is not sufficient to trigger the disease [97, 99]. A specific challenge is posed for diseases involving X-linked genes. When using hiPSCs to model X-linked developmental disorders or inherited conditions that undergo sex-specific modulation of penetrance (e.g., autism spectrum disorders), it is crucial to consider the course and status of X chromosome inactivation (XCI). XCI is a unique dosage compensation mechanism that occurs during early embryogenesis and enables equivalent expression of X-linked genes between male and female mammals [56]. Reprogramming of female cells can give rise to two different hiPSCs: XCI has been maintained as in the original somatic cell, or X-chromosomal reactivation (XCR) occurs followed by random mosaic or skewed XCI in the differentiation state, according to the pluripotent state (primed versus naïve) [31, 80, 123]. A third rare outcome is an abnormal partial XCR, in which differentiation does not yield a fully XCI [149]. The failure to precisely characterize XCI status can have significant consequences for the validity of hiPSC-based disease models and their implementation after genome editing approaches.
Among genome editing technologies, CRISPR represents the most electable approach in many cell and tissue types [18, 65, 66, 74, 155]. As mentioned earlier, targeted dsDNA cleavages are repaired through two different pathways: NHEJ and HDR. NHEJ results in insertion and deletion (indel) mutations and can be employed to insert a premature stop codon, resulting in knockout of the gene. This strategy has been recently used to study the deficiency of the KCNA5 gene, which leads to a lower beating rate and prolonged field potential durations of atrial cardiomyocytes [96]. The same approach could be used to restore the normal activity of genes, for instance deleting expanded CGG repeats of the FMR1 gene to rescue Fragile X syndrome [111]. The combination of CRISPR and the PiggyBac transposon systems enables the insertion of large modifications needed in cases of large deletions such as Duchenne muscular dystrophy and Huntington’s disease [86, 165].
HDR-mediated genome editing repairs DNA in a precise manner referring to the sister chromatid as template and can be used to introduce or correct specific disease-associated variants for disease modeling. Correction or introduction of disease-associated gene variants (1–20 nt) has recently been accomplished with the use of short single-stranded oligonucleotide donors (ssODNs) with homology arms of 30–200 nt [121, 168]. An advantage of ssODNs, besides their fast design and generation, is that they are less likely to integrate randomly into the genome in comparison to plasmids or linear dsDNA donors [83]. The ssODN strategy results in scarless editing, but the lack of selectable markers encoded within the donor increases the difficulty for identification of positive clones. Alternative to ssODNs, single-stranded rAAV DNA templates harbor the beneficial feature of providing long homology arms and resistance genes for increased recombination efficiencies with low rate of genomic integration. Moreover, CRISPRa/i approaches are useful in treating human diseases due to haplo-insufficiency or protein accumulation such as Parkinson and Alzheimer’s disease [57].
Overall, genome manipulation of hiPSCs has become highly efficient using CRISPR, especially for NHEJ-based pathways (80–90% with NHEJ vs < 10% with HDR). Nonetheless, Cas9 can bind off-target sites with mismatches, resulting in variable off-target activities [151]; Cas9 construct is either permanently integrated [24, 50] or removable with a subsequent reagent delivery and/or clonal selection step [164] to achieve scarless editing. Alternatively, non-integrating methods include Cas9 ribonucleoprotein (RNP) complexes, which are immediately active and rapidly degraded over a period of around 12 h [69], reducing the potential for off-target mutagenesis and re-targeting after successful HDR.
Repairing causative lesions in patient-derived iPSCs which are subsequently differentiated towards specific cell types might be used for cellular therapy treatment. Some strategies are promising, e.g., injection of autologous iPSC-derived dopaminergic neurons in the treatment of a chemically induced primate model of Parkinson’s disease [55]. Nevertheless, various technical hurdles and biological questions need to be addressed before the introduction of therapeutic use of CRISPR/Cas in humans.
Genome editing in animals
Genome editing and disease modelling in zebrafish
TALEN and CRISPR/Cas9 are meanwhile the standard tools for reverse genetics in zebrafish (Fig. 3). Originally, Cas9 and a single sgRNA were used to target a specific region within the gene of interest forming insertion/deletions (indels), premature stop codons, or frameshifts, which inactivate the generation of a functional gene product [26, 35, 60, 95]. Depending on the position of the premature stop, nonsense-mediated decay of transcribed RNA and production of small RNA fragments may trigger the expression of genes that can compensate for the mutation and mask the phenotype [44]. Such compensation mechanisms, which are also operational in mice, account for the discrepancy between morpholino knockdown and mutant phenotypes. The current consensus for generating a mutant therefore is to suppress the RNA transcription of the target gene completely, thereby avoiding the formation of RNA decay products and compensation. This is achieved by either deleting the full gene locus, or by removing the complete transcriptional or translational start regions using a combination of two sgRNAs. In general, control experiments suitable to uncover potential compensation do not target the gene but rather its expression, like CRISPRi or shRNA.
Domains of gene editing in animals. Whereas Cas9-mediated germline gene editing now becomes the standard technology for the generation of transgenic fish and rodents, larger mammals, like transgenic pigs are still generated by “conventional” technology. Pigs and dogs are important laboratory animals in the translational avenue to established somatic gene editing for clinical use
CRISPR/Cas9 is also used for knock-in approaches in zebrafish [14, 19, 70]. The knock-in can be utilized to generate specific—patient similar—mutations, or repair existing mutations [148]. The current limitations of this technique are the low efficiency (between 0.5 and 1%) and the additionally generated off-target mutations. Base-editing, for example by a fusion of dCas9 with cytidine deaminase, has a higher efficiency (10–30%) and greater specificity [171].
Genome editing and disease modelling in mice
Similar to zebrafish, also in mice, the CRISPR/Cas9 system replaced classical genetic engineering techniques to generate different genome-engineered transgenic mouse lines [52, 78, 89]. Cas9 is the most widely used nuclease due to the simple protospacer-adjacent motiv (PAM) 5′-NGG-3′ and the generation of double-strand breaks (DSB) compared to Cas12a (Cpf1), which has a PAM (5′-TTN-3′) that is T-rich, less frequent in the genome and creates staggered ends [28, 107]. Among the mutations generated are classical and conditional gene knock-outs (KOs), knock-ins (KIs) at specific genomic loci as well as point mutations or epitope insertions in desired gene regions. The efficiency of gene modification at specific loci using the NHEJ mechanism reaches almost 90% and the efficiency of KI using the HDR mechanism is up to 50%. To increase HDR frequency, different strategies to inhibit NHEJ and to enhance DNA repair mechanisms are implemented (for review, see [35]). Nevertheless, these rapidly evolving techniques still contain serious limitations exemplified in several KI experiments by the creation of conditional KO mouse alleles. All gene-targeting protocols are performed by direct injection of CRISPR/Cas9 components together with donor DNA template into fertilized oocytes. For KI experiments, comparatively long-donor DNA fragments (~ 600–1650 nt) are utilized employing either single-stranded (ssDNA) or double-stranded (dsDNA) templates. Efficiency of homologous donor DNA template integration into the locus is variable and correlates with template size and the locus. In general, longer DNA templates integrate less efficient than shorter ones.
Attention should be paid to the fact that most genome edited mice obtained from CRISPR/Cas9-modified zygotes (F0 generation) exhibited mosaic genotypes. The mosaic genotype can harbor subpopulations of germ cells derived from different DNA-editing events, and contain diverse copy numbers of DNA template integrations into the targeted loci [136]. The latter suggests that PCR amplification of short flanking genomic regions together with parts of the inserted artificial sequences, including the LoxP sites in template DNA, is the most efficient and reliable approach for the identification of F0 mice with a correctly targeted event. When the selected F0 founders were crossed with wild-type mice to obtain F1 offspring, animals harboring multiple head-to-tail integrations (MHTI) of the donor DNA template at the targeted locus can be detected frequently. These DNA template multiplications occur irrespectively of the size, nucleotide composition, or the utilization of dsDNA or ssDNA [136, 138].
Importantly, a commonly used PCR analysis method of heterozygous animals, employing locus-specific oligonucleotides located outside of the targeted homology region, would under this condition, in most cases, mistakenly indicate a single-copy integration event. Southern blot analysis is an efficient accompanying method to reliably identify the desired single-copy targeted events in F1 offspring. It is recommended to include two different specific restriction endonuclease sites flanking the LoxP regions. This will allow the detection of correctly targeted events by restriction fragment length polymorphisms (RFLP), using these restriction enzyme sites, and also detects the MHTI. In addition, a PCR approach with reverse orientation of DNA template-specific primers, qPCR, or the ddPCR (droplet digital PCR) can be alternative methods for detection of MHTI of the DNA template [136]. To exclude any genome alterations, sequencing of the entire locus is required.
Genome editing and disease modelling in large animals
Large animals are important model organisms to bridge between basic science studies and clinical applications. In the cardiovascular system, this has been exemplified for mutations of the dystrophin gene leading to Duchenne’s muscle dystrophy (DMD). A number of mouse models have demonstrated efficacy of an AAV-based CRISPR/Cas9 approach to edit the dystrophin gene. In the mdx mouse, excision of the mutated exon 23 suffices to enable expression of a shortened, but stable dystrophin gene [43, 88, 105, 146]. Functional assessment, though limited due to a mild phenotype of the mouse model, suggested improvement of the skeletal and heart muscles after local or systemic vector application.
Extending this evidence, Amoasii from the Olson lab [10] applied single-guide RNAs and SpCas9 AAV into DMD dogs (lacking exon 50), either intramuscularly (i.m.) or intravenously (i.v.), and found robust DMD expression at the injected sites (i.m.) and also in the heart (i.v. approach). Functional consequences of this approach though were not reported yet.
In a complementary transgenic pig model lacking exon 52 of the dystrophin allele, an intein-split version of SpCas9 [150] was used together with a vector containing a pair of gRNAs capable of excising exon 51 also using AAV technology [103]. The intein-split approach bypasses the packing limit of AAVs: two virus particles are generated which both carry a gRNA and the C-terminal and N-terminal, respectively, part of Cas9. Only after a double infection of a cell, a complete Cas9 is produced [150]. After i.m. injection, the intein-split-Cas9-gRNA approach was efficient in editing up to 78% of the muscle nuclei analyzed. Upon i.v. injection, 7% of the cardiac genomes were edited, resulting in a reduction of sudden cardiac death of the animals [103].
Delivery of CRISPR/Cas9 in somatic tissue
Besides off-target activity which will be discussed in the next section, the aspect of a safe and efficient delivery of the gene editing tools is crucial in a therapeutic context. Although a number of different approaches have been developed to facilitate direct Cas9 ribonucleoprotein complex delivery by utilizing nanoparticles, extracellular vesicles, or cell penetrating peptides (for review, see [22, 166]), many therapeutic approaches are relying on the delivery of genomic encoded tools via viral systems (Fig. 3). In this context, the AAV system is advantageous due to its capability to transduce both proliferating as well as post-mitotic cells, its diverse tissue tropism, its robust and prolonged expression levels, and its relatively low immunogenicity [13]. Nevertheless, the maximum packaging capacity of about 4.7 kb resembles a limitation for single vector approaches, especially if the relatively large Streptococcus pyogenes variant of Cas9 is used. To overcome this limitation, either the co-administration of a Cas9 virus plus a separate gRNA expressing virus or the utilization of a split-Cas9 system can be helpful [13].
The previously described therapeutic studies for DMD exemplify these different strategies: the group of Eric Olson applied a single cut approach to restore the reading frame of exon 52 deficient dystrophin by insertions or deletions (Indels) in the 5 prime region of exon 51, and partially also by enhanced skipping of exon 51. The group of Christian Kupatt follows a different approach utilizing the split-intein system (Fig. 4). Here, two distinct AAV constructs, each harboring one individual gRNA under the control of an U6 promoter and one half of the intein-fused SpCas9 nuclease under the control of a pol-II promoter, are co-injected. Upon co-expression in the same cell, the N- and C-terminal halves reconstitute and perform guided nuclease activity analogous to wild-type Cas9 [103]. This setup allows the delivery of SpCas9 together with two independent gRNAs and is only active in co-transduced cells. Instead of one gRNA located in exon 51, two gRNA in the intronic regions flanking exon 51 have been selected to precisely excise the exon and thereby restoring the reading frame. In general, these exon snipping approaches allow a more flexible design of the utilized gRNAs and thus enable a more stringent selection regarding predicted off-target activity. Furthermore, compared to full length Cas9 approaches, this system has the potential to operate under the combined control of two distinct, tissue and/or cell type-specific pol-II promoters, which further enhances not only the specificity but also the safety of the gene therapy approach. Apart from this, in general, every aspect of a gene therapeutic approach has to be optimized to achieve the highest level of safety. This includes an efficient and safe delivery of the tools, a specific expression of a Cas9—ideally restricted to the tissue/cells of interest and preferably a temporal restriction of the gene-editing event.
Split-Intein-AAV-System: due to the packaging limit of adeno-associated virus (AAV), the most-frequently used Cas9 genes together with two guide RNA cannot be transducted by a single AAV. In the Split-Intein-System, the cell is infected with two different viruses, both containing one part of Cas9 and one part of the Intein gene. Expression of both constructs yields a complete, enzymatically active Cas9
Analysis of CRISPR/Cas9 off-targets
Beside the thorough characterization of the targets, off-target events deserve closer attention. Programmable nucleases such as CRISPR/Cas9 are efficient to generate on-target genetic modifications; however, rigorous design of sgRNAs, Cas9, and delivery modes are necessary to minimize potential off-target genomic alterations and to ensure the integrity of the genome of manipulated cells (for review, see [62]). To facilitate the use of crRNA, representing 20 bp complementary sequence to the target genomic region and tracrRNA (trans-activating crRNA) representing a scaffold to bind Cas9 both have been combined into a sgRNA [66]. It has been reported that even 3–5 mismatches at the distal (5′) end of the protospacer sequence can be tolerated leading to DSB [46]. Thus, if sgRNAs are not properly designed, off-target effects might be induced.
To minimize off-target activity and increase specificity, truncated guide sequence at the distal 5′ end [45] was shown to be beneficial so is chemical modification at the 3′ end of gRNA [102]. In addition, a number of web tools have been designed for proper design of gRNAs taking also PAM sequences into account and to identify potential off-targets genome wide, e.g., CRISPRdirect [104], Cas9 design [90], and ChopChop [39, 101]
Based on the three-dimensional structure of Cas9, mutants (D1135E) have been generated which increase on-target specificity and recognize [71] different PAM sequences of different lengths, thereby increasing specificity. Furthermore, Cas9 nickase mutants have been developed in which the RuvC or HNH nuclease domain is inactivated resulting in juxtaposed single-stranded nicks [79], or a pair of catalytically inactive dCas9 nucleases each fused to FokI nuclease domain [26, 113]. Each of these approaches reduce off-target mutagenesis; however, they also have their limitations with respect to cleaving efficiency, e.g., double nicking requires two guides and truncated guides can reduce on-target efficiency, as well. Using structure-guided protein engineering in combination with unbiased whole-genome off-target analysis, enhanced efficiency variants of Streptococcus pyogenes Cas9 (eSpCas9) have been generated which exhibited reduced off-target and robust on-target effects [137].
Reduction of off-target mutagenesis can also be achieved via temporal expression of Cas9 taking advantage of a tetracycline responsive promoter [137], using split-Cas9 intein system [137], and by inactivating Cas9 through self-cleavage providing in addition Cas9-specific gRNA [137].
Different methods are available to search and inspect potential off-target sites. There are three main strategies available: (1) to determine sequences of potential off-target sites; (2) to determine potential DSBs genome wide; (3) to determine genome integrity.
The first straightforward strategy is to determine predicted off-target sites using software and assay the PCR products for mismatches by single-strand annealing and endonuclease treatment (e.g., T7EI, Surveyor nuclease assay). These techniques are most commonly used as they are easy to implement and cost effective. Alternatively, deep sequencing of exome genome wide (all protein coding regions) is performed to identify potential mutations in protein coding genes [148]. A more in-depth analysis is achieved by whole-genome sequencing (WGS) used in cell lines [139, 145, 156] and mice [64]. These techniques are powerful and allow to identify small indels but not large genomic rearrangements. Larger genomic rearrangements and CNVs need to be examined using alternative methods, e.g., fluorescence-in-situ-hybridisation [20].
Second, identifying potential DSBs by chromatin immunoprecipitation and pull-down of DNA fragments (ChIP-seq) have been employed in different flavors, e.g., using dCas9 to determine Cas9-binding sites [162]. Alternatively, strategies have been developed which label DSB by either streptavidin-biotinylated linkers [33] or by incorporating short phosphorylated double-stranded oligodeoxynucleotides (GUIDE-seq [152]). An elegant extension of these strategies is the DISCOVER-seq (discovery of in situ Cas off-targets and verification by sequencing) method that leverages on the recruitment of DNA repair factors in particular MRE11, which binds closely around the Cas9 cleavage site, to uncover Cas9 activity [161].
Additional technologies are useful to confirm the results obtained by CRISPR/Cas9: Despite having their own potential off-target effects, RNA interference with different siRNAs/shRNAs, the use of different (morpholino) antisense oligonucleotides, and LNA GapmeRs, as well as different pharmacological inhibitors should be taken into consideration to validate on-target effects seen by CRISPR/Cas9-mediated knockout or inhibition experiments and to further rule out the possibility of off-target effects. In the case of CRISPR/Cas9-mediated activation systems, plasmid- or viral-overexpression systems can be used for clarification. Moreover, as mentioned before in the section regarding clonal expansion, but not only concerning mixed populations, several clones of the control and the mutants should be characterized carefully to rule out off-target effects and to strictly define the limitations of the systems. Another way is to test directly the expression of genes which were predicted in the gRNA web design tools. For genome editing in mice, back crossing the transgenic mice generated by CRISPR/Cas9 could help to reduce the chance of off-target effects.
Thus, as with other genome and nucleic acid interfering techniques, we should be cautious with results obtained by CRISPR/Cas9. A broad set of tools is already available to reduce and detect off-targets and maintain robust on-target mutation. Selection of the strategy depends on the experimental setup, e.g., cell lines, animal models, preclinical gene therapy models, and whether an ex vivo or in vivo gene therapy approach is taken. In case of genome editing in model systems, potential off-targets can be reduced simply by back crossing wild-type animals to the F2 generation. In addition, at least two independently established mutants should be phenotypically characterized. For an in vivo gene therapy approach, rigorous experimental design and pretesting of gRNA, ideally in patient-derived cells, are required as well as applying high-end design, optimized endonucleases, optimized delivery strategies, and validation tools to minimize potential off-targets. Irrespective of all precaution measures, there is still an unforeseen risk of generation of by-stander mutations in the genome.
Considerations for clinical gene editing
Clinical translation and thus disease treatment are the ultimate aim of many gene therapy approaches. Several clinical studies with zinc-finger nucleases or later on TALENs have been initiated since 2009. They focused on a variety of conditions such as cancer, HIV, and hematological diseases with the outcomes still to be reported in most cases [84, 109]. In the last years, first clinical trials with CRISPR/Cas9 approaches have started to recruit patients (Fig. 5). Importantly, recent preliminary results from a clinical phase I trial showed that gene editing using CRISPR/Cas9 might be safe and feasible to apply [143]. T-cells from three patients with different types of advanced cancers were gene edited by CRISPR/Cas9 ex vivo using electroporation, resulting in ablation of three proteins that could inhibit the T-cells’ ability to target tumor cells. In a second step, a cancer-specific T-cell receptor transgene was expressed in these cells by lentiviral gene transfer to recognize a particular epitope on tumor cells. After administration into the respective donors, gene-edited T-cells were engrafted and persisted for at least 9 months without significant side-effects. As cancer continued to progress in all three patients, the question of efficiency of the gene-edited T-cells against advanced cancer remains open. In this particular trial, gene disruption efficiency was 15–45%, based on the techniques available at the time the study was approved (2016), whereas newer techniques allow now > 90% of gene disruption [122, 129]. Apart from the preliminary CRISPR/Cas9 clinical data, further gene editing studies in patients have been initiated aiming at elimination of mutations that lead to the development of cancer or hereditary diseases such as sickle-cell anemia, beta-thalassemia, or Leber congenital amaurosis [84]. The selection of human diseases currently treated with gene editing approaches, however, also illustrates the current bottleneck of clinical gene therapy: delivery of the CRISPR/Cas9 system and the sgRNA in patients.
While efficiency is already a major hurdle in clinical trials of gene therapy in general and in particular for cardiac diseases [63, 85], specificity of gene transfer is of decisive importance for gene editing approaches considering potential off-target effects such as chromosomal translocations and rearrangements with at least a theoretical oncogenic potential. Therefore, current studies in patients predominantly focus on ex vivo gene editing (i.e., cell therapy), for example in hematopoietic stem cells for sickle-cell anemia or T cells for novel cancer therapies, where the gene-edited cells can be reinfused into the body. In addition, ex vivo delivery allows a transient transfer of Cas9 and sgRNAs by electroporation that results in almost an absence of residual Cas9 activity in the cell product [143]. This might be important not only to limit the duration of Cas9 activity for safety reasons, but also to avoid immunological clearance of the gene-edited cells due to development of a humoral response to Cas9.
It is not surprising that the first clinical study using viral vectors for delivery of Cas9 and sgRNAs in vivo aims at treating a particular form of Leber congenital amaurosis [4], in which the coding sequence of the defective gene is too large to be packaged in a viral vector for the conventional overexpression. The eye is an immune privileged and easily accessible organ which allows efficient subretinal injection of AAV vectors as previously shown in a successful clinical study: a classical AAV-based gene replacement strategy revealed improved vision in patients with congenital amaurosis due to deficiency of a protein required for photoreceptor function [15].
While rapid clinical translation of the CRISPR/Cas9 technology is impressive, several limitations need to be overcome before gene editing approaches can be applied for cardiovascular diseases. Target cells such as cardiomyocytes require an efficient in vivo delivery system consisting of a suitable vector (for example, AAV vectors), but also a suitable application system for transvascular gene transfer such as coronary venous retroinfusion for cardiac transduction [63, 117]. Considering that any transvascular vector application results in some systemic spill-over, unwanted side-effects need to be prevented by confining expression of gene editing elements to the target tissue, for example, by use of tissue-specific promoters or alternative detargeting approaches [47, 63]. Alternatively, ex vivo gene editing therapies of whole organs (i.e., heart or lung), for which short time artificial organ support is possible, can be a first step towards clinical application in cardiac or pulmonary diseases.
Even if gene transfer into less immune privileged organs such as the heart would be efficient with viral vectors and appropriate delivery systems, it is not clear whether Cas9—a bacterial protein—will trigger an adaptive cellular immune response. Furthermore, also preexisting humoral reactivity to Cas9, which was previously detected in serum of healthy individuals, could affect sustained expression of Cas9 in transduced cells [27, 135, 158]. Thus, it might be necessary for in vivo gene editing in less immune privileged organs to control Cas9 gene expression using inducible promoters or switch to vector systems that enable an only transient Cas9 expression.
Beside the immunological effects of the Cas9 enzyme, a further limitation towards broad clinical translation of the CRISPR/Cas9 system might be the p53-mediated response to double-strand breaks induced by CRISPR/Cas9, leading to a principally but not widely proven enrichment for P53-deficient and thus more cancer-prone cells [53, 61]. Moreover, target recognition is prone to error, so that gene editing may introduce changes at partly unpredictable off-target sites due to similarities to the target recognition sequence. Even more alarming is that repair of double-strand breaks induced by gene editing approaches leads to large deletions, insertions, or complex rearrangement events involving the target site [77]. These unintended edits are detected consistently [6, 110, 133, 163, 173] and highlight the need for accurate genotype characterization, as these potentially have devastating outcomes in clinical tests. The repertoire of molecular, genetic, and next-generation-sequencing techniques to analyze (CRISPR/Cas9-mediated) changes on the genome is large and sufficient for verification of editing success and prevention of unwanted events. The opinion article by Burgio and Teboul [21] provides assistance and advice for the application of CRISPR/Cas9. Accurate predictions and the understanding of the whole range of possible editing outcomes are critical for the continuous success of CRISPR/Cas9.
Numerous approaches to increase specificity of gene correction have been undertaken focusing on optimization of the gRNA or the nucleases used for gene editing itself [59]. These improvements include the development of small molecule tunable Cas9 enzymes [36]. Also new variants of Cas nucleases, such as xCas9 and SpCas9-NG, enable the targeting of alternative PAM sequences and thus expand the range of genomic loci that can be edited [10, 143]. Fusion of nucleobase deaminase enzymes to catalytically inactive Cas variants makes it possible to accomplish mutual conversion among four bases [51, 74, 81, 172]. These so-called “base editors” modify base pairs at specific sites, thereby expanding the potential applications of the CRISPR/Cas system to correct disease-associated single nucleotide polymorphisms. Cytosine and adenine DNA base editors (CBE, ABE) and their approximate editing windows have been described in [120]. Both could be useful tools also to install or correct pathogenic point mutations. CBEs mutate C•G-to-T•A by binding to genomic target regions and R-loop formation. They bind to a target DNA sequence and form a single-stranded R-loop, and the covered cytosines are deaminated to form uracil bases. Uracil glycosylases are inhibited by the uracil glycosylase inhibitor domain and the Cas9 domain regulates DNA repair leading to an adenine opposite the uracil. ABEs mutate A•T-to-G•C by deaminating target deoxyadenosines to deoxyinosines, regulating the direct DNA repair to install a cytosine opposite the inosine nucleoside [12]. A recent development called “prime editing” showed less off-target editing without introducing double-strand breaks or donor DNA by fusing a catalytically inactive Cas to an engineered reverse transcriptase [11]. Editing individual bases in RNA offers also great potential in medicine. Adenosine conversion to inosine, which is generated by the adenosine deaminases from the ADAR family, has been shown to be a useful tool using both antisense and Cas13-guided RNA-targeting methods [120]. If and when these improvements (see also Fig. 1) may enter clinical trials is not known.
In summary, development of therapeutic gene editing approaches requires a careful design of the genome editing strategy including the most appropriate Cas variant, an appropriate vector system, and mode of vector delivery.
Clinical perspective of gene editing
Considering the fast pace of clinical translation of gene editing approaches and but also the many open safety questions, the question of the best way to clinical translation emerges. It is current consensus that clinical trials can be started now and that these should aim on advanced cancers or rare hereditary diseases, for which efficient gene delivery modes are well established. These early studies enable collection of safety data which is highly needed for further studies with improved gene editing tools.
It is important to stress that gene editing techniques—as all other advanced therapeutics—require continuous consideration of social, ethical, and regulatory issues. However, increasing commercial interests may cause conflicts of interest preventing unbiased dissemination of methods and results. Instead of being published in peer-reviewed journals, results of commercial research and trials executed by companies are rather disseminated in business statements and announcements on websites [25, 109]. However, full disclosure of methodologies and concise analysis of off-target effects will remain necessary to assess the impact of any novel gene editing approach and to establish trust for further clinical translations.
Although apparently technically feasible, there is broad consensus among stakeholders that gene editing needs to be restricted to somatic cells as there are significant ethical, scientific, and socio-economic concerns regarding germline genome modification [9, 30]. As for all gene therapy approaches, Germany and many other countries have issued a strict ban on application of techniques for the purpose of human germline modification.
Legislation of gene editing approaches in somatic cells follow those for gene therapy products. As ex vivo gene therapy requires reapplication of genetically modified cellular products into the patient, these products must also comply with both cell-based medicinal product and gene therapy product guidelines and regulation. Details regarding the regulations of gene therapy products are reviewed elsewhere [54]. In Germany, two studies investigating ex vivo somatic gene editing for treatment of beta-thalassemia (NCT03655678) [2] and sickle-cell anemia (NCT03745287) [3] have already been approved to be conducted.
To further improve safety of gene editing approaches, several organizations, including the World Health Organization, as well as recognized standards developing organizations such as the US National Institute of Standards and Technology Genome Editing Consortium, the US Pharmacopeia, and the International Organization for Standardization (ISO) formulate gene editing standards [9]. Such standards are urgently needed to address key concepts like off-target effects and their impact on tumor suppressors and oncogenes. In addition, national and supranational regulatory organizations such as the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) already provide guidance for the development of gene editing techniques for therapeutic modification of somatic cells.
While six gene therapy products have already been approved by the FDA and EMA since 2016, more than 2000 are in different stages of clinical approval including several gene editing approaches [1, 58, 84]. Considering the enormous development costs and high prices of current gene therapy products, socio-economic conflicts can be foreseen. While ex vivo gene editing of immune cells might be commercially exploited as cancer treatment, reimbursement for gene editing approaches focusing on rare diseases with individual mutations might become more challenging. Thus, reduction of costs for development and production of gene therapy products is necessary to allow individualized gene editing therapies for rare genetic cardiovascular diseases in the future.
In conclusion, within a few years, gene editing developed from a scientific concept into everyday research reality which now enters the clinic. This development is occurring with breathtaking speed, also owing to the progress in shuttle development, sequencing technology, and bioinformatics. Whereas gene editing just revolutionized basic science, it will soon revolutionize medicine. This will open up treatment avenues for fatal diseases, and cures for many inherent diseases.
Data availability
Does not apply.
Code availability
Does not apply.
References
(2017) Gene therapy clinical trials worldwide. Provided by the Journal of Gene Medicine. http://www.abedia.com/wiley/. Accessed 14 Apr 2020
(2018) NCT03655678: A Safety and Efficacy Study Evaluating CTX001 in Subjects With Transfusion-Dependent β-Thalassemia. https://clinicaltrials.gov/ct2/show/NCT03655678. Accessed 14 Apr 2020
(2019) NCT03745287: A Safety and Efficacy Study Evaluating CTX001 in Subjects With Severe Sickle Cell Disease. https://clinicaltrials.gov/ct2/show/NCT03745287. Accessed 14 Apr 2020
(2019) NCT03872479: Single Ascending Dose Study in Participants With LCA10. https://clinicaltrials.gov/ct2/show/NCT03872479. Accessed 14 Apr 2020
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F (2017) RNA targeting with CRISPR-Cas13. Nature 550:280–284. https://doi.org/10.1038/nature24049
Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ (2018) Large deletions induced by Cas9 cleavage. Nature 560:E8–E9. https://doi.org/10.1038/s41586-018-0380-z
Adli M (2018) The CRISPR tool kit for genome editing and beyond. Nat Commun 9:1911. https://doi.org/10.1038/s41467-018-04252-2
Agrotis A, Ketteler R (2015) A new age in functional genomics using CRISPR/Cas9 in arrayed library screening. Front Genet 6:300. https://doi.org/10.3389/fgene.2015.00300
Alliance for Regenartive Medicine (2019) ARM Gene Editing Task Force Therapeutic Developers' Statement of Principles. https://alliancerm.org/therapeutic-developers-statement-of-principles-2/. Accessed 14 Apr 2020
Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, Stathopoulou T-R, Massey C, Shelton JM, Bassel-Duby R, Piercy RJ, Olson EN (2018) Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362:86–91. https://doi.org/10.1126/science.aau1549
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157. https://doi.org/10.1038/s41586-019-1711-4
Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38:824–844. https://doi.org/10.1038/s41587-020-0561-9
Asokan A, Schaffer DV, Samulski RJ (2012) The AAV vector toolkit: poised at the clinical crossroads. Mol Ther 20:699–708. https://doi.org/10.1038/mt.2011.287
Auer TO, Duroure K, de Cian A, Concordet J-P, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24:142–153. https://doi.org/10.1101/gr.161638.113
Bainbridge JWB, Mehat MS, Sundaram V, Robbie SJ, Barker SE, Ripamonti C, Georgiadis A, Mowat FM, Beattie SG, Gardner PJ, Feathers KL, Luong VA, Yzer S, Balaggan K, Viswanathan A, de Ravel TJL, Casteels I, Holder GE, Tyler N, Fitzke FW, Weleber RG, Nardini M, Moore AT, Thompson DA, Petersen-Jones SM, Michaelides M, van den Born LI, Stockman A, Smith AJ, Rubin G, Ali RR (2015) Long-term effect of gene therapy on Leber’s congenital amaurosis. N Engl J Med 372:1887–1897. https://doi.org/10.1056/NEJMoa1414221
Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, Ansari R, Harper S, Jackson DA, McRae R, Pooley R, Wilkinson P, van der Meer D, Dow D, Buser-Doepner C, Bertotti A, Trusolino L, Stronach EA, Saez-Rodriguez J, Yusa K, Garnett MJ (2019) Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568:511–516. https://doi.org/10.1038/s41586-019-1103-9
Bellin M, Casini S, Davis RP, D’Aniello C, Haas J, Ward-van Oostwaard D, Tertoolen LGJ, Jung CB, Elliott DA, Welling A, Laugwitz K-L, Moretti A, Mummery CL (2013) Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J 32:3161–3175. https://doi.org/10.1038/emboj.2013.240
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512. https://doi.org/10.1126/science.1178811
Boel A, de Saffel H, Steyaert W, Callewaert B, de Paepe A, Coucke PJ, Willaert A (2018) CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis Model Mech. https://doi.org/10.1242/dmm.035352
Boroviak K, Fu B, Yang F, Doe B, Bradley A (2017) Revealing hidden complexities of genomic rearrangements generated with Cas9. Sci Rep 7:12867. https://doi.org/10.1038/s41598-017-12740-6
Burgio G, Teboul L (2020) Anticipating and identifying collateral damage in genome editing. Trends Genet 36:905–914. https://doi.org/10.1016/j.tig.2020.09.011
Campbell LA, Richie CT, Maggirwar NS, Harvey BK (2019) Cas9 ribonucleoprotein complex delivery: methods and applications for neuroinflammation. J Neuroimmune Pharmacol 14:565–577. https://doi.org/10.1007/s11481-019-09856-z
Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan G-C, Zhang F, Orkin SH, Bauer DE (2015) BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527:192–197. https://doi.org/10.1038/nature15521
Cao J, Wu L, Zhang S-M, Lu M, Cheung WKC, Cai W, Gale M, Xu Q, Yan Q (2016) An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res 44:e149. https://doi.org/10.1093/nar/gkw660
Capps B, Chadwick R, Joly Y, Mulvihill JJ, Lysaght T, Zwart H (2017) Falling giants and the rise of gene editing: ethics, private interests and the public good. Hum Genomics 11:20. https://doi.org/10.1186/s40246-017-0116-4
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong J-W, Xi JJ (2013) Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 23:465–472. https://doi.org/10.1038/cr.2013.45
Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, Behlke MA, Dejene B, Cieniewicz B, Romano R, Lesch BJ, Gomez-Ospina N, Mantri S, Pavel-Dinu M, Weinberg KI, Porteus MH (2019) Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 25:249–254. https://doi.org/10.1038/s41591-018-0326-x
Chaudhary K, Chattopadhyay A, Pratap D (2018) The evolution of CRISPR/Cas9 and their cousins: hope or hype? Biotechnol Lett 40:465–477. https://doi.org/10.1007/s10529-018-2506-7
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155:1479–1491. https://doi.org/10.1016/j.cell.2013.12.001
Cohen J (2019) The long shadow of a CRISPR scandal. Science 365:436. https://doi.org/10.1126/science.365.6452.436
Collier AJ, Panula SP, Schell JP, Chovanec P, Plaza Reyes A, Petropoulos S, Corcoran AE, Walker R, Douagi I, Lanner F, Rugg-Gunn PJ (2017) Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell 20:874-890.e7. https://doi.org/10.1016/j.stem.2017.02.014
Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358:1019–1027. https://doi.org/10.1126/science.aaq0180
Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, Pasero P, Rowicka M, Dikic I (2013) Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10:361–365. https://doi.org/10.1038/nmeth.2408
Cui Y, Xu J, Cheng M, Liao X, Peng S (2018) Review of CRISPR/Cas9 sgRNA design tools. Interdiscip Sci 10:455–465. https://doi.org/10.1007/s12539-018-0298-z
Danner E, Bashir S, Yumlu S, Wurst W, Wefers B, Kühn R (2017) Control of gene editing by manipulation of DNA repair mechanisms. Mamm Genome 28:262–274. https://doi.org/10.1007/s00335-017-9688-5
Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR (2015) Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 11:316–318. https://doi.org/10.1038/nchembio.1793
Diaz AA, Qin H, Ramalho-Santos M, Song JS (2015) HiTSelect: a comprehensive tool for high-complexity-pooled screen analysis. Nucleic Acids Res 43:e16. https://doi.org/10.1093/nar/gku1197
Diehl V, Wegner M, Grumati P, Husnjak K, Schaubeck S, Gubas A, Shah VJ, Langschied F, Kalousi A, Ebersberger I, Dikic I, Kaulich M (2020) Combinatorial CRISPR screening reveals functional buffering in autophagy. https://doi.org/10.1101/2020.07.28.201152. www.biorxiv.org/content/10.1101/2020.07.28.201152v1
Doench JG (2018) Am I ready for CRISPR? A user’s guide to genetic screens. Nat Rev Genet 19:67–80. https://doi.org/10.1038/nrg.2017.97
Dominguez AA, Lim WA, Qi LS (2016) Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17:5–15. https://doi.org/10.1038/nrm.2015.2
Dow LE, Fisher J, O’Rourke KP, Muley A, Kastenhuber ER, Livshits G, Tschaharganeh DF, Socci ND, Lowe SW (2015) Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 33:390–394. https://doi.org/10.1038/nbt.3155
Ehrke-Schulz E, Schiwon M, Leitner T, Dávid S, Bergmann T, Liu J, Ehrhardt A (2017) CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci Rep 7:17113. https://doi.org/10.1038/s41598-017-17180-w
El Refaey M, Xu L, Gao Y, Canan BD, Adesanya TMA, Warner SC, Akagi K, Symer DE, Mohler PJ, Ma J, Janssen PML, Han R (2017) In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res 121:923–929. https://doi.org/10.1161/CIRCRESAHA.117.310996
El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai S-L, Fukuda R, Gerri C, Giraldez AJ, Stainier DYR (2019) Genetic compensation triggered by mutant mRNA degradation. Nature 568:193–197. https://doi.org/10.1038/s41586-019-1064-z
Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10:741–743. https://doi.org/10.1038/nmeth.2532
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826. https://doi.org/10.1038/nbt.2623
Geisler A, Jungmann A, Kurreck J, Poller W, Katus HA, Vetter R, Fechner H, Müller OJ (2011) microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther 18:199–209. https://doi.org/10.1038/gt.2010.141
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451. https://doi.org/10.1016/j.cell.2013.06.044
Gjaltema RAF, Rots MG (2020) Advances of epigenetic editing. Curr Opin Chem Biol 57:75–81. https://doi.org/10.1016/j.cbpa.2020.04.020
González F, Zhu Z, Shi Z-D, Lelli K, Verma N, Li QV, Huangfu D (2014) An iCRISPR platform for rapid, multiplexable and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15:215–226. https://doi.org/10.1016/j.stem.2014.05.018
Grünewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK (2019) Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569:433–437. https://doi.org/10.1038/s41586-019-1161-z
Gurumurthy CB, Lloyd KCK (2019) Generating mouse models for biomedical research: technological advances. Dis Model Mech. https://doi.org/10.1242/dmm.029462
Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24:927–930. https://doi.org/10.1038/s41591-018-0049-z
Halioua-Haubold C-L, Peyer JG, Smith JA, Arshad Z, Scholz M, Brindley DA, MacLaren RE (2017) Regulatory considerations for gene therapy products in the US, EU, and Japan. Yale J Biol Med 90:683–693
Hallett PJ, Deleidi M, Astradsson A, Smith GA, Cooper O, Osborn TM, Sundberg M, Moore MA, Perez-Torres E, Brownell A-L, Schumacher JM, Spealman RD, Isacson O (2015) Successful function of autologous iPSC-derived dopamine neurons following transplantation in a non-human primate model of Parkinson’s disease. Cell Stem Cell 16:269–274. https://doi.org/10.1016/j.stem.2015.01.018
Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20:1848–1867. https://doi.org/10.1101/gad.1422906
Heman-Ackah SM, Bassett AR, Wood MJA (2016) Precision modulation of neurodegenerative disease-related gene expression in human iPSC-derived neurons. Sci Rep 6:28420. https://doi.org/10.1038/srep28420
High KA, Roncarolo MG (2019) Gene therapy. N Engl J Med 381:455–464. https://doi.org/10.1056/NEJMra1706910
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556:57–63. https://doi.org/10.1038/nature26155
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229. https://doi.org/10.1038/nbt.2501
Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, Randhawa R, Kulkarni T, Yang Z, McAllister G, Russ C, Reece-Hoyes J, Forrester W, Hoffman GR, Dolmetsch R, Kaykas A (2018) p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24:939–946. https://doi.org/10.1038/s41591-018-0050-6
Ishida K, Gee P, Hotta A (2015) Minimizing off-target mutagenesis risks caused by programmable nucleases. Int J Mol Sci 16:24751–24771. https://doi.org/10.3390/ijms161024751
Ishikawa K, Weber T, Hajjar RJ (2018) Human Cardiac Gene Therapy. Circ Res 123:601–613. https://doi.org/10.1161/CIRCRESAHA.118.311587
Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, Skarnes WC (2015) Off-target mutations are rare in Cas9-modified mice. Nat Methods 12:479. https://doi.org/10.1038/nmeth.3408
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. Elife 2:e00471. https://doi.org/10.7554/eLife.00471
Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F (2017) Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12:828–863. https://doi.org/10.1038/nprot.2017.016
Kato-Inui T, Takahashi G, Hsu S, Miyaoka Y (2018) Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 with improved proof-reading enhances homology-directed repair. Nucleic Acids Res 46:4677–4688. https://doi.org/10.1093/nar/gky264
Kim S, Kim D, Cho SW, Kim J, Kim J-S (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019. https://doi.org/10.1101/gr.171322.113
Kimura Y, Hisano Y, Kawahara A, Higashijima S (2014) Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 4:6545. https://doi.org/10.1038/srep06545
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485. https://doi.org/10.1038/nature14592
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen N, Zheng Z, Joung JK (2016) High-fidelity CRISPR-Cas9 variants with undetectable genome-wide off-targets. Nature 529:490–495. https://doi.org/10.1038/nature16526
Kohama Y, Higo S, Masumura Y, Shiba M, Kondo T, Ishizu T, Higo T, Nakamura S, Kameda S, Tabata T, Inoue H, Motooka D, Okuzaki D, Takashima S, Miyagawa S, Sawa Y, Hikoso S, Sakata Y (2020) Adeno-associated virus-mediated gene delivery promotes S-phase entry-independent precise targeted integration in cardiomyocytes. Sci Rep 10:15348. https://doi.org/10.1038/s41598-020-72216-y
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424. https://doi.org/10.1038/nature17946
Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD (2018) Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173:665-676.e14. https://doi.org/10.1016/j.cell.2018.02.033
Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R (2016) Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol 34:192–198. https://doi.org/10.1038/nbt.3450
Kosicki M, Tomberg K, Bradley A (2018) Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36:765–771. https://doi.org/10.1038/nbt.4192
Lanigan TM, Kopera HC, Saunders TL (2020) Principles of genetic engineering. Genes (Basel). https://doi.org/10.3390/genes11030291
Cong Le, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
Lessing D, Anguera MC, Lee JT (2013) X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet 14:85–110. https://doi.org/10.1146/annurev-genom-091212-153530
Levy JM, Yeh W-H, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, Liu DR (2020) Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng 4:97–110. https://doi.org/10.1038/s41551-019-0501-5
Li W, Xu H, Xiao T, Cong Le, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS (2014) MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554. https://doi.org/10.1186/s13059-014-0554-4
Li H, Beckman KA, Pessino V, Huang B, Weissman JS, Leonetti MD (2017) Design and specificity of long ssDNA donors for CRISPR-based knock-in 25. https://doi.org/10.1101/841098, www.biorxiv.org/content/10.1101/841098v1
Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X (2020) Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther 5:1. https://doi.org/10.1038/s41392-019-0089-y
Li C, Samulski RJ (2020) Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 21:255–272. https://doi.org/10.1038/s41576-019-0205-4
Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H, Yamamoto T, Yamanaka S, Hotta A (2015) Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep 4:143–154. https://doi.org/10.1016/j.stemcr.2014.10.013
Liu Y, Cao Z, Wang Y, Guo Y, Xu P, Yuan P, Liu Z, He Y, Wei W (2018) Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat Biotechnol. https://doi.org/10.1038/nbt.4283
Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351:400–403. https://doi.org/10.1126/science.aad5725
Low BE, Kutny PM, Wiles MV (2016) Simple, Efficient CRISPR-Cas9-mediated gene editing in mice: strategies and methods. Methods Mol Biol 1438:19–53. https://doi.org/10.1007/978-1-4939-3661-8_2
Ma M, Ye AY, Zheng W, Kong L (2013) A guide RNA sequence design platform for the CRISPR/Cas9 system for model organism genomes. Biomed Res Int 2013:270805. https://doi.org/10.1155/2013/270805
Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T (2015) Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A 112:3002–3007. https://doi.org/10.1073/pnas.1420024112
Ma H, Tu L-C, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T (2016) Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34:528–530. https://doi.org/10.1038/nbt.3526
Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA (2009) Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci USA 106:15768–15773. https://doi.org/10.1073/pnas.0906894106
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838. https://doi.org/10.1038/nbt.2675
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826. https://doi.org/10.1126/science.1232033
Marczenke M, Piccini I, Mengarelli I, Fell J, Röpke A, Seebohm G, Verkerk AO, Greber B (2017) Cardiac subtype-specific modeling of Kv1.5 ion channel deficiency using human pluripotent stem cells. Front Physiol 8:469. https://doi.org/10.3389/fphys.2017.00469
McClellan J, King M-C (2010) Genetic heterogeneity in human disease. Cell 141:210–217. https://doi.org/10.1016/j.cell.2010.03.032
Miles LA, Garippa RJ, Poirier JT (2016) Design, execution, and analysis of pooled in vitro CRISPR/Cas9 screens. FEBS J 283:3170–3180. https://doi.org/10.1111/febs.13770
Mitchell KJ (2012) What is complex about complex disorders? Genome Biol 13:237. https://doi.org/10.1186/gb-2012-13-1-237
Mohr S, Bakal C, Perrimon N (2010) Genomic screening with RNAi: results and challenges. Annu Rev Biochem 79:37–64. https://doi.org/10.1146/annurev-biochem-060408-092949
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42:W401–W407. https://doi.org/10.1093/nar/gku410
Moore R, Spinhirne A, Lai MJ, Preisser S, Li Y, Kang T, Bleris L (2015) CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res 43:1297–1303. https://doi.org/10.1093/nar/gku1326
Moretti A, Fonteyne L, Giesert F, Hoppmann P, Meier AB, Bozoglu T, Baehr A, Schneider CM, Sinnecker D, Klett K, Fröhlich T, Rahman FA, Haufe T, Sun S, Jurisch V, Kessler B, Hinkel R, Dirschinger R, Martens E, Jilek C, Graf A, Krebs S, Santamaria G, Kurome M, Zakhartchenko V, Campbell B, Voelse K, Wolf A, Ziegler T, Reichert S, Lee S, Flenkenthaler F, Dorn T, Jeremias I, Blum H, Dendorfer A, Schnieke A, Krause S, Walter MC, Klymiuk N, Laugwitz KL, Wolf E, Wurst W, Kupatt C (2020) Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med 26:207–214. https://doi.org/10.1038/s41591-019-0738-2
Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31:1120–1123. https://doi.org/10.1093/bioinformatics/btu743
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351:403–407. https://doi.org/10.1126/science.aad5143
Nihongaki Y, Kawano F, Nakajima T, Sato M (2015) Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol 33:755–760. https://doi.org/10.1038/nbt.3245
Nishimasu H, Cong Le, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O (2015) Crystal structure of Staphylococcus aureus Cas9. Cell 162:1113–1126. https://doi.org/10.1016/j.cell.2015.08.007
Ong SH, Li Y, Koike-Yusa H, Yusa K (2017) Optimised metrics for CRISPR-KO screens with second-generation gRNA libraries. Sci Rep 7:7384. https://doi.org/10.1038/s41598-017-07827-z
Papasavva P, Kleanthous M, Lederer CW (2019) Rare opportunities: CRISPR/Cas-based therapy development for rare genetic diseases. Mol Diagn Ther 23:201–222. https://doi.org/10.1007/s40291-019-00392-3
Parikh BA, Beckman DL, Patel SJ, White JM, Yokoyama WM (2015) Detailed phenotypic and molecular analyses of genetically modified mice generated by CRISPR-Cas9-mediated editing. PLoS ONE 10:e0116484. https://doi.org/10.1371/journal.pone.0116484
Park C-Y, Halevy T, Lee DR, Sung JJ, Lee JS, Yanuka O, Benvenisty N, Kim D-W (2015) Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep 13:234–241. https://doi.org/10.1016/j.celrep.2015.08.084
Pickar-Oliver A, Gersbach CA (2019) The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol 20:490–507. https://doi.org/10.1038/s41580-019-0131-5
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455. https://doi.org/10.1016/j.cell.2014.09.014
Polstein LR, Gersbach CA (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11:198–200. https://doi.org/10.1038/nchembio.1753
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. https://doi.org/10.1016/j.cell.2013.02.022
Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K, Singh R, Darzacq X, Yildiz A, Adli M (2017) Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun 8:14725. https://doi.org/10.1038/ncomms14725
Raake PW, Hinkel R, Müller S, Delker S, Kreuzpointner R, Kupatt C, Katus HA, Kleinschmidt JA, Boekstegers P, Müller OJ (2008) Cardio-specific long-term gene expression in a porcine model after selective pressure-regulated retroinfusion of adeno-associated viral (AAV) vectors. Gene Ther 15:12–17. https://doi.org/10.1038/sj.gt.3303035
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. https://doi.org/10.1038/nprot.2013.143
Rauch S, He C, Dickinson BC (2018) Targeted m6A reader proteins to study epitranscriptomic regulation of single RNAs. J Am Chem Soc 140:11974–11981. https://doi.org/10.1021/jacs.8b05012
Rees HA, Liu DR (2018) Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19:770–788. https://doi.org/10.1038/s41576-018-0059-1
Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34:339–344. https://doi.org/10.1038/nbt.3481
Roth TL, Puig-Saus C, Yu R, Shifrut E, Carnevale J, Li PJ, Hiatt J, Saco J, Krystofinski P, Li H, Tobin V, Nguyen DN, Lee MR, Putnam AL, Ferris AL, Chen JW, Schickel J-N, Pellerin L, Carmody D, Alkorta-Aranburu G, Del Gaudio D, Matsumoto H, Morell M, Mao Y, Cho M, Quadros RM, Gurumurthy CB, Smith B, Haugwitz M, Hughes SH, Weissman JS, Schumann K, Esensten JH, May AP, Ashworth A, Kupfer GM, Greeley SAW, Bacchetta R, Meffre E, Roncarolo MG, Romberg N, Herold KC, Ribas A, Leonetti MD, Marson A (2018) Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559:405–409. https://doi.org/10.1038/s41586-018-0326-5
Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, Kuoy E, Langerman J, Clark AT, Jaenisch R, Plath K (2017) Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20:87–101. https://doi.org/10.1016/j.stem.2016.10.006
Sanjana NE (2017) Genome-scale CRISPR pooled screens. Anal Biochem 532:95–99. https://doi.org/10.1016/J.AB.2016.05.014
Sanjana NE, Shalem O, Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. https://doi.org/10.1038/nmeth.3047
Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F (2016) High-resolution interrogation of functional elements in the noncoding genome. Science 353:1545–1549. https://doi.org/10.1126/science.aaf7613
Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JG (2018) Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9:5416. https://doi.org/10.1038/s41467-018-07901-8
Schuster A, Erasimus H, Fritah S, Nazarov PV, van Dyck E, Niclou SP, Golebiewska A (2019) RNAi/CRISPR screens: from a pool to a valid hit. Trends Biotechnol 37:38–55. https://doi.org/10.1016/j.tibtech.2018.08.002
Seki A, Rutz S (2018) Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med 215:985–997. https://doi.org/10.1084/jem.20171626
Selle K, Klaenhammer TR, Barrangou R (2015) CRISPR-based screening of genomic island excision events in bacteria. Proc Natl Acad Sci U S A 112:8076–8081. https://doi.org/10.1073/pnas.1508525112
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. https://doi.org/10.1126/science.1247005
Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR (2015) Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33:661–667. https://doi.org/10.1038/nbt.3235
Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, Hennighausen L (2017) CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun. https://doi.org/10.1038/ncomms15464
Shukla A, Huangfu D (2018) Decoding the noncoding genome via large-scale CRISPR screens. Curr Opin Genet Dev 52:70–76. https://doi.org/10.1016/J.GDE.2018.06.001
Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, Sauna ZE (2018) Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol Ther Methods Clin Dev 10:105–112. https://doi.org/10.1016/j.omtm.2018.06.006
Skryabin BV, Kummerfeld D-M, Gubar L, Seeger B, Kaiser H, Stegemann A, Roth J, Meuth SG, Pavenstädt H, Sherwood J, Pap T, Wedlich-Söldner R, Sunderkötter C, Schwartz YB, Brosius J, Rozhdestvensky TS (2020) Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9-mediated genome editing events. Sci Adv 6:eaax2941. https://doi.org/10.1126/sciadv.aax2941
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88. https://doi.org/10.1126/science.aad5227
Smirnov A, Fishman V, Yunusova A, Korablev A, Serova I, Skryabin BV, Rozhdestvensky TS, Battulin N (2020) DNA barcoding reveals that injected transgenes are predominantly processed by homologous recombination in mouse zygote. Nucleic Acids Res 48:719–735. https://doi.org/10.1093/nar/gkz1085
Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z (2014) Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15:12–13. https://doi.org/10.1016/j.stem.2014.06.011
Soldner F, Jaenisch R (2018) Stem cells, genome editing, and the path to translational medicine. Cell 175:615–632. https://doi.org/10.1016/j.cell.2018.09.010
Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R (2009) Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136:964–977. https://doi.org/10.1016/j.cell.2009.02.013
Spahn PN, Bath T, Weiss RJ, Kim J, Esko JD, Lewis NE, Harismendy O (2017) PinAPL-Py: a comprehensive web-application for the analysis of CRISPR/Cas9 screens. Sci Rep 7:15854. https://doi.org/10.1038/s41598-017-16193-9
Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, Kulikovskaya I, Gupta M, Chen F, Tian L, Gonzalez VE, Xu J, Jung I-Y, Melenhorst JJ, Plesa G, Shea J, Matlawski T, Cervini A, Gaymon AL, Desjardins S, Lamontagne A, Salas-Mckee J, Fesnak A, Siegel DL, Levine BL, Jadlowsky JK, Young RM, Chew A, Hwang W-T, Hexner EO, Carreno BM, Nobles CL, Bushman FD, Parker KR, Qi Y, Satpathy AT, Chang HY, Zhao Y, Lacey SF, June CH (2020) CRISPR-engineered T cells in patients with refractory cancer. Science. https://doi.org/10.1126/science.aba7365
Stojic L, Lun ATL, Mangei J, Mascalchi P, Quarantotti V, Barr AR, Bakal C, Marioni JC, Gergely F, Odom DT (2018) Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res 46:5950–5966. https://doi.org/10.1093/nar/gky437
Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E, Zhang F, Xu X, Soligalla RD, Chen F, Kim J, Kim NY, Liao H-K, Benner C, Esteban CR, Jin Y, Liu G-H, Li Y, Izpisua Belmonte JC (2014) Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 15:31–36. https://doi.org/10.1016/j.stem.2014.06.016
Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong Le, Zhang F, Vandenberghe LH, Church GM, Wagers AJ (2016) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351:407–411. https://doi.org/10.1126/science.aad5177
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. https://doi.org/10.1016/j.cell.2007.11.019
Tessadori F, Roessler HI, Savelberg SMC, Chocron S, Kamel SM, Duran KJ, van Haelst MM, van Haaften G, Bakkers J (2018) Effective CRISPR/Cas9-based nucleotide editing in zebrafish to model human genetic cardiovascular disorders. Dis Model Mech. https://doi.org/10.1242/dmm.035469
Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, Eilertson KE, Tsang P, Baba S, White MP, Sami S, Srivastava D, Conklin BR, Panning B, Yamanaka S (2012) Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell 11:91–99. https://doi.org/10.1016/j.stem.2012.05.019
Truong D-JJ, Kühner K, Kühn R, Werfel S, Engelhardt S, Wurst W, Ortiz O (2015) Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res 43:6450–6458. https://doi.org/10.1093/nar/gkv601
Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33:187–197. https://doi.org/10.1038/nbt.3117
Tsai SQ, Topkar VV, Joung JK, Aryee MJ (2016) Open-source guideseq software for analysis of GUIDE-seq data. Nat Biotechnol 34:483. https://doi.org/10.1038/nbt.3534
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC (2017) Defining a cancer dependency map. Cell 170:564-576.e16. https://doi.org/10.1016/J.CELL.2017.06.010
Tyagi S, Kumar R, Das A, Won SY, Shukla P (2020) CRISPR-Cas9 system: a genome-editing tool with endless possibilities. J Biotechnol 319:36–53. https://doi.org/10.1016/j.jbiotec.2020.05.008
Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435:646–651. https://doi.org/10.1038/nature03556
Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K (2014) Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15:27–30. https://doi.org/10.1016/j.stem.2014.04.020
Verma IC, Puri RD (2015) Global burden of genetic disease and the role of genetic screening. Semin Fetal Neonatal Med 20:354–363. https://doi.org/10.1016/j.siny.2015.07.002
Wagner DL, Amini L, Wendering DJ, Burkhardt L-M, Akyüz L, Reinke P, Volk H-D, Schmueck-Henneresse M (2019) High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med 25:242–248. https://doi.org/10.1038/s41591-018-0204-6
Wang T, Wei JJ, Sabatini DM, Lander ES (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84. https://doi.org/10.1126/science.1246981
Wegner M, Diehl V, Bittl V, de Bruyn R, Wiechmann S, Matthess Y, Hebel M, Hayes MG, Schaubeck S, Benner C, Heinz S, Bremm A, Dikic I, Ernst A, Kaulich M (2019) Circular synthesized CRISPR/Cas gRNAs for functional interrogations in the coding and noncoding genome. Elife. https://doi.org/10.7554/eLife.42549
Wienert B, Wyman SK, Richardson CD, Yeh CD, Akcakaya P, Porritt MJ, Morlock M, Vu JT, Kazane KR, Watry HL, Judge LM, Conklin BR, Maresca M, Corn JE (2019) Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364:286–289. https://doi.org/10.1126/science.aav9023
Wu X, Kriz AJ, Sharp PA (2014) Target specificity of the CRISPR-Cas9 system. Quant Biol 2:59–70. https://doi.org/10.1007/s40484-014-0030-x
Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, Zhu Z, Lin S, Zhang B (2013) Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 41:e141. https://doi.org/10.1093/nar/gkt464
Xie Y, Wang D, Lan F, Wei G, Ni T, Chai R, Liu D, Hu S, Li M, Li D, Wang H, Wang Y (2018) Author correction: an episomal vector-based CRISPR/Cas9 system for highly efficient gene knockout in human pluripotent stem cells. Sci Rep 8:17900. https://doi.org/10.1038/s41598-018-36738-w
Xu X, Tay Y, Sim B, Yoon S-I, Huang Y, Ooi J, Utami KH, Ziaei A, Ng B, Radulescu C, Low D, Ng AYJ, Loh M, Venkatesh B, Ginhoux F, Augustine GJ, Pouladi MA (2017) Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in huntington disease patient-derived induced pluripotent stem cells. Stem Cell Rep 8:619–633. https://doi.org/10.1016/j.stemcr.2017.01.022
Xu X, Wan T, Xin H, Li D, Pan H, Wu J, Ping Y (2019) Delivery of CRISPR/Cas9 for therapeutic genome editing. J Gene Med 21:e3107. https://doi.org/10.1002/jgm.3107
Xu CL, Ruan MZC, Mahajan VB, Tsang SH (2019) Viral delivery systems for CRISPR. Viruses. https://doi.org/10.3390/v11010028
Yang L, Guell M, Byrne S, Yang JL, de Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, Huang P-Y, Daley G, Church G (2013) Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41:9049–9061. https://doi.org/10.1093/nar/gkt555
Yang L, Yang JL, Byrne S, Pan J, Church GM (2014) CRISPR/Cas9-directed genome editing of cultured cells. Curr Protoc Mol Biol 107:31. https://doi.org/10.1002/0471142727.mb3101s107 (1.1-17)
Zamanighomi M, Jain SS, Ito T, Pal D, Daley TP, Sellers WR (2019) GEMINI: a variational Bayesian approach to identify genetic interactions from combinatorial CRISPR screens. Genome Biol 20:137. https://doi.org/10.1186/s13059-019-1745-9
Zhang Y, Qin W, Lu X, Xu J, Huang H, Bai H, Li S, Lin S (2017) Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat Commun 8:118. https://doi.org/10.1038/s41467-017-00175-6
Zhou C, Sun Y, Yan R, Liu Y, Zuo E, Gu C, Han L, Wei Y, Hu X, Zeng R, Li Y, Zhou H, Guo F, Yang H (2019) Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571:275–278. https://doi.org/10.1038/s41586-019-1314-0
Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DTW, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J (2015) Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 6:7391. https://doi.org/10.1038/ncomms8391
Acknowledgements
This position paper was written on behalf of the DGK-DZHK Taskforce Genome Editing with the following additional members who have contributed to the manuscript by critical discussion: Johannes Backs, Charlotta Behrens, Lucie Carrier, Friederike Cuello, Thomas Eschenhagen, Roger Foo, Mauro Giacca, Ralf Gilsbach, Arne Hansen, Denise Hilfiker-Kleiner, Rabea Hinkel, Michaela Kuhn, Malte Loos, Kristina Lorenz, Giulia Mearini, Patrick Most, Maksymilian Prondzynski, Angelika Schnieke, Katrin Schroeder, Steven Schulz, Thorsten Stafforst, Justus Stenzig, Malte Tiburcy, Thomas Voit, Kai Wollert, and Wolfram Hubertus Zimmermann. It has been approved by these scientists. The DGK commission on experimental cardiology as well as the DGK and the DZHK board consented to this manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. The initiative for this manuscript was supported by a grant from the DGK and the DZHK.
Author information
Authors and Affiliations
Contributions
All authors were actively involved in writing and composing subsection of the position paper.
Corresponding author
Ethics declarations
Conflicts of interest
The following conflict of interest do exist: Thomas Thum is founder and shareholder of Cardior Pharmaceuticals GmbH. Manuel Kaulich is co-founder, shareholder, and chief scientific officer of Vivlion GmbH. The following authors declare not to have a conflict of interest: Ralf P. Brandes, Anne Dueck, Stfan Engelhardt, Christian Kupatt, Maria Terese De Angelis, Matthias S. Leisegang, Ferdinand le Noble, Alessandra Moretti, Oliver J. Müller, Boris V. Skryabin, and Wolfgang Wurst.
Ethicas approval
Does not apply.
Consent to participate
All authors have actively participated in this work and give their consent.
Consent for publication
All authors have read the final version of the manuscript and consent to its publication.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brandes, R.P., Dueck, A., Engelhardt, S. et al. DGK and DZHK position paper on genome editing: basic science applications and future perspective. Basic Res Cardiol 116, 2 (2021). https://doi.org/10.1007/s00395-020-00839-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00395-020-00839-3