Abstract
Purpose
MYCN onco-gene amplification in neuroblastoma confers patients to the high-risk disease category for which prognosis is poor and more aggressive multimodal treatment is indicated. This retrospective study leverages machine learning techniques to develop a computed tomography (CT)–based model incorporating semantic and non-semantic features for non-invasive prediction of MYCN amplification status in pediatric neuroblastoma.
Methods
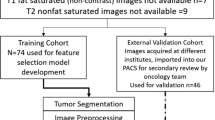
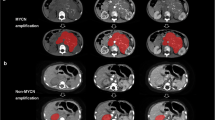
From 2009 to 2020, 54 pediatric patients treated for neuroblastoma at a specialized children’s hospital with pre-treatment contrast-enhanced CT and MYCN status were identified (training cohort, n = 44; testing cohort, n = 10). Six morphologic features and 107 quantitative gray-level texture radiomics features extracted from manually drawn volume-of-interest were analyzed. Following feature selection and class balancing, the final predictive model was developed with eXtreme Gradient Boosting (XGBoost) algorithm. Accumulated local effects (ALE) plots were used to explore main effects of the predictive features. Tumor texture maps were also generated for visualization of radiomics features.
Results
One morphologic and 2 radiomics features were selected for model building. The XGBoost model from the training cohort yielded an area under the receiver operating characteristics curve (AUC-ROC) of 0.930 (95% CI, 0.85–1.00), optimized F1-score of 0.878, and Matthews correlation coefficient (MCC) of 0.773. Evaluation on the testing cohort returned AUC-ROC of 0.880 (95% CI, 0.64–1.00), optimized F1-score of 0.933, and MCC of 0.764. ALE plots and texture maps showed higher “GreyLevelNonUniformity” values, lower “Strength” values, and higher number of image-defined risk factors contribute to higher predicted probability of MYCN amplification.
Conclusion
The machine learning model reliably classified MYCN amplification in pediatric neuroblastoma and shows potential as a surrogate imaging biomarker.



Similar content being viewed by others
Availability of data and material
Anonymized data is available upon request. Please contact the corresponding author.
Code availability
Available upon request. Please contact the corresponding author.
References
Swift CC, Eklund MJ, Kraveka JM, Alazraki AL (2018) Updates in diagnosis, management, and treatment of neuroblastoma. Radiographics 38:566–580
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71:7–33
Irwin MS, Naranjo A, Zhang FF et al (2021) Revised neuroblastoma risk classification system: a report from the children’s oncology group. J Clin Oncol JCO.21.00278
Yan P, Qi F, Bian L, Xu Y, Zhou J, Hu J, Ren L, Li M, Tang W (2020) Comparison of incidence and outcomes of neuroblastoma in children, adolescents, and adults in the United States: a surveillance, epidemiology, and end results (SEER) program population study. Med Sci Monit Int Med J Exp Clin Res 26:e927218-1-e927218-13
Morgenstern DA, Bagatell R, Cohn SL et al (2019) The challenge of defining “ultra-high-risk” neuroblastoma. Pediatr Blood Cancer 66:e27556
Morgenstern DA, Pötschger U, Moreno L et al (2018) Risk stratification of high-risk metastatic neuroblastoma: a report from the HR-NBL-1/SIOPEN study. Pediatr Blood Cancer 65:e27363
Cohn SL, Pearson ADJ, London WB et al (2009) The international neuroblastoma risk group (INRG) classification system: an INRG task force report. J Clin Oncol 27:289–297
Ladenstein RL, Poetschger U, Luksch R et al (2011) Busulphan-melphalan as a myeloablative therapy (MAT) for high-risk neuroblastoma: results from the HR-NBL1/SIOPEN trial. J Clin Oncol 29:2–2
Berthold F, Boos J, Burdach S et al (2005) Myeloablative megatherapy with autologous stem-cell rescue versus oral maintenance chemotherapy as consolidation treatment in patients with high-risk neuroblastoma: a randomised controlled trial. Lancet Oncol 6:649–658
Georgantzi K, Sköldenberg E, Janson ET, Jakobson Å, Christofferson R (2019) Diagnostic ultrasound-guided cutting needle biopsies in neuroblastoma: a safe and efficient procedure. J Pediatr Surg 54:1253–1256
Campagna G, Rosenfeld E, Foster J, Vasudevan S, Nuchtern J, Kim E, Commander S, Naik-Mathuria B (2018) Evolving biopsy techniques for the diagnosis of neuroblastoma in children. J Pediatr Surg 53:2235–2239
Hassan SF, Mathur S, Magliaro TJ et al (2012) Needle core vs open biopsy for diagnosis of intermediate- and high-risk neuroblastoma in children. J Pediatr Surg 47:1261–1266
Overman RE, Kartal TT, Cunningham AJ et al (2020) Optimization of percutaneous biopsy for diagnosis and pretreatment risk assessment of neuroblastoma. Pediatr Blood Cancer 67:e28153
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin J-C, Pieper S, Aerts HJWL (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Liu Z, Wang S, Dong D et al (2019) The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics 9:1303–1322
Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W, Beets-Tan R (2019) Radiogenomics: bridging imaging and genomics. Abdom Radiol 44:1960–1984
Gerlinger M, Rowan AJ, Horswell S et al (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366:883–892
Ambros PF, Ambros IM, Brodeur GM et al (2009) International consensus for neuroblastoma molecular diagnostics: report from the international neuroblastoma risk group (INRG) biology committee. Br J Cancer 100:1471–1482
Kuhn M (2008) Building predictive models in R using the caret package. J Stat Softw 28:1–26
Monclair T, Brodeur GM, Ambros PF et al (2009) The international neuroblastoma risk group (INRG) staging system: an INRG task force report. J Clin Oncol Off J Am Soc Clin Oncol 27:298–303
Avanzini S, Pio L, Erminio G et al (2017) Image-defined risk factors in unresectable neuroblastoma: SIOPEN study on incidence, chemotherapy-induced variation, and impact on surgical outcomes. Pediatr Blood Cancer. https://doi.org/10.1002/pbc.26605
Brisse HJ, McCarville MB, Granata C et al (2011) Guidelines for imaging and staging of neuroblastic tumors: consensus report from the international neuroblastoma risk group project. Radiology 261:243–257
Fedorov A, Beichel R, Kalpathy-Cramer J et al (2012) 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30:1323–1341
Zwanenburg A, Vallières M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295:328–338
De Jay N, Papillon-Cavanagh S, Olsen C, El-Hachem N, Bontempi G, Haibe-Kains B (2013) mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics 29:2365–2368
Lunardon N, Menardi G, Torelli N (2014) ROSE: A package for binary imbalanced learning. The R Journal 6:79–89
Apley DW, Zhu J (2020) Visualizing the effects of predictor variables in black box supervised learning models. J R Stat Soc Ser B Stat Methodol 82:1059–1086
Lancaster JL, Cykowski MD, McKay DR, Kochunov PV, Fox PT, Rogers W, Toga AW, Zilles K, Amunts K, Mazziotta J (2010) Anatomical global spatial normalization. Neuroinformatics 8:171–182
Chen T, He T, Benesty M, Khotilovich V, Tang Y, Cho H, Chen K, Mitchell R, Cano I, Zhou T, Li M, Xie J, Lin M, Geng Y, Li Y (2020) xgboost: Extreme Gradient Boosting R package version 1.2.0.1. https://CRAN.R-project.org/package=xgboost
John CR (2020) MLeval: Machine Learning Model Evaluation. R package version 0.3. https://CRAN.R-project.org/package=MLeval
Molnar C, Casalicchio G, Bischl B (2018) iml: An R package for interpretable machine learning. J Open Source Softw 3:786
Chen T, Guestrin C (2016) Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp 785–794. https://doi.org/10.1145/2939672.2939785
Alabi RO, Mäkitie AA, Pirinen M, Elmusrati M, Leivo I, Almangush A (2021) Comparison of nomogram with machine learning techniques for prediction of overall survival in patients with tongue cancer. Int J Med Inf 145:104313
Chen X, Wang H, Huang K, Liu H, Ding H, Zhang L, Zhang T, Yu W, He L (2021) CT-based radiomics signature with machine learning predicts MYCN amplification in pediatric abdominal neuroblastoma. Front Oncol 11:687884
Wu H, Wu C, Zheng H, Wang L, Guan W, Duan S, Wang D (2021) Radiogenomics of neuroblastoma in pediatric patients: CT-based radiomics signature in predicting MYCN amplification. Eur Radiol 31:3080–3089
Di Giannatale A, Di Paolo PL, Curione D, Lenkowicz J, Napolitano A, Secinaro A, Tomà P, Locatelli F, Castellano A, Boldrini L (2021) Radiogenomics prediction for MYCN amplification in neuroblastoma: a hypothesis generating study. Pediatr Blood Cancer 68:e29110
Otte J, Dyberg C, Pepich A, Johnsen JI (2021) MYCN function in neuroblastoma development. Front Oncol 10:624079
Traverso A, Wee L, Dekker A, Gillies R (2018) Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 102:1143–1158
Pavic M, Bogowicz M, Würms X et al (2018) Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol Stockh Swed 57:1070–1074
Zhang X, Zhong L, Zhang B, Zhang L, Du H, Lu L, Zhang S, Yang W, Feng Q (2019) The effects of volume of interest delineation on MRI-based radiomics analysis: evaluation with two disease groups. Cancer Imaging 19:89
Zwanenburg A, Leger S, Agolli L, Pilz K, Troost EGC, Richter C, Löck S (2019) Assessing robustness of radiomic features by image perturbation. Sci Rep 9:614
Trigg RM, Turner SD, Shaw JA, Jahangiri L (2020) Diagnostic accuracy of circulating-free DNA for the determination of MYCN amplification status in advanced-stage neuroblastoma: a systematic review and meta-analysis. Br J Cancer 122:1077–1084
Khodabakhshi Z, Mostafaei S, Arabi H, Oveisi M, Shiri I, Zaidi H (2021) Non-small cell lung carcinoma histopathological subtype phenotyping using high-dimensional multinomial multiclass CT radiomics signature. Comput Biol Med 136:104752
Jang K, Russo C, Di Ieva A (2020) Radiomics in gliomas: clinical implications of computational modeling and fractal-based analysis. Neuroradiology 62:771–790
Choi Y, Ahn KJ, Nam Y, Jang J, Shin N-Y, Choi HS, Jung S-L, Kim B (2019) Analysis of peritumoral hyperintensity on pre-operative T2-weighted MR images in glioblastoma: additive prognostic value of Minkowski functionals. PLoS ONE 14:e0217785
Campbell K, Naranjo A, Hibbitts E, Gastier-Foster JM, Bagatell R, Irwin MS, Shimada H, Hogarty M, Park JR, DuBois SG (2020) Association of heterogeneous MYCN amplification with clinical features, biological characteristics and outcomes in neuroblastoma: a report from the children’s oncology group. Eur J Cancer 133:112–119
Liang WH, Federico SM, London WB, Naranjo A, Irwin MS, Volchenboum SL, Cohn SL (2020) Tailoring therapy for children with neuroblastoma on the basis of risk group classification: past, present, and future. JCO Clin Cancer Inform 4:895–905
Moreno L, Guo D, Irwin MS et al (2021) A nomogram of clinical and biologic factors to predict survival in children newly diagnosed with high-risk neuroblastoma: an international neuroblastoma risk group project. Pediatr Blood Cancer 68:e28794
Alexandrov LB, Nik-Zainal S, Wedge DC et al (2013) Signatures of mutational processes in human cancer. Nature 500:415–421
Author information
Authors and Affiliations
Contributions
Conception and design of study: ET, PH. Acquisition of data: ET, PH, KM, PO. Analysis and/or interpretation of data: ET, JZ, SE, BP, AR. Drafting the manuscript: ET, BP. Revising the manuscript: ET, BP, KM, JZ, PH. All authors have reviewed and approved the final article.
Corresponding author
Ethics declarations
Ethics approval
This study is approved by SingHealth centralized institutional review board (reference number: 2015/2608).
Consent to participate
Informed consent was waived by the review board.
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tan, E., Merchant, K., KN, B.P. et al. CT-based morphologic and radiomics features for the classification of MYCN gene amplification status in pediatric neuroblastoma. Childs Nerv Syst 38, 1487–1495 (2022). https://doi.org/10.1007/s00381-022-05534-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00381-022-05534-3