Abstract
Purpose
To map current literature and provide an overview of upcoming future diagnostic and prognostic methods for upper tract urothelial carcinoma (UTUC), including translational medical science.
Methods
A scoping review approach was applied to search the literature. Based on the published literature, and the experts own experience and opinions consensus was reached through discussions at the meeting Consultation on UTUC II in Stockholm, September 2022.
Results
The gene mutational profile of UTUC correlates with stage, grade, prognosis, and response to different therapeutic strategies. Analysis of pathway proteins downstream of known pathogenic mutations might be an alternative approach.
Liquid biopsies of cell-free DNA may detect UTUC with a higher sensitivity and specificity than urinary cytology. Extracellular vesicles from tumour cells can be detected in urine and may be used to identify the location of the urothelial carcinoma in the urinary tract. 3D microscopy of UTUC samples may add information in the analysis of tumour stage. Chemokines and chemokine receptors were linked to overall survival and responsiveness to neoadjuvant chemotherapy in muscle-invasive bladder cancer, which is potentially also of interest in UTUC.
Conclusion
Current diagnostic methods for UTUC have shortcomings, especially concerning prognostication, which is important for personalized treatment decisions. There are several upcoming methods that may be of interest for UTUC. Most have been studied for urothelial carcinoma of the bladder, and it is important to keep in mind that UTUC is a different entity and not all methods are adaptable or applicable to UTUC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Upper tract urothelial carcinoma (UTUC) is a rare malignancy with an increasing incidence [1]. Its survival rates have been unaltered in recent decades [2]. The overall survival (OS) is substantially lower than the cancer-specific survival (CSS) [1], reflecting the fragile patient group with advanced age and comorbidities. Potentially curative radical nephroureterectomy (RNU) carries a 15–40% overall complication rate [3]. Kidney-saving surgery (KSS) with ureterorenoscopy (URS) carries a substantially lower overall complication rate (3.5% in a large study on URS for stone disease) [4] but an increased risk of ipsilateral and intravesical recurrence (IVR) [5,6,7]. Several studies have shown similar oncological outcomes for RNU and KSS with URS in low-grade low-volume UTUC [5, 8,9,10], emphasizing the need for personalized treatment, depending on tumour risk stratification, comorbidities, surgical risk and the patient’s preferences.
There has been an increased interest and improvements in prognostication for UTUC during the last decades. A comprehensive review on current diagnostic methods was published after Consultation on UTUC in 2018 [11]. Despite improvements, there are still shortcomings. Tumour stage is a strong prognostic marker, but it cannot be reliably assessed before RNU. Stage and grade correlate well, and grade can be assessed in URS biopsies, unfortunately with a risk of undergrading [12]. The accuracy of cytology depends on how the sample was acquired [13, 14]. The correlation between grade and prognosis differs with the different WHO grading systems [15], resulting in recent suggestions of a merged 4-tier grading version [16, 17]. The EAU risk stratification of low- and high-risk UTUC [18] is a useful tool to aid treatment decision-making, although to our knowledge, studies on how the EAU risk classification correlates to long-term CSS are lacking. However, despite RNU, approximately 7% of low-grade non-muscle-invasive UTUC patients and 50% (34–62%) of high-grade and/or muscle-invasive UTUC patients experience recurrence and die from UTUC [8, 19]. The EAU risk stratification has evolved over the years, and recent work suggests further improvement: less emphasis on size and multifocality [20], the addition of age, biopsy stage and tumour architecture [21], or focusing on grade assessed by URS specimens and stage assessed by CT/MRI [22].
Current methods struggle to identify the patients with true high-risk tumours, i.e., the patients with low- or high-risk UTUC who will die from their disease despite radical treatment. Due to the rarity of UTUC, there is a lack of randomized controlled studies and larger prospective studies. This current paper aims to give a descriptive overview of current literature and expert opinion on upcoming possible future methods for improved diagnostics and prognostication in UTUC. As a scoping review and expert opinion, this paper will not make statements to guide decision-making, but rather map the literature and ongoing research within the field.
Methods
The recurrent meetings Consultation on UTUC are dedicated to UTUC. The faculty in 2022 covered clinicians and researchers focusing on UTUC, including urology, pathology/cytology, clinical and oncological genetics, immunology, biochemistry, cell signaling and 3D microscopy. The presenting faculty members were invited to present their area of expertise, combined with a literature overview of their field within diagnostics and prognostication. The experts were instructed to search the research field in accordance with scoping reviews, including current publications in English on PubMed, Web of Science, and Embase. Recent abstracts and unpublished data relevant to each field were also discussed. The search was conducted before the meeting, and consensus of expert opinion was reached through discussions at the meeting Consultation on UTUC II in Stockholm, September 2022. No predefined search protocol was used, as the methods presented cover a wide field of different research disciplines and such a protocol might limit the overview. The methods presented here are not yet commercially available, but in different stages of development.
Possible upcoming diagnostic and prognostic methods discussed at the meeting
Analysis of the gene mutational profile of tumour tissue
There is a growing body of research on the molecular genetics of urothelial carcinoma (UC), with emphasis on the more common urothelial carcinoma of the bladder (UCB) [23, 24]. Investigations of the genomic landscape of pure UTUC have revealed that UTUC and UCB have similar genetic mutations but different mutational frequencies, tumour mutational burden (TMB) and expression patterns [25, 26]. This implies subtle biologic differences between UTUC and UCB, reflected in clinical behavior. Several studies on UTUC have shown associations between tumour mutational pattern and stage, grade and CSS, where FGFR3 mutations indicate low-grade good prognosis and TP53/MDM2 aberrations indicate high-grade and worse prognosis [25, 27, 28]. There are cases reported where the mutational profile correlated better with the clinical outcome than the histopathological assessment [28]. Bagrodia et al. found a high concordance in gene mutations between URS biopsies and RNU specimens, irrespective of where in the tumour the biopsy was taken [29].
There are several suggested molecular classification systems for UCB, combining gene expression and mutational patterns. These were merged into a consensus molecular classification of muscle-invasive bladder cancer (MIBC) by Kamoun in 2020 [24]. The classes differ regarding underlying oncogenic mechanisms, histological and clinical characteristics, infiltration by immune and stromal cells and association with survival. Compared to UCB, UTUC is characterized by a luminal papillary expression signature, a T-cell depleted immunophenotype and decreased mismatch repair (MMR) gene expression [30]. There is a suggested molecular classification of UTUC, where the cluster subtypes are prognostic of clinical outcomes [31]. There are studies investigating how different transcriptional subtypes of UCB respond to therapeutic strategies [23] and how genetic biomarkers can be used to predict the response to oncological treatments [32, 33].
Analysis of the gene mutational profile from liquid biopsies
Liquid biopsy is the sampling of nonsolid biological tissue such as blood, urine, or saliva. Liquid biopsy can be used to study circulating tumour cell DNA (ctDNA) or cell-free DNA (cfDNA) [34]. cfDNA is released from blood cells by apoptosis and necrosis and from tumours by the active release of cfDNA which can be freely circulating or in extracellular vesicles (EVs) and protein complexes [35]. The proportion of cfDNA in blood that originates from a tumour is low and depends on the type of cancer and tumour burden [35]. Hurdles in analysis to overcome are artefacts and biological confounders, such as other concomitant malignancies and clonal haematopoiesis of indeterminate potential (CHIP).
For UC, plasma, urine and in situ barbotage can be used for liquid biopsies. Potential uses are for diagnosis, treatment response monitoring, and early detection of relapse and disease progression. In a review by Green et al., ctDNA was detected in plasma and urine in 14–80% of patients with localized UCB and in 70–90% of patients with MIBC [36]. For UTUC, UTUC-specific tumour mutations could be identified in urinary sediment-derived DNA from samples of voided urine. In a study by Fuji et al., the sensitivity and specificity of cancer detection using this method outperformed those of urinary cytology [31]. cfDNA in urine can be used for the diagnosis of UTUC [37]. ctDNA in plasma was used for disease monitoring in a case report by Blumendeller et al. [38]. A recent study on UTUC by Nakano et al. [39] showed that a high fraction of preoperative ctDNA and sustained ctDNA postoperatively were associated with a worse prognosis. Of note in this study, most identified gene mutations were in TP53, of which 19% were CHIPs, i.e., non-tumour-relevant artefacts.
Pathway proteins
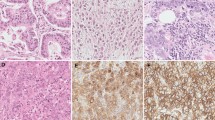
Not all gene mutations in a tumour are pathogenic, and pinpointing those that are drivers is a key issue in molecular diagnostics. A way to address this is to study the transcriptome, i.e., gene expression. A step further is to study proteomics, i.e., the proteins that are expressed by a cell. Pathway proteins are found intracellularly in the downstream signal transduction of ligands and receptors. Abnormalities in these pathways play key roles when a cell transform to a cancer cell, as uncontrolled cellular growth, invasiveness, and resistance to apoptosis [40]. Genes frequently mutated and/or overexpressed in UC include those involved in the RTK-Ras-PI3K pathway [31, 41]. A way to address abnormalities in these pathways is to study the transcriptome by measuring ligands and/or intracellular signaling pathway proteins. New techniques allow development of highly sensitive methods to quantify these low-abundance protein biomarkers. Aberrant concentrations of ligands or the intracellular pathway proteins ERK and AKT have been identified in breast [42] and colorectal cancer [42, 43] respectively. Unpublished test-of-concept studies have been done for UTUC and are presently further investigated.
3D microscopy
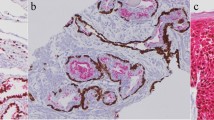
3D microscopy is a technique used to visualize three-dimensional structures within samples. Contrary to conventional histopathology, the sample is not sliced but rendered transparent to light, and the whole-mount sample is then imaged by a light-sheet microscope, creating a stack of images like a CT scan. Different structures within the sample can be visualized using conventional immunolabelling, and 3D rendering can be achieved with imaging software. The technique can be used for freshly frozen or paraffin-embedded samples, and imaging can be performed at a single-cell resolution level. Additionally, other processes of carcinogenesis, such as epithelial-to-mesenchymal transition (EMT) and angiogenesis, can be studied using this method. Tanaka et al. [44] used this method to visualize tumour heterogeneity. Focusing on tumour vascular patterns in UC samples and using the vessel marker CD34 in a multiparametric model, they could stage ≥ pT2 more accurately than conventional histopathology. Differences in vascular patterns in different stages of UTUC were also observed in a small pilot study on UTUC samples from RNU specimens [45]. The usefulness of this technique on small endoscopic biopsies is currently being evaluated. 3D imaging can also be used for single-cell RNA and protein expression analysis [46]. The location of cells expressing, for instance, PD-L1 can be shown and related to other structures within the tumour.
Cytokines and immunoprofile
The TNM staging system is used for the classification of cancers, but patients with similar stages may have different clinical outcomes. Recently, a new definition was developed, including the dynamic interaction between tumour cells and the immune system in all tumour stages. Different immune contexts have been associated with different clinical outcomes. Enrichment in cytotoxic T-cells, memory T-cells and B-cells is associated with prolonged survival [47]. T-cells are guided to their targets via certain chemokines and chemokine receptors. Abundance of the chemokine CXCL11 in tumour biopsies from MIBC was correlated with high numbers of tumour-infiltrating T-cells and response to neoadjuvant chemotherapy (NAC) [48]. Furthermore, the presence of a variant of the CXCL11 receptor, CXCR3alt, in combination with CXCL11 was associated with improved OS in MIBC and segregated NAC responding from non-responding patients prior to treatment. This demonstrates that immune signatures based on cytokines can reveal clinical information that is not covered by the TNM staging system [48]. This may potentially be highly relevant in UTUC since approximately 50% of UTUC patients with tumours suitable for adjuvant cisplatin-based combination chemotherapy are ineligible after RNU due to decreased renal function [49].
Extracellular vesicles
Extracellular vesicles (EVs) are exosomes (virus size) and microvesicles (bacterial size) that are excreted by cells and contain and protect proteins, mRNA and enzymes. They are probably functional elements, and the mechanism is conserved over species [50]. Cancer-derived EVs can induce angiogenesis and EMT and prepare the premetastatic niche [51]. EVs from cancer cells can inhibit the immune system, whereas EVs from dendritic cells can activate it, which opens the door for potential therapeutic strategies [50, 52, 53]. EVs are particularly interesting as biomarkers because they represent a snapshot of the status of the cell that excreted the EVs.
EVs can be identified in tissue samples or in liquid biopsies, where urine is of particular interest for UC. Both protein and micro-RNA (miRNA) profiles in EVs have shown potential as biomarkers in UCB. In a protein analysis of tissue-derived EVs in UCB, Eldh et al. found a malignant profile, with no differences in EVs between the previous tumour site and histologically normal distant bladder tissue [54]. Despite the absence of detectable tumours, the entire bladder released exosomes that may contribute to recurrence or metastasis. Hiltbrunner et al. [55] found that urine-derived EVs showed a carcinogenic metabolic profile, despite no detectable tumour. Studying urine-derived EVs from UCB patients in bladder urine (in contact with the tumour) and ureter urine (never in contact with the tumour), Eldh et al. found a UCB-specific protein signature exclusively found in bladder urine (not ureter urine) [54]. Certain EV-miRNAs have been shown to distinguish MIBC from non-muscle-invasive bladder cancer, where urine-derived EVs may serve as a source for miRNA analysis [56]. This needs to be further evaluated in a UTUC setting.
Discussion
As for other urological malignancies, modern management of UTUC has moved towards targeted treatments based on tumour risk classification, patient comorbidities, life expectancy and patient preferences. This is a delicate balance: not overtreating nor taking oncological risks. The key areas for improvement in the diagnostic workup of UTUC is to differentiate the true low-risk tumours from the true high-risk ones and to do so prior to treatment. Ideally, this test should also be as non-invasive as possible. Many diagnostic and prognostic tests for UTUC follow the same pattern of development: First the test is evaluated for the more common and more easily accessible UCB, and then for tumour samples from RNU specimens. RNU specimens form an important step, as they present the special features of UTUC, in combination with a reference standard for diagnosis and staging, which the test is to evaluate. The next step is to evaluate the test in URS samples and liquid biopsies. The methods described here are not yet available for commercial use and are in different stages of this development.
Analysis of the gene mutational profile of tumour tissue and liquid biopsies is a growing field, and much work have been done on both UCB and UTUC. Genetic markers have been identified and analyzed in different sample types (RNU specimens, URS biopsies, liquid biopsies) [25, 26, 29, 31, 37] and have been studied both in a pre-treatment diagnostic/prognostic setting and a monitoring setting [27, 38, 39]. For UTUC no single genetic marker has yet been identified, but studying patterns of gene mutations or gene expression seems to be the way forward. The distinction of relevant findings from artefacts remains challenging [35, 39]. Gene sequencing is getting more available and financially affordable. To study pathway proteins is a way to study the downstream effects of gene aberrations and their effects with a highly sensitive method. Work has been done on other malignancies [42, 43], for UTUC there are unpublished test-of-concept studies. However, the role of pathway proteins in UTUC needs further evaluation.
3D microscopy is a new methodology, but specimen dye and preparation are in some ways like conventional histopathology. An advantage is that the analysis is done by computer measurements and hence quantifiable. 3D microscopy has been evaluated for TURB-specimens for UCB and tumor biopsies from RNU-specimens for UTUC. Method development for URS biopsies is ongoing but not yet published.
Analysis of EVs has been studied for UCB and studies suggest that this method may be used to pin-point the location of UC in the urinary tract [54]. Naturally, this must be evaluated in an UTUC-setting. There are FDA-approved commercially available liquid biopsy exosome diagnostics of urine to detect prostate cancer, so a similar test could hypothetically be developed for UTUC, given that further studies identify what EV characteristics to look for regarding UTUC.
Analysis of chemokines and chemokine receptors has been linked to invasive UC and responsiveness to NAC [48]. If applicable also to UTUC, it would be beneficial for patients with aggressive UTUC since the opportunity to receive adjuvant cisplatin-based combination chemotherapy was lost after RNU in 49% of patients with high-risk UTUC [49]. Although no RCTs have been published, NAC has shown significant downstaging and survival benefits in UTUC patients compared to RNU alone [57]. The combined understanding of the genome, transcriptome, proteome and immune contexture of UTUC may help to identify which patients will benefit from adjuvant treatments in the future.
Most likely, none of the discussed techniques can entirely replace current tests, but they may be complementary and used as add-ons for specific questions in the diagnostic work-up and for prognostication, given that they reach clinical use. Since none of the tests are yet in clinical use, we refrain from analysis of cost and required expertise.
An arguable limitation of this paper is that we used no predefined search criteria. The rationale for this is partly because the involved fields are vastly different, and partly since the knowledge is scarce. This methodology potentially results in an incomplete coverage of the entire research field. We did not aspire to be all-covering but rather to explore the field of some upcoming techniques, to inspire further research on this topic.
Conclusions
Accurate risk stratification of UTUC before selecting treatment remains challenging but is crucial for personalized treatment and follow-up. At the Consultation on UTUC II in Stockholm 2022, upcoming, not yet clinically available methods for the diagnosis and prognostication of UTUC were discussed. The methods have different strengths and do in some cases complement each other. Most methods have first been studied on UCB, and it is important to keep in mind that UTUC has biological differences from UCB. All methods may not be adaptable or applicable to the specific characteristics of UTUC. Furthermore, the methods presented in this paper are in different stages of development and have different time spans to clinical application.
References
Almas B, Halvorsen OJ, Johannesen TB, Beisland C (2021) Higher than expected and significantly increasing incidence of upper tract urothelial carcinoma. A population based study. World J Urol 39:3385–3391
van Doeveren T, van der Mark M, van Leeuwen PJ, Boormans JL, Aben KKH (2021) Rising incidence rates and unaltered survival rates for primary upper urinary tract urothelial carcinoma: a Dutch population-based study from 1993 to 2017. BJU Int 128(3):343–351
Raman JD, Jafri SM (2016) Complications following radical nephroureterectomy. Curr Urol Rep 17(5):36
de la Rosette J, Denstedt J, Geavlete P, Keeley F, Matsuda T, Pearle M et al (2014) The clinical research office of the endourological society ureteroscopy global study: indications, complications, and outcomes in 11,885 patients. J Endourol 28(2):131–139
Cutress ML, Stewart GD, Wells-Cole S, Phipps S, Thomas BG, Tolley DA (2012) Long-term endoscopic management of upper tract urothelial carcinoma: 20-year single-centre experience. BJU Int 110(11):1608–1617
Guo RQ, Hong P, Xiong GY, Zhang L, Fang D, Li XS et al (2018) Impact of ureteroscopy before radical nephroureterectomy for upper tract urothelial carcinomas on oncological outcomes: a meta-analysis. BJU Int 121(2):184–193
Nowak Ł, Krajewski W, Chorbińska J, Kiełb P, Sut M, Moschini M et al (2021) The impact of diagnostic ureteroscopy prior to radical nephroureterectomy on oncological outcomes in patients with upper tract urothelial carcinoma: a comprehensive systematic review and meta-analysis. J Clin Med 10(18):4197
Grasso M, Fishman AI, Cohen J, Alexander B (2012) Ureteroscopic and extirpative treatment of upper urinary tract urothelial carcinoma: a 15-year comprehensive review of 160 consecutive patients. BJU Int 110(11):1618–1626
Gadzinski AJ, Roberts WW, Faerber GJ, Wolf JS Jr (2010) Long-term outcomes of nephroureterectomy versus endoscopic management for upper tract urothelial carcinoma. J Urol 183(6):2148–2153
Daneshmand S, Quek ML, Huffman JL (2003) Endoscopic management of upper urinary tract transitional cell carcinoma: long-term experience. Cancer 98(1):55–60
Fojecki G, Magnusson A, Traxer O, Baard J, Osther PJS, Jaremko G et al (2019) Consultation on UTUC, Stockholm 2018 aspects of diagnosis of upper tract urothelial carcinoma. World J Urol 37(11):2271–2278
Subiela JD, Territo A, Mercade A, Balana J, Aumatell J, Calderon J et al (2020) Diagnostic accuracy of ureteroscopic biopsy in predicting stage and grade at final pathology in upper tract urothelial carcinoma: Systematic review and meta-analysis. Eur J Surg Oncol 46(11):1989–1997
Messer J, Shariat SF, Brien JC, Herman MP, Ng CK, Scherr DS et al (2011) Urinary cytology has a poor performance for predicting invasive or high-grade upper-tract urothelial carcinoma. BJU Int 108(5):701–705
Malm C, Grahn A, Jaremko G, Tribukait B, Brehmer M (2017) Diagnostic accuracy of upper tract urothelial carcinoma: how samples are collected matters. Scand J Urol 51(2):137–145
Holmäng S, Johansson SL (2005) Urothelial carcinoma of the upper urinary tract: comparison between the WHO/ISUP 1998 consensus classification and WHO 1999 classification system. Urology 66(2):274–278
van der Kwast T, Liedberg F, Black PC, Kamat A, van Rhijn BWG, Algaba F et al (2021) International society of urological pathology expert opinion on grading of urothelial carcinoma. Eur Urol Focus. 8:438–446
van Rhijn BWG, Hentschel AE, Brundl J, Comperat EM, Hernandez V, Capoun O et al (2021) Prognostic Value of the WHO1973 and WHO2004/2016 classification systems for grade in primary Ta/T1 Non-muscle-invasive Bladder Cancer: a Multicenter European Association of Urology Non-muscle-invasive Bladder Cancer Guidelines Panel Study. Eur Urol Oncol 4(2):182–191
Rouprêt M, Burger M, Compérat E, Gontero P, Liedberg F, Masson-Lecomte A, Mostafid AH, Palou J, van Rhijn BWG, Sylvester R, Patient Advocates: Benedicte Gurses I, Wood R, Guidelines Associates: Capoun O, Cohen D, Dominguez-Escrig JL, Seisen T, Soukup V (2022) EAU Guidelines on Upper Tract Urothelial Carcinoma 2022
Rosiello G, Palumbo C, Knipper S, Pecoraro A, Luzzago S, Deuker M et al (2020) Contemporary conditional cancer-specific survival after radical nephroureterectomy in patients with nonmetastatic urothelial carcinoma of upper urinary tract. J Surg Oncol 121(7):1154–1161
Malm C, Grahn A, Jaremko G, Tribukait B, Brehmer M (2019) Predicting invasiveness and disease-specific survival in upper tract urothelial carcinoma: identifying relevant clinical tumour characteristics. World J Urol 37(11):2335–2342
Foerster B, Abufaraj M, Matin SF, Azizi M, Gupta M, Li WM et al (2021) Pretreatment risk stratification for endoscopic kidney-sparing surgery in upper tract urothelial carcinoma: an international collaborative study. Eur Urol 80(4):507–515
Katayama S, Mori K, Schuettfort VM, Pradere B, Mostafaei H, Quhal F et al (2022) Accuracy and clinical utility of a tumor grade- and stage-based predictive model in localized upper tract urothelial carcinoma. Eur Urol Focus 8(3):761–768
Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD et al (2017) Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171(3):540–56 e25
Kamoun A, de Reynies A, Allory Y, Sjodahl G, Robertson AG, Seiler R et al (2020) A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol 77(4):420–433
Sfakianos JP, Cha EK, Iyer G, Scott SN, Zabor EC, Shah RH et al (2015) Genomic characterization of upper tract urothelial carcinoma. Eur Urol 68(6):970–977
Moss TJ, Qi Y, Xi L, Peng B, Kim TB, Ezzedine NE et al (2017) Comprehensive genomic characterization of upper tract urothelial carcinoma. Eur Urol 72(4):641–649
Bagrodia A, Cha EK, Sfakianos JP, Zabor EC, Bochner BH, Al-Ahmadie HA et al (2016) Genomic biomarkers for the prediction of stage and prognosis of upper tract urothelial carcinoma. J Urol 195(6):1684–1689
Grahn A, Eisfeldt J, Malm C, ForoughiAsl H, Jaremko G, Tham E et al (2021) Genomic profile—a possible diagnostic and prognostic marker in upper tract urothelial carcinoma. BJU Int 79:S1072
Bagrodia A, Audenet F, Pietzak EJ, Kim K, Murray KS, Cha EK et al (2018) Genomic profile of urothelial carcinoma of the upper tract from ureteroscopic biopsy: feasibility and validation using matched radical nephroureterectomy specimens. Eur Urol Focus. 5:365–368
Robinson BD, Vlachostergios PJ, Bhinder B, Liu W, Li K, Moss TJ et al (2019) Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat Commun 10(1):2977
Fujii Y, Sato Y, Suzuki H, Kakiuchi N, Yoshizato T, Lenis AT et al (2021) Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer Cell 39(6):793-809 e8
Seiler R, Ashab HAD, Erho N, van Rhijn BWG, Winters B, Douglas J et al (2017) Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol 72(4):544–554
Liu D, Plimack ER, Hoffman-Censits J, Garraway LA, Bellmunt J, Van Allen E et al (2016) Clinical validation of chemotherapy response biomarker ERCC2 in muscle-invasive urothelial bladder carcinoma. JAMA Oncol 2(8):1094–1096
Heitzer E, Haque IS, Roberts CES, Speicher MR (2019) Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 20(2):71–88
Barbany G, Arthur C, Lieden A, Nordenskjold M, Rosenquist R, Tesi B et al (2019) Cell-free tumour DNA testing for early detection of cancer—a potential future tool. J Intern Med 286(2):118–136
Green EA, Li R, Albiges L, Choueiri TK, Freedman M, Pal S et al (2021) Clinical utility of cell-free and circulating tumor DNA in kidney and bladder cancer: a critical review of current literature. Eur Urol Oncol 4(6):893–903
Hayashi Y, Fujita K, Matsuzaki K, Matsushita M, Kawamura N, Koh Y et al (2019) Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci 110(5):1771–1779
Blumendeller C, Boehme J, Frick M, Schulze M, Rinckleb A, Kyzirakos C et al (2021) Use of plasma ctDNA as a potential biomarker for longitudinal monitoring of a patient with metastatic high-risk upper tract urothelial carcinoma receiving pembrolizumab and personalized neoepitope-derived multipeptide vaccinations: a case report. J Immunother Cancer. 9(1):e001406
Nakano K, Koh Y, Yamamichi G, Yumiba S, Tomiyama E, Matsushita M et al (2022) Perioperative circulating tumor DNA enables the identification of patients with poor prognosis in upper tract urothelial carcinoma. Cancer Sci 113(5):1830–1842
Orofiamma LA, Vural D, Antonescu CN (2022) Control of cell metabolism by the epidermal growth factor receptor. Biochim Biophys Acta Mol Cell Res 1869(12):119359
Hashimoto M, Fujita K, Tomiyama E, Fujimoto S, Adomi S, Banno E et al (2023) Immunohistochemical analysis of HER2, EGFR, and Nectin-4 expression in upper urinary tract urothelial carcinoma. Anticancer Res 43(1):167–174
Kjaer IM, Olsen DA, Brandslund I, Bechmann T, Jakobsen EH, Bogh SB et al (2020) Prognostic impact of serum levels of EGFR and EGFR ligands in early-stage breast cancer. Sci Rep 10(1):16558
Olsen DA, Thomsen CEB, Andersen RF, Madsen JS, Jakobsen A, Brandslund I (2020) Decreased concentrations of intracellular signaling proteins in colon cancer patients with BRAF mutations. Sci Rep 10(1):20113
Tanaka N, Kanatani S, Tomer R, Sahlgren C, Kronqvist P, Kaczynska D et al (2017) Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat Biomed Eng 1(10):796–806
Grahn A, Tanaka N, Uhlen P, Brehmer M (2019) Volumetric imaging: a potential tool to stage upper tract urothelial carcinoma. World J Urol 37(11):2297–2302
Tanaka N, Kanatani S, Kaczynska D, Fukumoto K, Louhivuori L, Mizutani T et al (2020) Three-dimensional single-cell imaging for the analysis of RNA and protein expression in intact tumour biopsies. Nat Biomed Eng 4(9):875–888
Bruni D, Angell HK, Galon J (2020) The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 20(11):662–680
Vollmer T, Schlickeiser S, Amini L, Schulenberg S, Wendering DJ, Banday V et al (2021) The intratumoral CXCR3 chemokine system is predictive of chemotherapy response in human bladder cancer. Sci Transl Med. https://doi.org/10.1126/scitranslmed.abb3735
Lane BR, Smith AK, Larson BT, Gong MC, Campbell SC, Raghavan D et al (2010) Chronic kidney disease after nephroureterectomy for upper tract urothelial carcinoma and implications for the administration of perioperative chemotherapy. Cancer 116(12):2967–2973
Veerman RE, GuclulerAkpinar G, Eldh M, Gabrielsson S (2019) Immune cell-derived extracellular vesicles—functions and therapeutic applications. Trends Mol Med 25(5):382–394
Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G et al (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18(6):883–891
Liu J, Wu S, Zheng X, Zheng P, Fu Y, Wu C et al (2020) Immune suppressed tumor microenvironment by exosomes derived from gastric cancer cells via modulating immune functions. Sci Rep 10(1):14749
Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D et al (2016) Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 5(4):e1071008
Eldh M, Mints M, Hiltbrunner S, Ladjevardi S, Alamdari F, Johansson M et al (2021) Proteomic profiling of tissue exosomes indicates continuous release of malignant exosomes in urinary bladder cancer patients, even with pathologically undetectable tumour. Cancers (Basel). 13(13):3242
Hiltbrunner S, Mints M, Eldh M, Rosenblatt R, Holmstrom B, Alamdari F et al (2020) Urinary exosomes from bladder cancer patients show a residual cancer phenotype despite complete pathological downstaging. Sci Rep 10(1):5960
Baumgart S, Meschkat P, Edelmann P, Heinzelmann J, Pryalukhin A, Bohle R et al (2019) MicroRNAs in tumor samples and urinary extracellular vesicles as a putative diagnostic tool for muscle-invasive bladder cancer. J Cancer Res Clin Oncol 145(11):2725–2736
Leow JJ, Chong YL, Chang SL, Valderrama BP, Powles T, Bellmunt J (2020) Neoadjuvant and adjuvant chemotherapy for upper tract urothelial carcinoma: a 2020 systematic review and meta-analysis, and future perspectives on systemic therapy. Eur Urol. 79:635–654
Acknowledgements
We are grateful to Boston Scientific, Apsu Medical, Olympus, Karl Storz Endoscope, Micron Med., Coloplast, BL Medical, Rocamed and and Stiftelsen Japanese-Swedish Research Foundation for supporting the Stockholm Consultation on UTUC meeting. We thank chief assistant Susanna Hertzberg and Stockholm Data Design for outstanding administrative support.
Funding
Open access funding provided by Karolinska Institute.
Author information
Authors and Affiliations
Contributions
AG: manuscript writing and editing. JAC: data collection and analysis, presentation, manuscript editing. SG: data collection and analysis, presentation, manuscript editing. JSM: data collection and analysis, presentation, manuscript editing. ET: data collection and analysis, presentation, manuscript editing. PU: data collection and analysis, presentation, manuscript editing. TV: data collection and analysis, presentation, manuscript editing. YE: manuscript editing. KT: manuscript editing. BT: manuscript editing. KZ: manuscript editing. PJSO: project co-ordination, editing of manuscript. MB: project co-ordination, collection and analysis of data, presentation, writing and editing of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Grahn: None. Eriksson: None. Gabrielsson: Scientific Advisory Board, Anjarium Biosciences and patent granted for Exosomes for targeting B-cells for therapy. Skov Madsen: None. Tham: None. Thomas: None. Uhlén: None. Vollmer: None. Zieger: None. Osther: Consultation for Olympus, speaker for Coloplast, Boston Scientific, Olympus. Brehmer: Consultation for Boston Scientific.
Ethical approval
This is a review article involving no studies on human participants and/or animals. Informed consent is not relevant.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Grahn, A., Coleman, J.A., Eriksson, Y. et al. Consultation on UTUC II Stockholm 2022: diagnostic and prognostic methods—what’s around the corner?. World J Urol 41, 3405–3411 (2023). https://doi.org/10.1007/s00345-023-04597-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00345-023-04597-4