Abstract
Salicylic acid (SA) plays a crucial role not only in defence against pathogen attacks, but also in abiotic stress responses. Recently, some key steps of SA signalling outlined the importance of redox state-dependent processes. This study explores the role of glutathione transferases (GSTs) in the transcriptional reprogramming of redox status-related genes in seven-day-old wild type and Atgst mutant Arabidopsis thaliana plants. The timing of redox changes, detected by the redox-sensitive green fluorescent protein (roGFP2), differed in wild type roots treated with 10 μM or 100 μM SA. Our results verified how the applied SA concentrations had different effect on the expression of oxidative stress- and redox-related genes, among them on the expression of AtGSTF8 and AtGSTU19 genes. Lower vitality and less negative EGSH values were specific characteristics of the Atgst mutants compared to the wild type plants throughout the experiment. Changes in the redox potential were only modest in the mutants after SA treatments. A slightly modified gene expression pattern was observed in control conditions and after 1 h of SA treatments in Atgst mutants compared to Col-0 roots. These data originating from the whole roots provide indirect evidence for the role of the investigated AtGSTF8 and AtGSTU19 isoenzymes in the transduction of the redox signal. Our results demonstrate that the investigated Arabidopsis GSTs have a role in maintaining the levels of reactive oxygen species- and redox homeostasis and are involved in transcriptional reprogramming in the roots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The phenolic compound salicylic acid (SA) is a plant hormone, which is involved in the complex network of growth responses, affected by developmental and environmental signals, that also regulate various physiological and biochemical processes in plants (Rivas-San Vicente and Plasencia 2011). One of its well-known effects is the induction of pathogenesis-related proteins and systemic acquired resistance (Uknes et al. 1992; Vlot et al. 2009). Immune reactions mediated by SA are regulated by nonexpressors of pathogenesis-related genes (NPR1-4) in Arabidopsis (Ding and Ding 2020). Exploring the NPR1 signalling mechanisms revealed its action requires upstream regulatory events, such as the changes in cellular redox status (Pokotylo et al. 2019). Normally, NPR1 is in an inactive state and forms oxidized oligomeric complexes in the cytosol via intermolecular disulphide bonds. Infection initiates a rapid change in the ion flux and in the levels of reactive oxygen species (ROS), leading to redox-dependent monomerization and translocation of NPR1 to the nucleus (Mou et al. 2003; Tada et al. 2008). NPRs and other transcription factors, such as TGA (TGACG motif-binding protein) or TGA box- and as-1-like elements containing proteins involved in SA signal transduction, are considered redox sensors for temporal control of gene expression (Blanco et al. 2009).
SA contributes to both plant protection against pathogen attack, and SA-mediated abiotic stress responses. Furthermore, its developmental role under different stresses and normal conditions has been recently highlighted (Miura and Tada 2014; Pasternak et al. 2019). Low concentrations of exogenous SA promote plant growth, but high concentrations of exogenous and endogenous SA have an inhibitory effect. However, the boundaries of these concentrations differ among plant species (Jirage et al. 2001; Tari et al. 2002; Horváth et al. 2015a; Pasternak et al. 2019). SA also influences root meristem structure and root system architecture in Arabidopsis in a concentration-dependent manner (Pasternak et al. 2005, 2019; Armengot et al. 2014; Horváth et al. 2015a; Zhao et al. 2015). Although the function of plant hormones and several transcription factors in the formation of root meristem has previously been described, the regulatory roles of ROS and redox status have only emerged in recent years. The central role of ROS in the gene regulatory network in root development was demonstrated by Mase and Tsukagoshi (2021). Plants are constantly subjected to different stresses that significantly increase ROS levels, leading to redox imbalance (Hasanuzzaman et al. 2020).
Jiang and coworkers explored the redox profile of Arabidopsis root using cytoplasmic redox-sensing green fluorescent protein (c-roGFP1). They revealed a shift in redox potential in the longitudinal zones of root tips (Jiang et al. 2016). More oxidizing redox status and higher ROS levels were detected in the roots of Arabidopsis glutathione transferase mutants (Atgstf8 and Atgstu19) compared to the wild types, while the size of the mutants’ meristem was shorter (Horváth et al. 2019).
In higher plants, glutathione transferases (GSTs) are grouped into ten classes, of which four (tau, phi, dehydroascorbate reductase, and lambda) are specific to plants. A widely accepted concept states that this protein superfamily evolved to mitigate the effects of oxidative stress, but distinct classes participate in partially specific reactions. Using glutathione (GSH) as a cosubstrate, specifically the most abundant tau (GSTU) and phi (GSTF) isoenzymes may diminish the GSH pool, thereby influencing various redox-dependent processes (Marrs 1996; Cummins et al. 2011). GSTs can also bind to different hormones such as auxins and cytokinins at their ligand-binding sites (Moons 2005). Moreover, several GSTs have been identified among SA-binding proteins (Tian et al. 2012; Manohar et al. 2015). SA alters the enzymatic activity of Arabidopsis GSTF8, GSTF9, GSTF10, and GSTF11 (Tian et al. 2012). Since it was suggested that many of SA's effects are mediated through SA-binding proteins whose enzymatic activities are altered by the binding of SA (Manohar et al. 2015), these proteins may deserve special attention. The expression of AtGSTF8 (At2g47730) is induced by short-term treatments with SA (Sappl et al. 2004; Uquillas et al. 2004), ethylene, and H2O2 (Wagner et al. 2002) and was proposed to be a marker gene for early stress and defence responses (Thatcher et al. 2015). AtGSTF8 isoenzyme is mainly localized in the cytosol (the other splice variant contains a chloroplast-targeting peptide signal) and, similarly to AtGSTU19, can use a wide range of substrates during GSH conjugation activity (Thatcher et al. 2007; Dixon et al. 2009; Krajewski et al. 2013). Under control conditions, AtGSTF8 and AtGSTU19 (At1g78380) are expressed almost in the whole Arabidopsis plant, but especially in specific parts and tissues of the root, such as the stele, endodermis, epidermal atrichoblasts, lateral root cap, and root hair zone (Dixon et al. 2010). The cytosolic AtGSTU19 is one of the most abundant GST isoenzymes involved in the stress- and hormonal responses of Arabidopsis. Its accumulation can be detected after SA treatment together with AtGSTF8, AtGSTF7, and AtGSTU24 proteins (Wagner et al. 2002; Sappl et al. 2004, 2009; Dixon et al. 2009; Xu et al. 2016). AtGSTU19 expression is induced by biotic stress (Wagner et al. 2002; Otulak-Koziel et al. 2022), NaCl, SA, and H2O2 as well (Sappl et al. 2009; Horváth et al. 2015a, 2019, 2020). The overexpression of the AtGSTU19 gene increased abiotic stress tolerance by strengthening ROS scavenging activity or maintaining ROS homeostasis by the elevation of antioxidant enzyme activities (Xu et al. 2016). Due to their GSH-conjugating activity, GSTs are pivotal in the cellular detoxification processes, and via their glutathione peroxidase or dehydroascorbate reductase (DHAR) activities, they can influence the redox status of GSH and ascorbic acid.
As an antioxidant, GSH is involved in ROS processing and limits the lifetime of oxidative signals (Diaz-Vivancos et al. 2015). GSH can also regulate the expression of genes by modulating the redox state of proteins and transcription factors, but this mechanism is yet poorly understood (Dietz 2014; He et al. 2018). Thiol redox biochemistry plays a crucial role in cellular processes, thus preserving the GSH pool, and the relatively high levels of reduced glutathione compared to the oxidized glutathione levels (GSH: GSSG ratio) are essential factors. The maintenance of a highly negative redox potential of glutathione (around—310 mV) is achieved through continuous reduction of glutathione disulphide by glutathione reductase (GR) (Aller et al. 2013). A useful tool for imaging thiol redox potential is a redox-sensitive green fluorescent protein (roGFP2) expressed in plants, which allows monitoring the redox status in selected parts via microscopy (Meyer et al. 2007; Schwarzländer et al. 2008).
In previous experiments, we demonstrated that supplementing the root media of five-week-old Arabidopsis plants with 10−6 M and 10−5 M SA for two weeks alleviated the adverse effects of one-week-long NaCl stress (Horváth et al. 2015a). Interestingly, due to increased antioxidant activities, H2O2 and lipid peroxidation marker malondialdehyde levels were similar to those measured in untreated controls after three weeks of treatment. The activity of superoxide dismutase, guaiacol peroxidase, glutathione peroxidase, GST, and GR enzymes was elevated. Among the investigated GST-coding genes, the transcript levels of AtGSTU19 and AtGSTU24 increased most at the end of the long-term experiments (Horváth et al. 2015a). Further studies showed that 10−5 M SA treatment caused a temporary shift in redox potential towards a less negative redox state, but after two and three weeks, it was close to untreated controls (Csiszár et al. 2018).
It is well documented that high endogenous or exogenous SA concentrations induce ROS accumulation, but less information is available about the changes in the redox state during exogenously applied lower SA concentrations and its effects on plant redox status. In this paper, we applied the cytosolic roGFP2 redox probe to monitor the changes in different root regions after applying 10 and 100 μM of SA. We aimed to investigate the relationships between ROS, redox status, and salicylic acid-induced transcriptional reprogramming. The potential roles of AtGSTF8 and AtGSTU19 as redox transducers in these processes were investigated by introducing Atgstf8 and Atgstu19 mutants into our experiments, which have elevated ROS levels and altered redox potentials in their roots compared to the wild type (Horváth et al. 2019).
Materials and Methods
Plant Material and Growth Conditions
We used the plant lines from our previous study (Horváth et al. 2019), namely the Columbia (Col-0) ecotype of Arabidopsis thaliana (L.) as a wild type control and Atgstf8 (At2g47730) and Atgstu19 (At1g78380) T-DNA insertional lines. The seedlings were grown under controlled conditions (100 μmol m−2 s−1 photon flux density, 12/12 day/night period, 70% relative humidity, 21 ℃) on solid half-strength Murashige and Skoog medium (½ MS, Duchefa Biochemie; Murashige and Skoog 1962). The seedlings were treated with SA in liquid medium, as described previously (Horváth et al. 2019). Briefly, seven-day-old Arabidopsis plants were placed into a liquid medium containing 10 μM or 100 μM of SA. The treatment solutions were uniformly adjusted to a pH of 5.6 before use. Although some changes were observed in ROS levels and in root vitality even after 30 min (data not shown), but fluorescent microscopic and gene expression analyses were performed after 1, 3, and 24 h long treatments. The experiments were performed in at least two repetitions and ten replicates (n = 10). The statistical analysis was performed as described previously (Horváth et al. 2019).
Detection of Cell Vitality, Superoxide Radical, and H2O2 Levels in Roots Using Fluorescent Microscopy
Fluorescent staining of roots was performed as described previously (Horváth et al. 2019). For cell vitality determination, seedlings were placed in fluorescein diacetate (FDA) solution (10 μM FDA in 10 mM/50 mM MES/KCl buffer, pH 6.15) for 15 min and washed with buffer. FDA is a cell membrane permeable substrate for non-specific esterases, widely used as a probe for vitality. By using FDA, the intracellular esterase activity which is required to activate its fluorescence and membrane integrity to retain the fluorescent probe inside the cell, was measured according to McCabe and Leaver (2000) and Feigl et al. (2019). By using FDA and propidium iodide (PI) double staining (Ambastha et al. 2020), we demonstrated that similar conclusion could be drawn related to the vitality of the cells from the two methods (data are not shown). Superoxide radical (O2•−) was visualized with dihydroethidium (DHE) by placing the seedlings in DHE solution (10 μM DHE in 10 mM, Tris–HCl buffer, pH 7.4) for 30 min at 37 °C. Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) dye was used for hydrogen peroxide (H2O2) detection since it reacts with H2O2 in the presence of horseradish peroxidase to produce the fluorescent compound resorufin (reviewed in Gomes et al. 2005). Seedlings were incubated in Amplex Red solution (50 μM Amplex Red prepared in 50 mM sodium phosphate buffer, pH 7.5) for 30 min. Measurements were performed with the same microscope and settings as in our previous study (Horváth et al. 2019). Microscopic studies were carried out using a Zeiss Axiowert 200 M microscope (Carl Zeiss, Jena, Germany) equipped with a high-resolution digital camera (Axiocam HR, HQ CCD, Carl Zeiss, Jena, Germany) and filter set 10 (exc.: 535–585 nm, em.: 600–655 nm) for FDA, filter set 9 (exc.: 450–490 nm, em.: 515–∞ nm) for DHE, or filter set 20HE (exc.: 546/12, em.: 607/80) for Amplex Red was used. The intensity of fluorescence was quantified using Axiovision Rel. 4.8 software in the proximal meristem (PM), transitional, and elongation zones (TZ and EZ, respectively) in a circle with 50 μm radius. The zones were specified according to Jiang et al. (2016). PM was defined as cells between the quiescent centre and the first isodiametric cortical cell. Isodiametric cortical cells constitute the TZ, and EZ is originated from cells which lose their isodiametric shape and elongate (Verbelen et al. 2006; Perilli and Sabatini 2010; Jiang et al. 2016). Measurement sites of fluorescent staining are shown in Supplementary Figure S1.
Redox Potential Determination by Ratiometric Measurements of GRX1-roGFP2
Fluorescence measurements were performed as described in Horváth et al. (2019), using a confocal laser scanning microscope (Olympus Fluoview FV1000, Olympus Life Science Europe GmbH, Hamburg, Germany) with 405 and 485 nm excitation wavelengths. Fluorescence was detected between 505 and 530 nm emission wavelengths. Each experiment included a calibration with 5 mM Aldrithiol™ − 4 and 10 mM dithiothreitol to fully oxidize and reduce the roGFP2, respectively. Redox potential was calculated as described by Schwarzländer et al. (2008):
where the midpoint potential of roGFP2 (E0roGFP2) is − 272 mV at 30 °C and pH 7; the gas constant (R) is 8.315 J K−1 mol−1; the absolute temperature (T) is 298.15 K; the number of transferred electrons (z) is 2; the Faraday constant (F) is 96,485 C mol−1. The degree of oxidation (OxD) of roGFP2 (OxDroGFP2 values) and the equation for OxDroGFP2 calculation can be found in Supplementary Table S1. The sites of redox potential analysis performed by the confocal laser scanning microscope are shown in Supplementary Figure S1.
High-Throughput Quantitative Real-Time Polymerase Chain Reaction (PCR) Measurements
High-throughput quantitative real-time PCR (HT-qPCR, Avidin Ltd.) assays were used to detect expression of the selected Arabidopsis thaliana GST- and other stress-related genes in the roots of seedlings. Roots were collected from approximately 120 seedlings using a scalpel to separate roots from shoots. RNA purification from 50 mg of root tissues was performed according to the manufacturer’s instructions using Quick-RNA Miniprep Kit (Zymo Research). Additional DNase digestion (Thermo Scientific) and purification was applied (RNA Clean and Concentrator-25 Kit, Zymo Research), and cDNA was synthesized from 1 µg of total RNA using RevertAid reverse transcriptase (Thermo Scientific) and random hexamers. For HT-qPCR analysis, 22 gene sequences were selected. The selected genes for gene expression analysis and primer sequences are described in the Supplementary material (Supplementary Tables S2 and S3). A single product-specific melting temperature peak was found in every case following melting curve analysis. Data were normalized to the expression levels of glyceraldehyde-3-phosphate dehydrogenase C2 (GAPDH2, At1g13440) and monensin sensitivity1, a SAND family protein (MON1, At2g28390) genes since these genes showed constant expression in our experiments (Czechowski et al. 2005; Papdi et al. 2008). The relative transcript levels in Arabidopsis thaliana wild type control root samples were taken as 1, and 2−∆∆Ct values were presented in the diagrams.
Results
The Vitality of Root Zones Showed a Rapid, Transitional Decrease to SA Treatments
We investigated the vitality of one-week-old seedlings to analyse how exogenously applied SA at different doses (10 or 100 µM of SA) affects Arabidopsis roots Fluorescence intensity measured in the proximal meristem (PM), transition zone (TZ), and elongation zone (EZ) by FDA staining revealed SA-induced concentration- and time-dependent changes in both wild type and mutant plants (Fig. 1).
Vitality in the proximal meristem (PM), transition- (TZ), and elongation zones (EZ) of seven-day-old Arabidopsis thaliana Col-0, Atgstf8, and Atgstu19 mutants after 1, 3, and 24 h of treatment with 10 or 100 μM of salicylic acid. Data are means ± SE, n = 10. Columns with different letters are significantly different at p ≤ 0.05, determined by Duncan’s test
In Col-0 plants, 1 h of lower, 10 µM of SA treatments slightly reduced vitality (Figs. 1 and 2). A further decrease in FDA fluorescence was measured in EZ after 24 h but not in PM and TZ. The 100 µM of SA treatment reduced vitality after 1 h, followed by an additional decrease after 3 h. Not only time-, but also root zone-dependent changes were observed. Wild type roots had higher FDA fluorescence in PM and TZ after 24 h of 100 µM of SA treatments, but EZ showed even lower vitality than after 3 h of treatments compared to the untreated root regions (Fig. 1).
Representative images of fluorescent analysis of vitality and redox status in seven-day-old Arabidopsis thaliana Col-0, Atgstf8, and Atgstu19 insertional mutants after 1 h of treatment with 10 and 100 μM of salicylic acid (see more details in Materials and methods). The redox status is represented as the ratio of fluorescence intensities (I405/I488) based on the corresponding colour scale, where black indicates a lower I405/I488 ratio (reduced), and white colour indicates a higher I405/I488 ratio (oxidized). I488: the intensity at 488 nm; I405: the intensity at 405 nm. Scale bar = 100 μm (Color figure online)
The vitality of Atgstf8 mutants was similar to Col-0 plants, but Atgstu19 mutants had a lower FDA fluorescence under control conditions for almost all time periods and root zones throughout the experiments. Ten μM of SA decreased the vitality of EZ in Atgstf8 and Atgstu19 mutants after 3 h and in TZ and EZ after 24 h of treatment. All mutants showed lower vitality after 1 h of 100 μM of SA treatments compared to their own controls, and further reductions were measured after 3 h. Similarly to Col-0 plants, we observed recovered vitality after 24 h in the PM and TZ of Atgstf8 roots but not in the Atgstu19 mutants, which showed remarkably lower values (Fig. 1).
SA Treatments Caused Time- and Concentration-Dependent Changes in Redox Potential
An in vivo redox probe (roGFP) was applied to detect the redox state of different root apical meristem (RAM) zones. The redox-dependent fluorescence signals from GRX1-roGFP2-expressing Col-0, Atgstf8, and Atgstu19 mutants were monitored and compared.
The most negative redox potential (from − 305 mV to − 311 mV) was detected in control Col-0 plants (Figs. 2 and 3). The lower concentration of SA treatments (10 μM) caused a higher shift in the redox potential towards more oxidized values after 1 and 3 h of treatment in contrast with the effects of 100 μM of SA in wild type plants. However, glutathione redox potential (EGSH) of Col-0 became less negative after 24 h of 100 μM SA treatment (by 13 mV [− 292.44 mV] in PM and 8 mV [− 303.41 mV] in EZ), in comparison to 10 μM of SA treatment, which caused 11 mV (− 294.10 mV) and 6 mV (− 304.81 mV) shifts in the PM and EZ, respectively.
Calculated redox potential values of roots expressing cytosolic GRX1-roGFP2 in primary roots of seven-day-old Arabidopsis thaliana Col-0, Atgstf8, and Atgstu19 mutants. Data are means ± SE, n = 10. Columns with different letters are significantly different at p ≤ 0.05, determined by Duncan’s test. PM proximal meristem, TZ transition zone, EZ elongation zone
In general, Atgst mutants showed a less negative redox state in RAM than the untreated Col-0 plants under control conditions (by 7–30 mV) and after SA treatments (by 4–26 mV). In contrast to Col-0 plants, the redox potential was more negative in Atgst mutants after SA treatments compared to their own untreated controls; moreover, they remained more oxidized over time (Figs. 2, 3). In Atgstf8 mutants, 10 μM and 100 μM of SA-induced changes in EGSH after 1 h of treatment. These changes were less definite after 3 h. However, the redox potential values of SA-treated roots were more negative in all root zones after 24 h of treatment (by 20–24 mV after 10 μM SA and 11–16 mV after 100 μM SA, respectively) in the Atgstf8 mutant compared to their control. In Atgstu19, the most significant decrease in the redox potential was measured in the EZ after 3 h (by 6 mV and 5 mV after 10 μM and 100 μM SA, respectively) and in the PM after 24 h (by 6 mV and 11 mV after 10 μM and 100 μM SA, respectively).
Mutation in GSTs Alters the ROS Levels and Changes in Roots After SA Treatments
Since different changes were observed in the vitality and redox potential of wild type and Atgst mutants after SA treatment, a detailed analysis of ROS levels was performed. The levels of superoxide radical (O2•−) dependent DHE fluorescence were slightly higher in Atgst mutants (especially in Atgstf8 under control conditions) compared to the Col-0 plants. Superoxide radical levels remained unaltered in the roots of investigated plants after 1 h of treatment (Fig. 4; Supplementary Fig. S2). However, after SA treatments, significantly higher O2•− levels were detected in Atgst mutants compared to the Col-0 plants after 3 h (in TZ and EZ) and 24 h (in PM and TZ) of SA treatments (Fig. 4).
The superoxide radical (dihydroethidium fluorescence) levels in the proximal meristem (PM), transition- (TZ), and elongation zones (EZ) of seven-day-old Arabidopsis thaliana Col-0, Atgstf8, and Atgstu19 mutants after 1, 3, and 24 h of treatment with 10 or 100 μM of salicylic acid. Data are means ± SE, n = 10. Columns with different letters are significantly different at p ≤ 0.05, determined by Duncan’s test
SA treatments induced time- and concentration-dependent changes in the H2O2-dependent resorufin fluorescence intensities; however, some fluctuation in the H2O2 levels was observed over time. While the accumulation of H2O2 was detected in all investigated root zones of wild type plants after 1 and 24 h of SA treatment, minor deviations were detected in the SA-treated roots after 3 h. The higher (100 μM) SA treatment induced a more noticeable accumulation of H2O2 in all investigated root zones after 1 and 24 h than 10 μM of SA treatment, but the highest levels were detected in the TZ of Col-0 after 24 h. The changes detected in H2O2 levels were similar in Atgst mutants and wild type roots. However, the SA treatments resulted in even higher H2O2 levels in the roots of mutants than in wild type plants after 1 and 3 h (Fig. 5 and Supplementary Fig. S3).
The hydrogen peroxide-dependent fluorescence (resorufin fluorescence) in the proximal meristem (PM), transition- (TZ), and elongation zones (EZ) of seven-day-old Arabidopsis thaliana Col-0, Atgstf8, and Atgstu19 mutants after 1 h of treatment with 10 or 100 μM of salicylic acid. Data are means ± SE, n = 10. Columns with different letters are significantly different at p ≤ 0.05, determined by Duncan’s test
HT-qPCR Analyses Revealed Modified Expression Levels of Certain Glutathione- and Stress-Related Genes After One Hour of SA Treatment
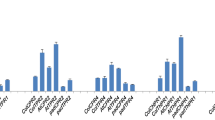
To investigate the effects of rapid changes in redox status and H2O2 levels within the roots, we analysed the expression patterns of some GSTs and other oxidative stress-inducible genes in wild type and Atgst mutants. The expression of 22 selected genes was investigated in whole roots after 1 h of 10 and 100 μM of SA treatment using high-throughput quantitative real-time PCR (HT-qPCR; Fig. 6 and Supplementary Fig. S4). More details of the selection of genes can be found in Supplementary Table S3. The expression of GSTF8 and GSTU19 was significantly lower in Atgstf8 and Atgstu19 mutants, respectively. Among the investigated genes, the highest induction was detected in WRKY38 after 1 h of treatment, similarly affected by 10 and 100 μM of SA concentration (Fig. 6). In addition, some elevation was detected in the transcript levels of WRKY46, GSTU7, GSH1, HSFA8, GSTF8, ZAT10, UPB1, NAC032, and OXI1 after applying 10 μM SA on Col-0 plants. A partially different set of genes were induced in the presence of 100 μM of SA compared to 10 μM of SA treatments. Also, higher relative transcript levels were observed at WRKY38, GSTU7, GSTU24, NAC032, GSTU19, AKR4C9, GSTU25, WRKY46, OXI1, HSFA8, GPXL6, GSTF8, RRTF1, ZAT10, and GSH1 genes compared to controls. Decreased expression was detected in the case of WRKY28 and MYC2 genes in 100 μM of SA-treated roots (Fig. 6 and Supplementary Fig. S4).
The expression rate of Arabidopsis thaliana glutathione transferase and selected stress-related genes. Relative transcript amounts of genes were determined by high-throughput quantitative RT-PCR under control conditions (C) and after 1 h under 10 and 100 μM salicylic acid treatments in wild type (Col-0) and mutant (Atgstf8 and Atgstu19) Arabidopsis roots. Expression of genes was normalized first by referencing the average transcript amount of glyceraldehyde-3-phosphate dehydrogenase C2 (GAPDH2, At1g13440) and monensin sensitivity1 (MON1, At2g28390) genes, then by the average transcript amount of each gene in wild type control plants. 2−∆∆Ct values are presented in the diagrams. Data are means ± SE and the presented data are the average of two biological replicates. Columns with different letters are significantly different at p ≤ 0.05, determined by Duncan’s test. n.s. not significant
Under control conditions, in Atgstf8 mutants, GSH1 showed slightly higher, while WRKY38 showed lower expression in contrast to the Col-0 plants. The knockdown mutation of GSTU19 resulted in moderately reduced transcript levels of GR1, GSTU24, GSTU25, and AKR4C9 genes, where only ZAT10 showed elevated expression compared to the wild type. After SA treatments, different changes were detected in the gene expression of Atgstf8 and Atgstu19 compared to Col-0 plants. The exogenous SA treatments, especially at 100 μM concentration, slightly (GSH1, GSTU7, HSFA8, and GPXL6) or significantly (GSTU24 and GSTU25) elevated the transcription of investigated genes in both mutants compared to the Col-0 roots. Furthermore, GSTU19, OXI1, and WRKY38 genes in Atgstf8 and AKR4C9, ZAT10, WRKY28, and WRKY46 in Atgstu19 showed significantly higher expression levels. Other sets of genes demonstrated lower transcript amounts in mutants compared to wild type plants after SA treatments, including WRKY46 and NAC032 in Atgstf8 and GR1 and GSTU7 in Atgstu19 (Fig. 6).
Discussion
SA is a promising natural compound which can be applied exogenously to enhance stress tolerance in plants; however, several studies have confirmed concentration-dependent opposing effects (Tari et al. 2002; Horváth et al. 2007; Szepesi et al. 2009; Rivas-San Vicente and Plasencia 2011; Pál et al. 2013; Horváth et al. 2015a, b; Szalai et al. 2016). In our previous studies, we demonstrated that 10−4 M (100 μM) SA treatment decreased the growth of five-week-old Arabidopsis, leading to the death of plants, but 10−5 M (10 μM) SA pre-treatment promoted the growth and induced hardening, alleviating the effects of subsequently applied salt treatments (Horváth et al. 2015a). Earlier results suggested that concentrations lower than 50 μM of SA have regulatory roles in the development of Arabidopsis roots, while higher concentrations act as stress hormones (Pasternak et al. 2019). However, other components and molecular mechanisms are suspected to be involved in the complex SA signalling as well. The current short-term experiments verified that exogenously applied 100 μM SA treatments decreased the vitality of seven-day-old Arabidopsis wild type roots. At the same time, 10 μM SA induced only minor changes in all investigated root zones, where vitality of the roots did not differ significantly from controls after 1 and 3 h (Figs. 1, 2).
It was reported that SA directly affects NPR1 (SA receptor and transcriptional regulator) by binding and indirectly inducing redox changes in the cells, leading to NPR1 monomerization (Ding et al. 2018; He et al. 2018). Cysteine residues are sensitive targets of H2O2, and diverse effects can be attributed to changes in their oxidized status (reviewed in He et al. 2018 and Foyer 2020). Recently, Wu et al. (2020) identified the first known cell-surface H2O2 receptor hydrogen-peroxide-induced Ca2+ increase1 (HPCA1) in plants. Similarly, for NPR proteins and interacting TGA transcription factors, the redox conditions of HPCA1 are crucial for its activation. HPCA1 kinase is activated by oxidation of two special cysteine residues in the presence of extracellular H2O2, which in turn induces Ca2+ influx in stomata and other cell types. This extracellular H2O2 can trigger Ca2+ influxes between cells and activate pathogen resistance or other stress acclimation processes in the entire plant (Evans et al. 2016). In the present study, elevated H2O2 levels were detected after 1 h of treatment with 100 μM SA in wild type roots; however, fluctuations were observed in all root zones over time, suggesting a tight control of H2O2 levels in SA response (Fig. 5). Using the roGFP2 redox probe to study the in vivo effects of SA treatments on redox status of GSH revealed that both SA concentrations resulted in a less negative redox potential in wild type Arabidopsis roots. However, the timing of these changes differed. The lower concentrations of SA (10 μM) elevated EGSH after 1 h of treatment in all root zones, but the redox potential remained more oxidized in the presence of 100 μM of SA compared to 10 μM SA. It showed further oxidation in TZ of Col-0 roots after 24 h (Figs. 2, 3). Early increments in H2O2 levels and/or redox potential values were detected after 1 h of treatment, which could cause rapid induction of GSTs, GPXL6, and oxidative stress-induced genes, such as ANAC032, AKR4C9, HSFA8, and OXI1 in 100 μM SA-treated roots (Figs. 5, 6). H2O2-induced expression of AtANAC032 and AtAKR4C9 in Arabidopsis seedlings has been previously reported by Claeys et al. (2014). The schematic model of SA-induced changes in the roots of Col-0 plants after 1 h of treatment, along with the supposed interaction between GSH redox state and H2O2 levels, supported by our results and by data found in the literature, are summarized in Fig. 7.
Schematic model summarizing the results presented in this study. Gene expression changes after 1 h of salicylic acid (SA) treatment in Col-0 plants, and their supposed regulation by SA, glutathione (GSH) redox status, and H2O2 levels are depicted in this figure. (An upward arrow indicates increase; the thickness of the arrows refers to the extent of the changes; blue arrows: 10 μM SA-induced changes, red arrows: 100 μM SA-induced changes, black arrows: both 10 and 100 μM SA treatment-induced changes) (Color figure online)
AtGSTF8 (earlier called GST6), together with AtGSTU7 (GST25), were identified as early SA response genes and characterized as genes involved in detoxification or defence against stress (Uquillas et al. 2004; Blanco et al. 2005). Moreover, the expression of AtGSTF8 is induced by a wide range of stressors, thus its expression is considered as a marker gene for early stress and defence responses. AtGSTF8 promoter fused to a luciferase reporter in forward genetic studies found mutants with altered SA responses (Sappl et al. 2009; Gleason et al. 2011). In our experiments, SA treatment induced the expression of the AtGSTF8 gene in a concentration-dependent manner. According to our results, the SA-binding function of AtGSTF8 in SA response is more important than its glutathione transferase enzyme activity.
In our previous experiments, the Atgstu19 mutants were characterized by lower vitality, higher H2O2 levels, and less negative redox state in RAM compared to wild type plants under salt stress (Horváth et al. 2019). Furthermore, Atgstu19 mutants showed lower GST activities than wild type plants. Despite this, Atgstu19 mutants can compensate the effect of the mutations by elevating the non-enzymatic antioxidant levels, a modified GSH redox status, and elevated DHAR and GR enzyme activities (Horváth et al. 2020). In the current study, we estimated the role of AtGSTU19 in early SA responses and redox-related salicylic acid-induced transcriptional reprogramming in Arabidopsis roots. Several cis-acting regulatory sequences involved in stress responses or hormone regulation can be found in the 5′ promoter region of the AtGSTU19 gene (Horváth et al. 2019), which may explain the increased expression of the AtGSTU19 gene in Col-0 plants after 1 h of SA treatment (Fig. 6). Besides the lower vitality and higher EGSH values of Atgstu19 mutants compared to wild type plants (Figs. 1, 3), several AtGSTs increased expression profiles were observed (Fig. 6), which corroborates with our earlier results about how plants compensate the effects of mutations in AtGSTU19 by activating other processes (Horváth et al. 2020).
Our results supported the role of AtGSTU19, especially in early responses to above 50 µM of SA. While lower transcript levels were observed in several redox-related genes (GR1, GSTU24, GSTU25, and AKR4C9), the expression of the abiotic stress marker gene, AtZAT10, was elevated in the roots of untreated mutants, suggesting a less negative redox state affected cell processes in control roots (Fig. 6). Several studies have shown that AtZAT10 overexpression, knockout, and RNAi mutants displayed enhanced resistance to drought-, osmotic-, and salt stresses, highlighting its dual role in response to environmental stresses (Sakamoto et al. 2004; Mittler et al. 2006). Recent studies found that AtZAT10 is part of the regulatory network responsible for jasmonic acid-induced growth control by repressing auxin biosynthesis, transport, and signalling (Sakamoto et al. 2004; Zhang et al. 2020). Although the results from the current study do not confirm the direct role of AtGSTU19 in SA perception or signalling, the modified redox processes and gene expression patterns suggested its involvement in SA responses, which may exceed its antioxidant functions. However, estimating its role in SA signalling requires further investigations.
In conclusion, this study showed that during early SA-induced responses, the vitality of the root tips decreased; however, the regained FDA fluorescence indicated a recovered vitality after 24 h of treatment in wild type Arabidopsis plants. After 1 h of SA treatment, increased H2O2 levels and GSH-dependent redox potential values were measured, demonstrating a more oxidized redox environment in the cells. These changes led to altered gene expression in several cases. Investigating the role of AtGSTs in the SA response of the roots by studying roGFP2-expressing Atgst mutants revealed that lower vitality and higher EGSH values characterized these mutants compared to wild type plants. Only modest changes were found in the redox potential values in mutants following SA treatments. We observed modified gene expression patterns in the Atgst mutants not just in control conditions, but even after applying 1 h of SA treatment compared to the Col-0 roots. Our results may verify the varying effects of applied SA concentrations on the expression of oxidative stress- and redox-related genes, among them the expression of AtGSTF8 and AtGSTU19 genes. The enhanced expression of redox-related genes in Atgst mutants may be the result of either direct antioxidant roles or their involvement in redox signalling. These data, originating from the whole roots, provided indirect evidence for the role of AtGSTF8 and AtGSTU19 isoenzymes within the transduction of the redox signal and strengthened the potential SA binding functions of AtGSTF8.
References
Aller I, Rouhier N, Meyer AJ (2013) Development of roGFP2-derived redox probes for measurement of the glutathione redox potential in the cytosol of severely glutathione-deficient rml1 seedlings. Front Plant Sci 4:506. https://doi.org/10.3389/fpls.2013.00506
Ambastha V, Friedmann Y, Leshem Y (2020) Laterals take it better–emerging and young lateral roots survive lethal salinity longer than the primary root in Arabidopsis. Sci Rep 10(1):1–11. https://doi.org/10.1038/s41598-020-60163-7
Armengot L, Marquès-Bueno MM, Soria-Garcia A, Müller M, Munné-Bosch S, Martínez MC (2014) Functional interplay between protein kinase CK2 and salicylic acid sustains PIN transcriptional expression and root development. Plant J 78(3):411–423. https://doi.org/10.1111/tpj.12481
Blanco F, Garretón V, Frey N, Dominguez C, Perez-Acle T, Van der Straeten D, Jordana X, Holuigue L (2005) Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol Biol 59(6):927–944. https://doi.org/10.1007/s11103-005-2227-x
Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME et al (2009) Early genomic responses to salicylic acid in Arabidopsis. Plant Mol Biol 70(1–2):79–102. https://doi.org/10.1007/s11103-009-9458-1
Claeys H, Van Landeghem S, Dubois M, Maleux K, Inzé D (2014) What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol 165(2):519–527. https://doi.org/10.1104/pp.113.234641
Csiszár J, Brunner S, Horváth E, Bela K, Ködmön P, Riyazuddin R et al (2018) Exogenously applied salicylic acid maintains redox homeostasis in salt-stressed Arabidopsis gr1 mutants expressing cytosolic roGFP1. Plant Growth Regul 86(2):181–194. https://doi.org/10.1007/s10725-018-0420-6
Cummins I, Dixon DP, Freitag-Pohl S, Skipsey M, Edwards R (2011) Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab Rev 43(2):266–280. https://doi.org/10.3109/03602532.2011.552910
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17. https://doi.org/10.1104/pp.105.063743
Diaz-Vivancos P, de Simone A, Kiddle G, Foyer CH (2015) Glutathione—linking cell proliferation to oxidative stress. Free Radic Biol Med 89:1154–1164. https://doi.org/10.1016/j.freeradbiomed.2015.09.023
Dietz KJ (2014) Redox regulation of transcription factors in plant stress acclimation and development. Antioxid Redox Sign 21:1356–1372. https://doi.org/10.1089/ars.2013.5672
Ding P, Ding Y (2020) Stories of salicylic acid: a plant defense hormone. Trends Plant Sci 25(6):549–565. https://doi.org/10.1016/j.tplants.2020.01.004
Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X et al (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173(6):1454–1467. https://doi.org/10.1016/j.cell.2018.03.044
Dixon DP, Hawkins T, Hussey PJ, Edwards R (2009) Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot 60(4):1207–1218. https://doi.org/10.1093/jxb/ern365
Dixon DP, Skipsey M, Edwards R (2010) Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71(4):338–350. https://doi.org/10.1016/j.phytochem.2009.12.012
Evans MJ, Choi WG, Gilroy S, Morris RJ (2016) A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol 171:1771–1784. https://doi.org/10.1104/pp.16.00215
Feigl G, Molnár Á, Szőllősi R, Ördög A, Törőcsik K, Oláh D et al (2019) Zinc-induced root architectural changes of rhizotron-grown B. napus correlate with a differential nitro-oxidative response. Nitric Oxide 90:55–65. https://doi.org/10.1016/j.niox.2019.06.003
Foyer CH (2020) Making sense of hydrogen peroxide signals. Nature 578:518–519. https://doi.org/10.1038/d41586-020-00403-y
Gomes A, Fernandes E, Lima JL (2005) Fluorescence probes used for detection of reactive oxygen species. J Biochem Biophys Methods 65(2–3):45–80. https://doi.org/10.1016/j.jbbm.2005.10.003
Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Harvey Millar A, Singh KB (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. PNAS 108(26):10768–10773. https://doi.org/10.1073/pnas.1016060108
Hasanuzzaman M, Bhuyan MHMB, Parvin K, Bhuiyan TF, Anee TI, Nahar K et al (2020) Regulation of ROS metabolism in plants under environmental stress: a review of recent experimental evidence. Int J Mol Sci 21(22):8695. https://doi.org/10.3390/ijms21228695
He H, Van Breusegem F, Mhamdi A (2018) Redox-dependent control of nuclear transcription in plants. J Exp Bot 69(14):3359–3372. https://doi.org/10.1093/jxb/ery130
Horváth E, Szalai G, Janda T (2007) Induction of abiotic stress tolerance by salicylic acid signaling. J Plant Growth Regul 26(3):290–300. https://doi.org/10.1007/s00344-007-9017-4
Horváth E, Brunner S, Bela K, Papdi C, Szabados L, Tari I et al (2015a) Exogenous salicylic acid-triggered changes in the glutathione transferases and peroxidases are key factors in the successful salt stress acclimation of Arabidopsis thaliana. Funct Plant Biol 42:1129–1140. https://doi.org/10.1071/FP15119
Horváth E, Csiszár J, Gallé Á, Poór P, Szepesi Á, Tari I (2015b) Hardening with salicylic acid induces concentration-dependent changes in abscisic acid biosynthesis of tomato under salt stress. J Plant Physiol 183:54–63. https://doi.org/10.1016/j.jplph.2015.05.010
Horváth E, Bela K, Holinka B, Riyazuddin R, Gallé Á, Hajnal Á et al (2019) The Arabidopsis glutathione transferases, AtGSTF8 and AtGSTU19 are involved in the maintenance of root redox homeostasis affecting meristem size and salt stress sensitivity. Plant Sci 283:366–374. https://doi.org/10.1016/j.plantsci.2019.02.005
Horváth E, Bela K, Gallé Á, Riyazuddin R, Csomor G, Csenki D et al (2020) Compensation of mutation in Arabidopsis glutathione transferase (AtGSTU) genes under control or salt stress conditions. Int J Mol Sci 21:2349. https://doi.org/10.3390/ijms21072349
Jiang K, Moe-Lange J, Hennet L, Feldman LJ (2016) Salt stress affects the redox status of Arabidopsis root meristems. Front Plant Sci 7:81. https://doi.org/10.3389/fpls.2016.00081
Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J (2001) Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J 26(4):395–407. https://doi.org/10.1046/j.1365-313X.2001.2641040.x
Krajewski MP, Kanawati B, Fekete A, Kowalski N, Schmitt-Kopplin P, Grill E (2013) Analysis of Arabidopsis glutathione-transferases in yeast. Phytochemistry 91:198–207. https://doi.org/10.1016/j.phytochem.2012.04.016
Manohar M, Tian M, Moreau M, Park SW, Choi HW, Fei Z et al (2015) Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front Plant Sci 5:777. https://doi.org/10.3389/fpls.2014.00777
Marrs KA (1996) The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol 47:127–158. https://doi.org/10.1146/annurev.arplant.47.1.127
Mase K, Tsukagoshi H (2021) Reactive oxygen species link gene regulatory networks during arabidopsis root development. Front Plant Sci. https://doi.org/10.3389/fpls.2021.660274
McCabe PF, Leaver CJ (2000) Programmed cell death in cell cultures. Programmed cell death in higher plants. Springer, Dordrecht, pp 115–124
Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP et al (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52:973–986. https://doi.org/10.1111/j.1365-313X.2007.03280.x
Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S et al (2006) Gain-and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580(28–29):6537–6542. https://doi.org/10.1016/j.febslet.2006.11.002
Miura K, Tada Y (2014) Regulation of water, salinity, and cold stress responses by salicylic acid. Front Plant Sci 5:4. https://doi.org/10.3389/fpls.2014.00004
Moons A (2005) Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam Horm 72:155–202. https://doi.org/10.1016/S0083-6729(05)72005-7
Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113(7):935–944. https://doi.org/10.1016/S0092-8674(03)00429-X
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Otulak-Kozieł K, Kozieł E, Horváth E, Csiszár J (2022) AtGSTU19 and AtGSTU24 as moderators of the response of Arabidopsis thaliana to Turnip mosaic virus. Int J Mol Sci 23(19):11531. https://doi.org/10.3390/ijms231911531
Pál M, Szalai G, Kovács V, Gondor OK, Janda T (2013) Salicylic acid-mediated abiotic stress tolerance. In: Hayat S, Ahmad A, Alyemeni M (eds) Salicylic acid. Springer, Dordrecht, pp 183–247
Papdi C, Ábrahám E, Joseph MP, Popescu C, Koncz C, Szabados L (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147:528–542. https://doi.org/10.1104/pp.108.116897
Pasternak T, Rudas V, Potters G, Jansen MA (2005) Morphogenic effects of abiotic stress: reorientation of growth in Arabidopsis thaliana seedlings. Environ Exp Bot 53(3):299–314. https://doi.org/10.1016/j.envexpbot.2004.04.009
Pasternak T, Groo EP, Kazantsev FV, Teale W, Omelyanchuk N, Kovrizhnykh V et al (2019) Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol 180(3):1725–1739. https://doi.org/10.1104/pp.19.00130
Perilli S, Sabatini S (2010) Analysis of root meristem size development. Plant developmental biology. Humana Press, Totowa, pp 177–187
Pokotylo I, Kravets V, Ruelland E (2019) Salicylic acid binding proteins (SABPs): the hidden forefront of salicylic acid signalling. Int J Mol Sci 20(18):4377. https://doi.org/10.3390/ijms20184377
Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot 62(10):3321–3338. https://doi.org/10.1093/jxb/err031
Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K et al (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136(1):2734–2746. https://doi.org/10.1104/pp.104.046599
Sappl PG, Onate-Sanchez L, Singh KB, Millar AH (2004) Proteomic analysis of glutathione S-transferases of Arabidopsis thaliana reveals differential salicylic acid-induced expression of the plant-specific phi and tau classes. Plant Mol Biol 54(2):205–219. https://doi.org/10.1023/B:PLAN.0000028786.57439.b3
Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Harvey Millar A et al (2009) The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J 58(1):53–68. https://doi.org/10.1111/j.1365-313X.2008.03761.x
Schwarzländer M, Fricker MD, Müller C, Marty L, Brach T, Novak J et al (2008) Confocal imaging of glutathione redox potential in living plant cells. J Microsc 231:299–316. https://doi.org/10.1111/j.1365-2818.2008.02030.x
Szalai G, Pál M, Árendás T, Janda T (2016) Priming seed with salicylic acid increases grain yield and modifies polyamine levels in maize. Cereal Res Commun 44(4):537–548. https://doi.org/10.1556/0806.44.2016.038
Szepesi Á, Csiszár J, Gémes K, Horváth E, Horváth F, Simon ML et al (2009) Salicylic acid improves acclimation to salt stress by stimulating abscisic aldehyde oxidase activity and abscisic acid accumulation, and increases Na+ content in leaves without toxicity symptoms in Solanum lycopersicum L. J Plant Physiol 166(9):914–925. https://doi.org/10.1016/j.jplph.2008.11.012
Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C et al (2008) Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321(5891):952–956. https://doi.org/10.1126/science.1156970
Tari I, Csiszár J, Szalai G, Horváth F, Pécsváradi A, Kiss G et al (2002) Acclimation of tomato plants to salinity stress after a salicylic acid pretreatment. Acta Biol Szeged 46:55–56
Thatcher LF, Carrie C, Andersson CR, Sivasithamparam K, Whelan J, Singh KB (2007) Differential gene expression and subcellular targeting of Arabidopsis glutathione S-transferase F8 is achieved through alternative transcription start sites. J Biol Chem 282(39):28915–28928. https://doi.org/10.1074/jbc.M702207200
Thatcher LF, Kamphuis LG, Hane JK, Onate-Sanchez L, Singh KB (2015) The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS ONE 10(5):e0126978
Tian M, von Dahl CC, Liu PP, Friso G, van Wijk KJ, Klessig DF (2012) The combined use of photoaffinity labeling and surface plasmon resonance-based technology identifies multiple salicylic acid-binding proteins. Plant J 72(6):1027–1038. https://doi.org/10.1111/tpj.12016
Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S et al (1992) Acquired resistance in Arabidopsis. Plant Cell 4:645–656. https://doi.org/10.1105/tpc.4.6.645
Uquillas C, Letelier I, Blanco F, Jordana X, Holuigue L (2004) NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol Plant Microbe Interact 17(1):34–42. https://doi.org/10.1094/MPMI.2004.17.1.34
Verbelen JP, Cnodder TD, Le J, Vissenberg K, Baluška F (2006) The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signal Behav 1(6):296–304. https://doi.org/10.4161/psb.1.6.3511
Vlot AC, Dempsey DMA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206. https://doi.org/10.1146/annurev.phyto.050908.135202
Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49(5):515–532. https://doi.org/10.1023/A:1015557300450
Wu F, Chi Y, Jiang Z, Xu Y, Xie L, Huang F et al (2020) Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578(7796):577–581. https://doi.org/10.1038/s41586-020-2032-3
Xu J, Tian YS, Xing XJ, Peng RH, Zhu B, Gao JJ et al (2016) Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol Plant 156(2):164–175. https://doi.org/10.1111/ppl.12347
Zhang N, Zhao B, Fan Z, Yang D, Guo X, Wu Q et al (2020) Systematic identification of genes associated with plant growth–defense tradeoffs under JA signaling in Arabidopsis. Planta 251(2):43. https://doi.org/10.1007/s00425-019-03335-8
Zhao X, Wang J, Yuan J, Wang XL, Zhao QP, Kong PT et al (2015) NITRIC OXIDE-ASSOCIATED PROTEIN1 (AtNOA1) is essential for salicylic acid-induced root waving in Arabidopsis thaliana. New Phytol 207(1):211–224. https://doi.org/10.1111/nph.13327
Acknowledgements
We thank Dr. F. Ayaydin and I. Kelemen for their help using the confocal microscope in the Cellular Imaging Laboratory of BRC, Szeged. We especially thank Prof. Dr. A. Meyer for the c-GRX1-roGFP2-harboring plasmids and Dr. G. Rigó for the GV3101 Agrobacterium strain. We are grateful to Ádám Hajnal for his critical reading of the manuscript and to Mrs. Erzsébet Porkoláb for her excellent technical assistance.
Funding
Open access funding provided by University of Szeged. This study was supported by the Hungarian National Research, Development and Innovation Office [Grant Numbers: NKFIH K 125265, PD 131884, and PD 131909]. OA was supported by the University of Szeged Open Access Fund, Grant Number: 5796.
Author information
Authors and Affiliations
Contributions
EH: contributed to Conceptualization, Methodology, Formal analysis, Investigation, Visualization, Writing—original draft, Writing—review & editing, Funding acquisition; KB: contributed to Conceptualization, Investigation, Writing—review & editing, Funding acquisition; KK: contributed to Investigation, Visualization; NF: Investigation; RR: Investigation, Writing—review & editing; ÁG: contributed to Conceptualization, Writing—review & editing; LGP: contributed to Conceptualization, Supervision; JC: contributed to Conceptualization, Methodology, Resources, Supervision, Writing—original draft, Writing—review & editing, Funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Handling Editor: Naeem Khan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Horváth, E., Bela, K., Kulman, K. et al. Glutathione Transferases are Involved in Salicylic Acid-Induced Transcriptional Reprogramming. J Plant Growth Regul 42, 4497–4510 (2023). https://doi.org/10.1007/s00344-023-10915-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-023-10915-2