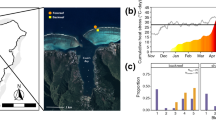
Abstract
Coral reefs are increasingly threatened by heat stress events leading to coral bleaching. In 2016, a mass bleaching event affected large parts of the Great Barrier Reef (GBR). Whilst bleaching severity and coral mortality are usually monitored throughout major bleaching events, other health indicators, such as changes in microbial partners, are rarely assessed. We examined the impact of the 2016 bleaching event on the composition of the microbial communities in the coral Pocillopora acuta at Havannah Island Pandora reef, separated by 12 km on the inshore central GBR. Corals experienced moderate heat stress (3.6 and 5.3 degree heating weeks), inducing major bleaching (30–60%) at the coral community level. Samples were partitioned according to Symbiodiniaceae densities into three bleaching severity categories (mild, moderate, and severe). Whilst Symbiodiniaceae densities were similar at both reef locations, sequencing of the Symbiodiniaceae ITS2 and prokaryotic 16S rRNA genes revealed that microbial communities were significantly different between reefs, but not according to bleaching severity. Symbiodiniaceae composition was dominated by the genus Cladocopium with low abundances of Durusdinium detected in moderately and severely bleached colonies at both sites, despite site-specific ITS2 profiles. Bacterial communities were dominated by Proteobacteria and were almost entirely lacking the common Pocilloporid associate Endozoicomonas regardless of bleaching severity. Strikingly, only 11.2% of the bacterial Amplicon Sequencing Variants (ASVs) were shared between sites. This reef specificity was driven by 165 ASVs, mainly from the family Rhodobacteraceae. Comparison with previous studies suggests that the moderate heat stress experienced on the central GBR in 2016 caused the near-complete absence of Endozoicomonas. Symbiodiniaceae and bacteria (particularly Rhodobacteraceae) can be vertically transmitted in P. acuta, and larval propagation can be spatially restricted for this brooding species. Our results demonstrate that, unlike bleaching severity, location-specific factors and species-specific life history traits might have been paramount in shaping the P. acuta microbiome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reef-building corals are symbiotic animals, often referred to as “holobionts” composed of an animal (cnidarian) host, eukaryotic algae of the family Symbiodiniaceae as well as other protists, fungi, prokaryotes and viruses (Zilber-Rosenberg and Rosenberg 2008). The fine-tuned relationships between the various partners ensure the health of the holobiont under optimal environmental conditions (Rosenberg et al. 2007; Bourne et al. 2016; Vanwonterghem and Webster 2020). One of the best characterised marine symbioses occurs between the cnidarian host and the Symbiodiniaceae, whose taxonomy has only recently been resolved (LaJeunesse et al. 2018). Whilst the coral provides carbon and nitrogen to sustain the growth of photosynthetic Symbiodiniaceae, the algal symbiont transfers photosynthates and essential nutrients to the host to support daily metabolic energy demands (Muscatine and Porter 1977). In parallel, the presence and role of coral-associated prokaryotic communities have been increasingly investigated over the last two decades (see Bourne et al. 2016 for an extensive review), revealing that reef-building corals harbour host-specific, diverse and distinct prokaryotic communities in their surface mucus layer, tissues and skeleton (Rohwer et al. 2002; Koren and Rosenberg 2006; Pernice et al. 2020; Pogoreutz et al. 2021). Bacteria have been frequently implicated in coral fitness, with shifts in microbial communities occurring concomitantly with disease or more generally declining health (Bourne et al. 2009, 2013; Glasl et al. 2016). Within the coral tissues, prokaryotes play key roles in nitrogen cycling (Lema et al. 2012, 2016; Ceh et al. 2013; Cardini et al. 2015; Messer et al. 2017) and have been linked to carbon and sulphur cycling (Raina et al. 2009; Robbins et al. 2019). In addition, whilst the coral host can acquire essential nutrients heterotrophically or from the Symbiodiniaceae, it may also benefit from bacterial biosynthesis of the compounds it cannot synthesise itself, including antimicrobial compounds, amino acids and vitamins (Raina et al. 2016; Robbins et al. 2019; Modolon et al. 2020). The complex relationships within the coral holobiont illustrate how symbiotic partners efficiently cooperate under optimal environmental conditions (Voolstra and Ziegler 2020), but also how fragile the holobiont can be upon stress, and in particular thermal stress. Many corals live close to their upper thermal limit (Hoegh-Guldberg 1999) and during periods of heat stress, the symbiosis between the host and Symbiodiniaceae breaks down as a result of shifting metabolism, and an accumulation of reactive oxygen and nitrogen species within the tissue (Jones et al. 1998; Suggett and Smith 2020; Rädecker et al. 2021). Whilst the ultimate trigger of dysbiosis has not yet been elucidated, it results in the expulsion of the dense Symbiodiniaceae community into the environment, leaving behind the transparent coral tissue and white calcareous skeleton, a phenomenon referred to as coral bleaching (Glynn 1983; Lesser 1997).
Extreme or prolonged thermal stress can cause death of the coral colony within weeks of the peak heat stress (Hughes et al. 2017). Conversely, bleaching can be followed by survival of the colony and recovery of the symbiotic partners after moderate heat stress, if temperatures cool below normal summer maximum thresholds (Brown 1997). Changes in the Symbiodiniaceae community composition within the coral host are often implicated in the survival of corals following bleaching. This can result from “background” thermal-tolerant Symbiodiniaceae genotypes (typically from the Durusdinium genus) becoming dominant (Berkelmans and Van Oppen 2006), or from the uptake of more thermal-tolerant Symbiodiniaceae from the environment (Boulotte et al. 2016). Bacterial communities have also been reported to shift in response to bleaching events, but these shifts can have deleterious outcomes such as increases in potentially pathogenic Vibrio populations and bacterial virulence genes (Bourne et al. 2008; Littman et al. 2011; Ziegler et al. 2017).
The spatial footprint of extreme thermal anomalies and the severity of coral bleaching has increased at a faster rate than predicted only a decade ago (Hoegh-Guldberg et al. 2007; Hughes et al. 2017, 2019). Over the last 30 years, coral reefs have experienced three global mass bleaching events in 1998, 2010 and 2015–2017 (Eakin et al. 2019), which impacted large areas of coral reefs worldwide, including the largest coral reef ecosystem, Australia’s Great Barrier Reef (GBR). Additional bleaching events have occurred on the GBR in 2002, and 2020 (Berkelmans et al. 2004; Hughes et al. 2017, 2019) but the 2016 bleaching event was particularly widespread as only 8.9% of the 1,156 reefs surveyed escaped any form of bleaching (Hughes et al. 2017). In addition, the percentage of reefs experiencing extreme levels of bleaching was four times higher than in 1998 and 2002. The levels of accumulated heat stress during bleaching are often measured using the National Oceanic and Atmospheric Administration (NOAA) Degree Heating Week product (DHW; °C-week) which can be defined as the accumulation of temperature anomalies exceeding the threshold set by the historical climatological maximum of the monthly mean sea surface temperature (SST) over a rolling 12-week period (Skirving et al. 2020). This remote sensing tool provides predictive forecasting as heat waves develop, and hindcasting comparisons of heat stress levels across thermal ranges. In summer 2016, 4 °C-weeks resulted in 30–40% of bleached corals along the GBR (Hughes et al. 2017), regarded as a severe level of bleaching during past events on the GBR in 1998 and 2002 (Hughes et al. 2018). Whilst the Southern GBR reefs escaped high levels of bleaching, the Central GBR reefs generally experienced moderate bleaching (between ~ 2.5 and 5 °C-weeks).
Despite the profound impact of bleaching events on coral reefs and the crucial role of microbes for coral health, several aspects of the response of microbial communities to coral bleaching are under-studied. For instance, it is well-documented that different coral colonies of a given species, exposed to similar levels of thermal stress at the same location can exhibit varying degrees of bleaching (Swain et al. 2016; Green et al. 2019); however, we do not know if this mosaic of bleaching phenotypes observed at the colony-level is reflected in the microbiome. To address this question, we used the emerging model species Pocillopora acuta, a reef-building coral with well characterised ecology, reproduction and microbiome (Torda et al. 2013a; Schmidt-Roach et al. 2014a, b; Nakajima et al. 2018; Damjanovic et al. 2020b; Ros et al. 2021). Previous studies have revealed that the microbiomes of Pocillopora species are generally stable under changing environmental conditions (Pogoreutz et al. 2018; Epstein et al. 2019), with species such as P. acuta often hosting site-specific microbiomes (Torres et al. 2020), perhaps due to the brooding reproductive strategy of the host (Torda et al. 2013a). In this study, we characterised the impacts of a range of bleaching severities on the P. acuta microbiome within two reefs of the Palm Island archipelago on the Central GBR, following the 2016 mass bleaching event.
Material and methods
Temperature data
Seawater temperature measurements were retrieved for Pandora Reef (18°48′54.9″S 146°25′52.7″E) and Havannah Island (18°50′22.6″S 146°32′02.1″E) from the Australian Institute of Marine Science (AIMS) Sea Water Temperature Observing System temperature logger program (AIMS 2017). These two reefs (hereafter referred to as “Havannah” and “Pandora”) are located 12 km apart in the GBR Marine Park within the Palm Island Group (Fig. 1). Data for each site were collected from July 2015 to December 2016, capturing the warming rate and accumulation of heat stress during the 2016 bleaching event. Remote sensing SST data and DHW thermal stress index data across the entire GBR were collected from the satellite observations from the NOAA Coral Reef Watch Version 3.1 for the 2016 summer heat wave (Skirving et al. 2020). Maps of site locations, community bleaching severity and spatial footprint of thermal heat stress (NOAA DHW) were generated using QGIS v. 3.12.
Spatial footprint of the maximum annual accumulated thermal stress in 2016 measured using the NOAA Coral Reef Watch v3.1 degree heating week (DHW) product including the latitudinal variation in Maximum Monthly Mean (MMM) SST climatology (black isolines); orange dot represents results of in-water bleaching surveys at Havannah Island Reef (HAV) and Pandora Reef (PAN). Coloured scale represents max DHW (a) throughout the Great Barrier Reef and (b) surrounding the sampled reef locations
In-water community coral bleaching surveys
To assess whether corals of Havannah and Pandora reefs had experienced similar levels of bleaching at the community level, in-water bleaching surveys of the reef communities around the two sites were conducted at two depths (2 m and 5 m) on the 4th and 6th of April 2016, respectively. Five 10 × 1 m belt transects were used with observers capturing 1 m × 1 m square continuous images along each transect with an Olympus TG-4 camera in a housing using an Inon UWL-H100 28 mm Wide Conversion lens and a Coral Watch colour card (Siebeck et al. 2006) for colour correction and white balancing at the start of each transect. Observers counted each coral colony > 5 cm in diameter within each 1 m × 1 m image, recorded genus level identification and assigned a categorical bleaching score for each individual hard and soft coral colony, based upon the proportion of living coral tissue visibly bleached white. Individual colony bleaching severity was determined using a categorical bleaching score previously established (Gleason 1993; Marshall and Baird 2000). The following individual colony categories were used, based on the total area of the living tissue visibly bleached white: (1) No bleaching or (2) Pale; (3) 1–50% tissue bleached white; (4) 51–99% bleached white; (5) 100% bleached white with or without intense fluorescence; (6) partial or complete mortality of the colony (recently dead regions of tissue). Survey methods used to assess reef community bleaching severity aligned with and contributed to the 2016 Australian National Bleaching Taskforce GBR Survey response (Hughes et al. 2017). Accordlingly, bleaching severity categories at the reef community level were based on the percentage of corals bleached as follows: no bleaching (0% bleached corals); minor bleaching (1–10%), moderate (10–30%); major (30–60%); severe (> 60%) (Berkelmans et al. 2004) (Supplementary Material).
Coral sample collection
Coral fragments were collected from 22 colonies at Pandora on the 4th April 2016 and from 25 colonies at Havannah on the 12th April 2016 at the end of the summer peak heat stress (Fig. 2) using GBRMPA permit G16/38488.1. Colonies were randomly selected, and all samples were collected at 5 m depth. A small branch from each colony (> 5 cm) was cut using sterilised stainless steel bone cutters, snap-frozen in liquid nitrogen, transported to AIMS and stored at − 80 °C until further processing.
Daily average water temperature from July 2015 to October 2016 at each sampling site. Slope data (SL) were collected at 5.6 m at Pandora and 6.0 m at Havannah; reef flat (FL) data were collected at 2.0 m (Havannah only). The solid black line represents in-water climatology from Pandora reef SL based on continuous seawater temperature data from 2007 to 2012 as daily average. Grey fill represents daily standard deviation. Reference lines represent 5 km pixel centred on Pandora reef from the NOAA CRW v3.1 Maximum Monthly Mean (MMM) Sea Surface Temperature climatology and the upper thermal limit 1 °C above the MMM value (Degree Heating Weeks + 1 Threshold) at which point daily hotspot values begin to accumulate Degree Heating Weeks (DHW) as an indicator of thermal stress for coral reefs
Tissue sample preparation and Symbiodiniaceae density
Frozen samples were thawed on ice, rinsed in 0.02 μm filtered seawater (FSW) and placed into sterile zip-lock bags. A high-pressure air gun was used to remove the coral tissue from the calcium carbonate skeleton into 10–15 mL of 0.02 μm FSW. The coral blastate was decanted into a 50 mL Falcon tube and its volume topped to 15 mL for all samples with 0.02 μm FSW, prior to vortexing using a bench top homogeniser (Bio-Gen PRO200, PRO Scientific, USA). Aliquots of 2 mL each were kept at − 20 °C for DNA extraction whilst a 1 mL aliquot was preserved in 2% formaldehyde for subsequent Symbiodiniaceae cell counts using a Neubauer haemocytometer (Fitt et al. 2000). Cell counts were achieved by adding 7 μL of the preserved aliquot onto the haemocytometer and conducting at least four replicate counts to achieve a coefficient of variance of < 10%.
Coral skeletons were bleached and dried to standardise the tissue parameters to surface area of each individual coral branch. To estimate surface area, each coral branch was dipped twice into 60 °C paraffin wax following an established protocol (Veal et al. 2010). Briefly, coral skeletons were dipped once in paraffin wax to seal the porous skeleton and weighed. Corals were then dipped a second time to estimate the surface area of the coral branch. PVC cylinders across the full-size range of coral branches were also double-dipped to fit a > 99% confidence linear curve to standardise paraffin weight gain to a known surface area.
Pocillopora acuta colony bleaching severity
P. acuta colonies were categorised for statistical analysis of the microbial communities based on Symbiodiniaceae density counts. Bleaching categories were compared to the healthiest colony within the sample set: (1) Mild bleaching (0–30% decrease in symbiont cell density); (2) Moderate (30–70% decrease in cell density) and (3) Severe (> 70% decrease in symbiont cell density).
DNA extraction and host species identification
The MOBIO Power Plant Pro DNA Isolation kit was used to extract DNA from each sample (50 mg of pelleted tissue homogenate) including two bead-beating steps of 40 s at 4 ms−1 (MP Biomedicals FastPrep). To further confirm species identification, we conducted molecular typing of the samples following previously established methodology (Torda et al. 2013b) with minor modifications as follows: the BIO-X-ACT Taq Polymerase from Bioline was used at 0.08 U/μL in reactions containing 1 X buffer, 1 mM DNTPs (0.25 mM each), 3 mM MgCl2, 0.5 μL of Hi-Specificity additive, 0.5 μM of each primer (Pdam-F AAGAAGATTCGGGCTCGTTT and Pdam-R CGCCTCCTCTACCAAGACAG), and 2 μL of 1:100 diluted DNA in a total 10 μL PCR reaction. Cycling started with 5 min at 95 °C, followed by 32 cycles of 30 s at 94 °C, 60 s at 58 °C and 60 s at 72 °C, and a long extension of 10 min at 72 °C. PCR products were then subjected to Alu1 restriction enzyme digestion without purification for 1 h at 37 °C, using 5 μL of PCR product with 0.25 μL (2.5 U) of Alu1 enzyme and 1 μL of 10 × NE CutSmart Buffer (New England BioLabs) in a 10 μL reaction. Five μL of the digested products were run on 4% TBE agarose gels (using SFR Super Fine Resolution Low Melt Agarose, VWR) for 2 h at 150 Volts and 4 °C. Banding patterns were scored manually and independently by two individuals and confirmed that all samples included in this study were P. acuta, as per Torda et al. (2013b).
DNA sequencing of microbial taxonomic markers
Sequencing was performed using Illumina MiSeq 300 bp paired-end chemistry at the Australian Centre for Ecogenomics (ACE) at the University of Queensland. The ITS2 region was amplified using primers ITS2_F (GTGAATTGCAGAACTCCGTG) and ITS2_R (CCTCCGCTTACTTATATGCTT) (Pochon et al. 2001). Each PCR reaction contained 12.5 μl of Q5 hot start high-fidelity 2 × master mix (M0494L), 1.25 μl of each primer (10 μM) and water to reach 25 μL total volume. Cycling conditions were 98 °C for 2 min, 98 °C for 10 s, 25 cycles of 55 °C for 30 s, 72 °C for 30 s and 72 °C for 2 min. Primers 27F (AGAGTTTGATCMTGGCTCAG) and 519R (GWATTACCGCGGCKGCTG) were used for 16S rRNA gene amplification (Suzuki and Giovanni 1996; Marchioro et al. 2020). PCR reactions contained 1X PCR buffer, 1 mM dNTPs, 3 mM MgCl2, 0.5 μL of Hi-Specificity additive, 0.4 μM of each primer, 0.08U.μl−1 of Q5 High-Fidelity DNA polymerase and 2 μl of DNA template for a total of 10 μl of PCR reaction. PCR cycling was conducted with 5 min at 95 °C, 35 cycles of 30 s at 94 °C, 45 s at 53 °C and 60 s at 72 °C, and an extension of 10 min at 72 °C. Three samples (SA8730 from Pandora mild bleaching, SA8686 from Pandora severe bleaching and SA8815 from Havannah mild bleaching) resulted in poor ITS2 rDNA sequencing but presented otherwise high numbers of 16S rRNA gene reads and sensible Symbiodiniaeae densities. Therefore, considering the relatively low sample size, these sample were included in all 16S rRNA gene sequence analyses. Symbiodiniaceae ITS2 profiling generated an average of 3,680 reads per sample, with a minimum of 1,357 reads and a maximum of 239,319 reads, and 16S rRNA gene sequencing generated between 10,303 and 86,995 reads, with an average of 36,354.
Bioinformatics
ITS2 raw reads were submitted to the SymPortal platform (Hume et al. 2019) for remote analysis (https://symportal.org/). This approach allowed us to retain intragenomic information and identify Defining Intragenomic Variants (DIVs) as well as ITS2 profiles. ITS2 profiles are built using the presence as well as the abundance of the DIVs identified. The name of each profile reflects the proportion of each DIV within. As an example, profile C1d.C1.C42.2 would be mostly composed of C1d-type sequences, with C1-type sequences and C42.2-type sequences present in decreasing proportions. Absolute abundances were used for subsequent analysis. Visualisation of 16S rRNA gene read quality using Fastqc as well as the demux plug-in of QIIME2 (Bolyen et al. 2019) revealed that reverse reads were of poor quality and hence only forward reads were retained and analysed following the QIIME2 pipeline. The DADA2 plug-in was used to remove primers and reads shorter than 200 bp, dereplicate reads, truncate high-quality reads (quality score > 30) to 200 bp and filter out chimaeras and singletons. The feature-classifier plug-in was used to train a naïve-Bayes classifier using the Silva database 132 (Quast et al. 2012) based on the 27F-519R primer pair region. The Amplicon Sequencing Variants (ASVs) table was filtered to remove all eukaryotic, mitochondrial and chloroplast ASVs from the dataset.
Statistical analysis of DNA sequencing data
ITS2 sequencing data recovered from the SymPortal platform were transformed into relative abundances and fourth-root transformed using PRIMER. A Bray–Curtis dissimilarity matrix was built prior to testing the effect of bleaching severity and location using PERMANOVA (10,000 permutations). The number of reads originating from 16S rRNA gene sequencing was visualised as rarefaction curves using the package vegan in R (Oksanen et al. 2017; R Development Core Team 2017) to confirm sufficient sequencing. Data were rarefied to the sample with the lowest number of reads (10,303) to calculate richness and diversity indices in R. Richness data were log10-transformed to meet homogeneity of variances and the normality of residues assumption, prior to performing ANOVA to test the effect of bleaching severity and location (Tables S1-S5). Shannon diversity index data were normal and homogeneous, and no transformation was necessary prior to ANOVA testing (Tables S6-S9). Statistical testing of the effect of bleaching severity and location on richness and Shannon diversity index was conducted with ANOVA. The effects of bleaching severity and reef were assessed on the overall bacterial communities (ASVs) with PERMDISP and PERMANOVA (10,000 permutations) in PRIMER on a Bray–Curtis dissimilarity matrix of a square-root transformed rarefied and non-rarefied dataset. Since rarefaction did not change the outcome of the PERMANOVA, all reads were retained in the final results to avoid biases of the rarefaction process (McMurdie and Holmes 2014). Similarly, as visualisation of the beta-diversity on a Principal Coordinate Analysis plot on PRIMER was identical on rarefied and non-rarefied data, the non-rarefied data were retained for Fig. 3. Based on initial statistical results, a Venn diagram was built to assess the percentage of ASVs shared between reefs, and a SIMPER analysis was conducted on the non-rarefied Bray–Curtis dissimilarity matrix to identify the ASVs explaining 20% of the cumulative variation between the two reefs. DESeq2 was used to identify ASVs that were significantly different between Havannah and Pandora reefs. Finally, a PERMANOVA was conducted with 10,000 permutations using ITS2 profiles as a factor. All data wrangling and visualisation were performed in R using the package tidyverse and ggplot2, respectively (Wickham and Chang 2015; Wickham et al. 2019).
Characterisation of the Symbiodiniaceae communities in P. acuta at Havannah and Pandora reefs across bleaching severities. Each bar in panel a represents the same sample in panel b. (a) Symbiodiniaceae cell density (× 106 cells.cm−2). (b) ITS2 profiles. Relative abundance is expressed as a percentage. Each horizontal bar represents a sample
Results
Thermal stress and coral bleaching in 2016 at Havannah and Pandora
Levels of thermal stress at the central GBR study sites in 2016 were considered moderate (between 2.5 and 5 °C-weeks) with severe levels of heat stress exceeding 6 °C-weeks restricted to the sector located North of Cooktown (Fig. 1a). Historical summer maximum temperatures are similar (using NOAA MMM CoralTemp v3.1; Skirving et al. 2020) at Havannah and Pandora reefs (28.86 °C and 28.95 °C, respectively). Pandora accumulated higher levels of heat stress up to 5.3 °C-weeks at the time of sampling, compared to 3.6 °C-weeks at Havannah, which corresponded to moderate heat stress (Fig. 1b). In-water survey further revealed that this level of stress induced major bleaching at the community level for corals living in both Havannah and Pandora reefs, with 30–60% of hard and soft corals bleached (Fig. 1b). Frequency of severe individual colony bleaching (colony bleaching category 5; 100% white) was low within the bleached communities at both reefs (0–1.0% and 9.3–17.9% of the hard and soft corals within the belt transects, at Pandora and Havannah, respectively) (Supplementary Material). Recent bleaching-related mortality (colony bleaching category 6) was also low at both study sites (0% and 0.3–1.2% mortality of hard and soft corals at Pandora and Havannah, respectively).
Maximum temperatures at both reef locations exceeded the historical maximum summer climatology by more than 1 °C for prolonged periods in January, February and March, with an extended summer period throughout the entire month of March, when temperatures typically begin to cool (Fig. 2). Maximum temperature anomalies were recorded on the slope of Pandora reef (Fig. 2).
Symbiodiniaceae communities
Symbiodiniaceae densities were used to attribute bleaching severity categories to individual colonies (see Methods section, Supplementary text and Fig. S1). Symbiodiniaceae densities (Fig. 3a) ranged between 0.2 and 1.1 million cells.cm−2 in Havannah-sourced corals and between 0.07 and 1 million cells.cm−2 in Pandora-sourced corals. Severe bleaching corresponded with colonies exhibiting < 0.4 million cells.cm−2 densities, moderate bleaching to colonies with densities comprised between 0.4 and 0.6 million cells.cm−2, whilst mild bleaching corresponded with corals presenting > 0.6 million cells.cm−2 Symbiodiniaceae densities.
The bleaching category assignments resulted in a total of 10 and 6 mildly bleached corals from Havannah and Pandora, respectively, 9 moderately bleached samples from both locations, as well as 6 and 7 severely bleached samples from Havannah and Pandora, respectively. These categories are those used in the remainder of this article unless otherwise stated.
Symbiodiniaceae communities were represented by 15 distinct profiles across Havannah and Pandora, including 10 containing the genus Cladocopium, three Durusdinium and two Symbiodinium (Fig. 3b). The two Symbiodinium (A-dominated profiles) were found at 0.1% and 0.5% in a single moderately bleached Pandora sample and we hereafter concentrate on the 13 remaining ITS2 profiles.
Symbiodiniaceae from the Cladocopium C1d-type dominated the communities at both reefs. However, 75% of the profiles were specific to a reef, with 5 found exclusively in Pandora and 5 exclusively in Havannah. Only 3 profiles (all in the genus Cladocopium) were shared between reefs. Consistent with these observations, location had a significant effect on the distribution of the Symbiodiniaceae profiles (PERMANOVA, F1,38 = 3.7916, p = 0.0027, Table S10), but bleaching severity did not and there was no interaction between the two factors (PERMANOVA, Table S10). Furthermore, bleaching severity did not significantly affect Symbiodiniaceae communities at each reef independently (PERMANOVA, Tables S11-S12).
The vast majority of samples only harboured a single profile, with only 5 samples harbouring a second profile (including all 3 Durusdinium profiles), all at a relative abundance of 1–4% within a sample. The mildly bleached corals only harboured Cladocopium profiles at Pandora, and the two Durusdinium profiles found at that location were symbionts of a moderately bleached coral and a severely bleached coral (1.2% and 3.7%, respectively). At Havannah, the only Durusdinium profile was found in a moderately bleached coral at 3% relative abundance.
The most common profile at Havannah (C1d.C1bl.C42.2.C1) was found in 4 mildly bleached corals, as well as 3 severely bleached and two moderately bleached corals. Conversely, the most common profile at Pandora (C1d.C42.2.C1.C1k.C1b) was found in 4 moderately bleached samples, two mildly bleached and two severely bleached corals.
Bacterial communities
A total of 5,584 ASVs (all belonging to bacterial phyla) were found in Havannah samples and 4,154 in Pandora samples. Richness and diversity (assessed with the Shannon index) of the bacterial communities were not significantly different between reefs or across bleaching severity categories, and there was no significant interaction between the location and bleaching severity factors (Fig. 4 and Tables S13-S14). Within each reef, richness was not statistically affected by bleaching severity (Tables S15-16), and nor was diversity (Tables S17-S18).
Diversity of the bacterial communities in P. acuta at Havannah and Pandora reefs across bleaching severity. (a) Richness expressed as the mean number of ASVs (± standard error). (b) Shannon index expressed as mean (± standard error). Data were rarefied with all samples containing 10,303 ASVs for both panels
The bacterial communities of P. acuta were dominated by Proteobacteria, in particular Alpha- and Gammaproteobacteria (Fig. 5a). Representatives of the phyla Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes, Planctomycetes and Spirochaetes were also found at abundances > 1%. The most abundant families at Havannah were the Rhodobacteraceae (with an average of 27.4–39.9% across the different bleaching categories), Marinobacteraceae (13.4–17.5%), Sphingomonadaceae (7.3–10.1%), Pseudomonadaceae (4.3–6.0%) and Cyanobiaceae (3.1–6.2%). Microbial communities of P. acuta at Pandora also exhibited high relative abundances of Rhodobacteraceae (11.1–27.4%), Marinobacteraceae (13.8–24.3%), Cyanobiaceae (7.3–14.9%) and Sphingomonadaceae (1.2–4.2%), as well as high proportions of Alcanivoracaceae (2.9–4.0%). The family Endoizoicomonadaceae which comprises the Endozoicomonas genus was found at low abundances only (< 1%), at both sites.
Characterisation of the bacterial communities of P. acuta at Havannah and Pandora reefs across bleaching categories. (a) Mean relative abundance at the family level for families representing > 1% of the total community. Ac: Actinobacteria, Ba: Bacteroidetes, Ch: Chloroflexi, Cy: Cyanobacteria, Fi: Firmicutes, Pl: Planctomycetes, Sp: Spirochetes, α: Alphaproteobacteria, γ: Gammaproteobacteria. Distribution of the samples on Principal Coordinates Analysis for both reefs (b), Havannah only (c), and Pandora only (d)
Bacterial communities of P. acuta partitioned according to reef (PERMANOVA, F1,41 = 4.744, p = 0.0001, Table S19), but not bleaching severity and no interaction was found between these two factors (PERMANOVA, Table S19) (Fig. 5b). When analysed separately, microbial communities were not significantly impacted by bleaching severity at Havannah nor at Pandora (PERMANOVA, Tables S20-S21, Fig. 5c, d). The combination of PCO1 and PCO2 explained 24.1% of the total variability at Havannah and 41.1% at Pandora. Dispersion of the samples did not significantly differ between sampling sites (PERMDISP, Table S22), nor bleaching severity at each of the reefs (PERMDISP, Tables S23-S24). Finally, Symbiodiniaceae ITS2 profiles did not have a significant effect on the prokaryotic communities of both reefs (PERMANOVA, Tables S25-S26).
In light of these results, further analyses were conducted to identify the bacterial groups responsible for this site-specific pattern. Only 11.2% of ASVs were shared between reefs (Venn diagram, not shown) and 32 ASVs explained 20% of the variability between reefs according to SIMPER analysis (Table 1). A total of 165 ASVs exhibited significantly different abundances between reefs (Supplementary Material). The taxonomy of 29 of these ASVs was not resolved at the family level but the remaining 136 ASVs were members of 62 families. Of particular interest were families Rhodobacteraceae, Sphingomonadaceae, Pirellulaceae, Marinobacteraceae and Cyanobiaceae (Fig. 6). The family containing the highest number of significantly different ASVs was Rhodobacteraceae with 13 ASVs, representing 9.6% of the differentially abundant ASVs taxonomically resolved at the family level, followed by Sphingomonadaceae with 8 ASVs and Pirellulaceae with 7 ASVs (5.8% and 5.1%, respectively) (Fig. 6a, b). When considering relative abundances, the 13 Rhobacteraceae ASVs together represented 19.4% of the bacterial communities at Havannah and 4.0% at Pandora, whilst the 8 Sphingomonadaceae ASVs represented 7% of the communities at Havannah and 1.8% of Pandora. The 6 significantly different Marinobacteraceae ASVs and the 4 significantly different Cyanobiaceae ASVs showed the opposite pattern: they respectively, represented 15.4% and 10.3% of the communities at Pandora but only 4.8% and 3.8% at Havannah. Finally, two Endozoicomonas ASVs were significantly different between reefs, and their combined mean relative abundances amounted to 0.2% at Havannah but to a negligible quantity at Pandora (< 0.0001%).
Significant differences within bacterial communities between Havannah and Pandora reefs. (a) Mean relative abundances of each significantly different ASV were calculated for each reef, then summed according to families. Data shown for sum > 0.1% relative abundance. Numbers on the side of the plot indicate the number of ASVs represented by the bubble in each family depicted (blank = 1 single ASV). (b) Mean relative abundance of the significantly different ASVs within 5 major families (see text). Families are colour-coded with the same colours in both panels
Discussion
Coral-associated microbial communities underpin the fitness of their host and have been shown to shift in response to environmental disturbances and geographic location. Here, we show that the composition of both Symbiodiniaceae and prokaryotic communities associated with P. acuta did not respond to bleaching severity during the 2016 global mass bleaching event. Rather, microbial communities differed substantially between two inshore reefs located only 12 km apart on the GBR, highlighting the importance of fine-scale sampling when investigating the consequences of large-scale heat stress events on host-associated microbes.
Increasingly severe bleaching did not induce drastic shifts in Symbiodiniaceae communities
Most P. acuta samples comprised a single Symbiodiniaceae ITS2 profile, consistent with previous reports (Damjanovic et al. 2020a). The Symbiodiniaceae community composition was dominated by Cladocopium and the most abundant profiles were C1d-dominant and C42-dominant, which was also consistent with P. acuta colonies from nearby Orpheus Island (Epstein et al. 2019). The most abundant profile at Havannah was C1d.C1bl.C42.2.C1, which is closely related to the most dominant ITS2 profile of P. acuta collected at nearby Magnetic Island (Damjanovic et al. 2020b).
Three Symbiodiniaceae profiles in the thermally tolerant genus Durusdinium were detected, all restricted to moderately and severely bleached samples. The two Durusdinium profiles detected at Pandora were 3 times more abundant in the severely bleached sample than in the moderately bleached sample. Whilst our small sample size precludes any strong conclusion, these findings constitute a low-lying signal of symbiont shuffling or shifting in corals having experienced heat stress, as was previously found in multiple species, including Pocillopora damicornis (LaJeunesse et al. 2009; Boulotte et al. 2016). As samples for the current study were collected only during the bleaching event (as opposed to prior or after), it remains uncertain whether these represent new genotypes acquired through symbiont shifting or if they were present prior to the bleaching event and if so, at which proportion. However, Durusdinium were completely absent from 10 P. acuta colonies sampled every month over the course of 8 months at Orpheus Island, except for a single colony in July 2016 (Epstein et al. 2019). It is therefore more likely that the Durusdinium detected in the present study represent newly acquired genotypes in response to heat stress.
In corals, the rare Symbiodiniaceae biosphere has been described as any genotype found at relative abundances lower than 1% (Boulotte et al. 2016). Here, we show that P. acuta experiencing 3 to 5 DHW can host heat-tolerant Symbiodiniaceae at relative abundances more than threefold higher than the rare biosphere threshold. Whilst rare taxa are often overlooked in microbial ecology studies, they have been shown to play crucial roles in natural ecosystems, including during environmental changes (Shade et al. 2014; Chen et al. 2020). Increased relative abundance of Durusdinium after a bleaching event has been linked to increased thermal tolerance of the coral holobiont (Berkelmans and Van Oppen 2006; Silverstein et al. 2015). Therefore, whilst changes in Symbiodiniaceae communities can come at the cost of other physiological traits, such as reduced growth, photophysiology and calcification (Little et al. 2004; Cantin et al. 2009; Pettay et al. 2015), it is possible that the colonies hosting Durusdinium Symbiodiniaceae may increase their tolerance to future bleaching events following major disturbance events such as the 2016 mass bleaching. Given that Symbiodiniaceae from the rare biosphere can shift up to 18 months post-bleaching (Boulotte et al. 2016), our findings call for greater and repeated temporal sampling of bleached colonies.
Near-complete lack of Endozoicomonas regardless of bleaching severity
Our results corroborate previous findings that heat stress does not impact the overall bacterial community composition of P. acuta (Epstein et al. 2019). The bacterial communities associated with Pocillopora verrucosa were also highly stable throughout bleaching induced by excessive dissolved organic carbon (Pogoreutz et al. 2018) and upon transplantation of coral colonies across environmental gradients (Ziegler et al. 2019). Nine families (namely Alcanivoracaceae, Chlorobiaceae, Flavobacteraceae, Francisellaceae, Hyphomonadaceae, Pseudomonadaceae, Rhodobacteraceae, Rhodothermaceae and Sphingomonadaceae) dominated the P. acuta microbial communities, in accordance with previous studies investigating P. acuta collected at Orpheus Island within the Palm Island archipelago throughout 2016 and at Magnetic Island in March 2017 (Epstein et al. 2019; Damjanovic et al. 2020b). However, it is important to note that DNA sequencing detects genetic information regardless of the viability or activity state of the cells, and that bacterial composition does not capture how these microbes are functionally contributing to the holobiont under stress.
The major deviation from previous reports of the Pocillopora-associated bacterial community was the very low relative abundance of Endozoicomonas reported here (< 1%) (Pogoreutz et al. 2018; Epstein et al. 2019; Ziegler et al. 2019; Voolstra and Ziegler 2020). Rarefaction curves indicated sufficient sequencing depth and read numbers were comparable with previous studies at nearby locations (Epstein et al. 2019). In addition, the primers used here can detect coral-associated Endozoicomonas (Glasl et al. 2019; Marchioro et al. 2020). Therefore, the reduced representation of Endozoicomonas may have been due to the fact that the onset of coral bleaching represents a tipping point for this bacterial species in P. acuta. Indeed, all corals from the present study exhibited some level of bleaching, with 3.5–5.6 °C-weeks and an absence of Endozoicomonas regardless of bleaching severity, whereas P. acuta, which experienced 2 °C-weeks and no bleaching within the same archipelago, still harboured normal levels of Endozoicomonas (Epstein et al. 2019).
Supporting this hypothesis is the fact that reduced abundances of Endozoicomonas were found in Acropora millepora after a bleaching event on the GBR (Bourne et al. 2008) and that such declines have often been correlated with degraded or stressful environmental conditions across geographic locations. For instance, A. millepora living in CO2 seep-acidified waters of Papua New Guinea presented a 50% reduction in relative abundance of Endozoicomonas compared to individuals sampled at a nearby pristine site (Morrow et al. 2015), and unhealthy Porites astreoides sampled in Belize displayed decreased abundances of Endozoicomonas compared to healthy individuals (Meyer et al. 2014). The consequences of these declines on coral health are unclear, as the functional role of Endozoicomonas within corals has remained elusive. However, genomic data and functional prediction tools have suggested that the Endozoicomonas-coral association involves amino acids metabolism and nutrient cycling (Morrow et al. 2015; Neave et al. 2017; Robbins et al. 2019). Recent empirical evidence demonstrated that Endozoicomonas acroporae isolated from Acropora sp. can degrade the sulphur compound dimethylsulphoniopropionate (DMSP) and use it as part of its central carbon metabolism (Tandon et al. 2020). Given that dinoflagellates and the coral host are major producers of DMSP (Broadbent and Jones 2004; Raina et al. 2013), the near-complete absence of Endozoicomonas in the present study might have aggravated any nutrient imbalance already at play in the holobiont and further deteriorated the host’s health.
The microbial signature of P. acuta is site-specific
Unlike bleaching severity, location strongly constrained both the Symbiodiniaceae and bacterial communities of P. acuta living only 12 km apart. A global scale study examining the microbiome of Pocillopora damicornis found that geographic location mattered less than thermal regimes in structuring the microbial communities (Brener-Raffalli et al. 2018). However, as noted previously, the present study examined bleached samples and showed differences in microbial communities compared to samples of the same species collected at the same location under milder thermal stress (Epstein et al. 2019).
Whilst it is impossible to rule out that a difference of 1.7 °C-weeks between Havannah and Pandora is responsible for the contrasting microbial community composition, other factors could have played an important structuring role. Indeed, finer scale studies have shown that the microbiome of a single coral species can vary across reef regions (Hernandez-Agreda et al. 2016) or across habitats within a given reef (Glasl et al. 2017). Similarly, Acropora hyacinthus and Acropora spathulata from Rib Reef and Davies Reef (mid-shelf Great Barrier Reef), separated by 80 km, were found to diverge in the community composition of both Symbiodiniaceae and bacteria (Epstein et al. 2019). Within the Palm Island archipelago, location was an important factor structuring prokaryotic communities of Acropora coral species separated by only 40 km (Littman et al. 2009).
Despite the smaller geographic scale discussed in the present study, a combination of ecological and oceanographic factors might explain the strong reef specificity we detected. P. acuta is a brooding species, which implies limited dispersal of larvae before settlement, compared to a broadcast spawning species (Torda et al. 2013a). In addition, P. acuta exhibits both sexual and asexual reproductive modes (Torda et al. 2013a; Schmidt-Roach et al. 2014b; Gélin et al. 2017; Nakajima et al. 2018). Whilst populations in the Palm Island archipelago preferentially use sexual reproduction, particular sites within the Palm Islands received a greater proportion of recruits than others when examined over a period of two years (Torda et al. 2013a). In other words, self-recruitment can be more prevalent in certain areas of a single reef (Torda et al. 2013a), perhaps due to water currents direction (King and Wolanski 1996; Luick et al. 2007) or differences in wave energy and the frequency or intensity of physical disturbances (Gélin et al. 2017). A lack of connectivity was found in P. acuta colonies living 5 km apart in the Philippines (Torres et al. 2020), which is much shorter than the distance between Havannah and Pandora reefs, demonstrating how spatially restricted larval propagation can be for this species.
Crucially, vertical transmission of both Symbiodiniaceae and bacteria occurs in P. acuta (Baird et al. 2009; Damjanovic et al. 2020b). In particular, the bacterial family Rhodobacteraceae was one of the two most abundant families shared between parents and offspring of P. acuta (Damjanovic et al. 2020b). Therefore, the fact that this family contributed most to site-driven differences in the present study, both in the number of significantly different ASVs and in their relative abundance, strongly suggests that vertical transmission combined with limited larval dispersal played a role in the high reef specificity.
Taken together, our results demonstrate that, unlike bleaching severity, reef site shaped the microbial communities of P. acuta. Converging signals suggest that the onset of bleaching might represent a tipping point for the bacterial communities, and that factors such as local oceanic currents and host life history traits are responsible for the detected differences in microbial community composition, both eukaryotic and prokaryotic, between Havannah and Pandora reefs.
References
AIMS (2017) AIMS Sea Water Temperature Observing System (AIMS Temperature Logger Program), https://doi.org/10.25845/5b4eb0f9bb848, accessed 13-Apr-2021.
Baird AH, Guest JR, Willis BL (2009) Systematic and Biogeographical Patterns in the Reproductive Biology of Scleractinian Corals. Ann Rev Ecol Evol Syst 40:551–571
Berkelmans R, Van Oppen MJ (2006) The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’for coral reefs in an era of climate change. Proc Royal Soc b: Biological Sciences 273:2305–2312
Berkelmans R, De’ath G, Kininmonth S, Skirving WJ (2004) A comparison of the 1998 and 2002 coral bleaching events on the Great Barrier Reef: spatial correlation, patterns, and predictions. Coral Reefs 23:74–83
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Titus Brown C, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Zhenjiang Zech X, Zaneveld JR, Zhang Y, Zhu Q, Rob Knight J, Caporaso G (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Boulotte NM, Dalton SJ, Carroll AG, Harrison PL, Putnam HM, Peplow LM, van Oppen MJH (2016) Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J 10:2693–2701
Bourne D, Iida Y, Uthicke S, Smith-Keune C (2008) Changes in coral-associated microbial communities during a bleaching event. ISME J 2:350–363
Bourne DG, Morrow KM, Webster NS (2016) Insights into the Coral Microbiome: Underpinning the Health and Resilience of Reef Ecosystems. Annu Rev Microbiol 70:317–340
Bourne DG, van der Zee MJJ, Botté ES, Sato Y (2013) Sulfur-oxidizing bacterial populations within cyanobacterial dominated coral disease lesions. Environ Microbiol Rep 5:518–524
Bourne DG, Garren M, Work TM, Rosenberg E, Smith GW, Harvell CD (2009) Microbial disease and the coral holobiont. Trends Microbiol 17:554–562
Brener-Raffalli K, Clerissi C, Vidal-Dupiol J, Adjeroud M, Bonhomme F, Pratlong M, Aurelle D, Mitta G, Toulza E (2018) Thermal regime and host clade, rather than geography, drive Symbiodinium and bacterial assemblages in the scleractinian coral Pocillopora damicornis sensu lato. Microbiome 6:1–13
Broadbent AD, Jones GB (2004) DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar Freshw Res 55:849–855
Brown B (1997) Coral bleaching: causes and consequences. Coral Reefs 16:S129–S138
Cantin NE, van Oppen MJH, Willis BL, Mieog JC, Negri AP (2009) Juvenile corals can acquire more carbon from high-performance algal symbionts. Coral Reefs 28:405–414
Cardini U, Bednarz VN, Naumann MS, van Hoytema N, Rix L, Foster RA, Al-Rshaidat MMD, Wild C (2015) Functional significance of dinitrogen fixation in sustaining coral productivity under oligotrophic conditions. Proc Royal Soc B: Biological Sciences 282:20152257
Ceh J, Kilburn MR, Cliff JB, Raina JB, van Keulen M, Bourne DG (2013) Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecol Evol 3:2393–2400
Chen Q-L, Ding J, Zhu D, Hu H-W, Delgado-Baquerizo M, Ma Y-B, He Ji-Z, Zhu Y-G (2020) Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biol Biochem 141:107686
Damjanovic K, Blackall LL, Menéndez P, van Oppen MJH (2020a) Bacterial and algal symbiont dynamics in early recruits exposed to two adult coral species. Coral Reefs 39:189–202
Damjanovic K, Menéndez P, Blackall LL, van Oppen MJ (2020b) Mixed-mode bacterial transmission in the common brooding coral Pocillopora acuta. Environ Microbiol 22:397–412
Eakin CM, Sweatman HP, Brainard RE (2019) The 2014–2017 global-scale coral bleaching event: insights and impacts. Coral Reefs 38:539–545
Epstein HE, Torda G, van Oppen MJ (2019) Relative stability of the Pocillopora acuta microbiome throughout a thermal stress event. Coral Reefs 38:373–386
Fitt WK, McFarland FK, Warner ME, Chilcoat GC (2000) Seasonal patterns of tissue biomass and densities of symbiotic dinoflagellates in reef corals and relation to coral bleaching. Limnol Oceanogr 45:677–685
Gélin P, Fauvelot C, Mehn V, Bureau S, Rouzé H, Magalon H (2017) Superclone expansion, long-distance clonal dispersal and local genetic structuring in the coral Pocillopora damicornis type β in Reunion Island South Western Indian Ocean. PloS one 12:e0169692
Glasl B, Herndl GJ, Frade PR (2016) The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J 10:2280–2292
Glasl B, Smith CE, Bourne DG, Webster NS (2019) Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. PeerJ 7:e6377
Glasl B, Bongaerts P, Elisabeth NH, Hoegh-Guldberg O, Herndl GJ, Frade PR (2017) Microbiome variation in corals with distinct depth distribution ranges across a shallow–mesophotic gradient (15–85 m). Coral Reefs 36:447–452
Gleason M (1993) Effects of disturbance on coral communities: bleaching in Moorea. French Polynesia Coral Reefs 12(3–4):193–201
Glynn PW (1983) Extensive’bleaching’and death of reef corals on the Pacific coast of Panama. Environ Conserv 10:149–154
Green RH, Lowe RJ, Buckley ML, Foster T, Gilmour JP (2019) Physical mechanisms influencing localized patterns of temperature variability and coral bleaching within a system of reef atolls. Coral Reefs 38(4):759–771
Hernandez-Agreda A, Leggat W, Bongaerts P, Ainsworth TD (2016) The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. MBio. https://doi.org/10.1128/mBio.00560-16
Hoegh-Guldberg O (1999) Climate change, coral bleaching and the future of the world’s coral reefs. Ma Freshw Res 50:839–866
Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, Knowlton N, Eakin CM, Iglesias-Prieto R, Muthiga N, Bradbury RH, Dubi A, Hatziolos ME (2007) Coral Reefs Under Rapid Climate Change and Ocean Acidification. Science 318:1737–1742
Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R, Bridge TC, Butler IR, Byrne M, Cantin NE, Comeau S, Connolly SR, Cumming GS, Dalton SJ, Guillermo Diaz-Pulido C, Eakin M, Figueira WF, Gilmour JP, Harrison HB, Heron SF, Hoey AS, Hobbs J-PA, Hoogenboom MO, Kennedy EV, Kuo C-Y, Lough JM, Lowe RJ, Liu G, McCulloch MT, Malcolm HA, McWilliam MJ, Pandolfi JM, Pears RJ, Pratchett MS, Schoepf V, Simpson T, Skirving WJ, Sommer B, Torda G, Wachenfeld DR, Willis BL, Wilson SK (2017) Global warming and recurrent mass bleaching of corals. Nature 543:373–377
Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, Baird AH, Baum JK, Berumen ML, Bridge TC, Claar DC, Mark Eakin C, Gilmour JP, Graham NAJ, Harrison H, Hobbs J-PA, Hoey AS, Hoogenboom M, Lowe RJ, McCulloch MT, Pandolfi JM, Pratchett M, Schoepf V, Torda G, Wilson SK (2018) Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359:80–83
Hughes TP, Kerry JT, Connolly SR, Baird AH, Eakin CM, Heron SF, Hoey AS, Hoogenboom MO, Jacobson M, Liu G, Pratchett MS, Skirving W, Torda G (2019) Ecological memory modifies the cumulative impact of recurrent climate extremes. Nat Clim Change 9:40–43
Hume BC, Smith EG, Ziegler M, Warrington HJ, Burt JA, LaJeunesse TC, Wiedenmann J, Voolstra CR (2019) SymPortal: A novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol ecology resources 19:1063–1080
Jones RJ, Hoegh-Guldberg O, Larkum AW, Schreiber U (1998) Temperature-induced bleaching of corals begins with impairment of the CO2 fixation mechanism in zooxanthellae. Plant Cell Environ 21:1219–1230
King B, Wolanski E (1996) Tidal current variability in the Central Great Barrier Reef. J Mar Syst 9:187–202
Koren O, Rosenberg E (2006) Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Microbiol 72:5254–5259
LaJeunesse TC, Smith RT, Finney J, Oxenford H (2009) Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’event. Proc Royal Soc b: Biological Sciences 276:4139–4148
LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR (2018) Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol 28(2570–2580):e2576
Lema KA, Willis BL, Bourne DG (2012) Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl Environ Microbiol 78:3136–3144
Lema KA, Clode PL, Kilburn MR, Thornton R, Willis BL, Bourne DG (2016) Imaging the uptake of nitrogen-fixing bacteria into larvae of the coral Acropora millepora. ISME J 10:1804–1808
Lesser MP (1997) Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 16:187–192
Little AF, van Oppen MJH, Willis BL (2004) Flexibility in algal endosymbioses shapes growth in reef corals. Science 304:1492–1494
Littman R, Willis BL, Bourne DG (2011) Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ Microbiol Rep 3:651–660
Littman RA, Willis BL, Pfeffer C, Bourne DG (2009) Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol Ecol 68:152–163
Luick JL, Mason L, Hardy T, Furnas MJ (2007) Circulation in the Great Barrier Reef Lagoon using numerical tracers and in situ data. Cont Shelf Res 27:757–778
Marchioro GM, Glasl B, Engelen AH, Serrão EA, Bourne DG, Webster NS, Frade PR (2020) Microbiome dynamics in the tissue and mucus of acroporid corals differ in relation to host and environmental parameters. PeerJ 8:e9644
Marshall P, Baird A (2000) Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19(2):155–163
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531
Messer LF, Brown MV, Furnas MJ, Carney RL, McKinnon AD, Seymour JR (2017) Diversity and activity of diazotrophs in Great Barrier Reef surface waters. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00967
Meyer JL, Paul VJ, Teplitski M (2014) Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS One 9:e100316
Modolon F, Barno AR, Villela HDM, Peixoto RS (2020) Ecological and biotechnological importance of secondary metabolites produced by coral-associated bacteria. J Appl Microbiol 129:1441–1457
Morrow KM, Bourne DG, Humphrey C, Botté ES, Laffy P, Zaneveld J, Uthicke S, Fabricius KE, Webster NS (2015) Natural volcanic CO2 seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J 9:894–908
Muscatine L, Porter JW (1977) Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454–460
Nakajima Y, Chuang P-S, Ueda N, Mitarai S (2018) First evidence of asexual recruitment of Pocillopora acuta in Okinawa Island using genotypic identification. PeerJ 6:e5915
Neave MJ, Mitchell CT, Apprill A, Voolstra C (2017) Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci Rep 7:40579
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre, P, McGlinn D, Ohara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2017) Vegan: community, Ecology Package. R package version 2.4-5
Pernice M, Raina J-B, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR (2020) Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J 14:325–334
Pochon X, Pawlowski J, Zaninetti L, Rowan R (2001) High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Mar Biol 139:1069–1078
Pettay DT, Wham DC, Smith RT, Iglesias-Prieto R, LaJeunesse TC (2015) Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc Natl Acad Sci USA 112:7513–7518
Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR (2018) Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol 8:2240–2252
Pogoreutz C, Voolstra CR, Rädecker N, Weis V, Cardenas A, Raina J-B (2021) The coral holobiont highlights the dependence of cnidarian animal hosts on their associated microbes. In TCG. Bosch, MG, Hadfield (eds.), Cellular Dialogues in the Holobiont (pp. 91–118). CRC Press
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
R Development Core Team (2017) R: A Language and Environment for Statistical Computing (Vienna, Austria). R Foundation for Statistical Computing.
Rädecker N, Pogoreutz C, Gegner HM, Cárdenas A, Roth F, Bougoure J, Guagliardo P, Wild C, Pernice M, Raina JB, Meibom A (2021) Heat stress destabilizes symbiotic nutrient cycling in corals. Proc Natl Acad Sci USA 118(5):e2022653118
Raina J-B, Tapiolas D, Willis BL, Bourne DG (2009) Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75:3492–3501
Raina J-B, Tapiolas D, Motti CA, Foret S, Seemann T, Tebben J, Willis BL, Bourne DG (2016) Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ 4:e2275
Raina J-B, Tapiolas DM, Forêt S, Lutz A, Abrego D, Ceh J, Seneca FO, Clode PL, Bourne DG, Willis BL, Motti CA (2013) DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502:677–680
Robbins SJ, Singleton CM, Chan CX, Messer LF, Geers AU, Ying H, Baker A, Bell SC, Morrow KM, Ragan MA, Miller DJ, Forêt S, Voolstra CR, Tyson GW, Bourne DG (2019) A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat Microbiol 4:2090–2100
Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10
Ros M, Suggett DJ, Edmondson J, Haydon T, Hughes DJ, Kim M, Guagliardo P, Bougoure J, Pernice Ma, Raina J-B, Camp EF (2021) Symbiont shuffling across environmental gradients aligns with changes in carbon uptake and translocation in the reef-building coral Pocillopora acuta. Coral Reefs 40:595–607
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007) The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362
Schmidt-Roach S, Miller KJ, Lundgren P, Andreakis N (2014a) With eyes wide open: a revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zool J Linn Soc 170:1–33
Schmidt-Roach S, Johnston E, Fontana S, Jury CP, Forsman Z (2014b) Daytime spawning of Pocillopora species in Kaneohe Bay, Hawai ‘i. Galaxea, Journal of Coral Reef Studies 16:11–12
Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N, Gilbert Jack A (2014) Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. MBio. https://doi.org/10.1128/mBio.01371-14
Siebeck U, Marshall N, Klüter A, Hoegh-Guldberg O (2006) Monitoring coral bleaching using a colour reference card. Coral Reefs 25:453–460
Silverstein RN, Cunning R, Baker AC (2015) Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob Change Biol 21:236–249
Skirving W, Marsh B, De La Cour J, Liu G, Harris A, Maturi E, Erick Geiger C, Eakin M (2020) CoralTemp and the Coral Reef Watch Coral Bleaching Heat Stress Product Suite Version 31. Remote Sens 12:3856
Suggett DJ, Smith DJ (2020) Coral bleaching patterns are the outcome of complex biological and environmental networking. Glob Change Biol 26:68–79
Suzuki MT, Giovanni SJ (1996) Biased caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630
Swain TD, Vega-Perkins JB, Oestreich WK, Triebold C, DuBois E, Henss J, Baird A, Siple M, Backman V, Marcelino L (2016) Coral bleaching response index: a new tool to standardize and compare susceptibility to thermal bleaching. Glob Change Biol 22(7):2475–2488
Tandon K, Lu C-Y, Chiang P-W, Wada N, Yang S-H, Chan Y-F, Chen PY, Chang HY, Chiou YJ, Chou MS, Chen WM (2020) Comparative genomics: Dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J 14:1290–1303
Torda G, Lundgren P, Willis BL, van Oppen MJH (2013a) Genetic assignment of recruits reveals short- and long-distance larval dispersal in Pocillopora damicornis on the Great Barrier Reef. Mol Ecol 22:5821–5834
Torda G, Schmidt-Roach S, Peplow LM, Lundgren P, van Oppen MJ (2013) A rapid genetic assay for the identification of the most common Pocillopora damicornis genetic lineages on the Great Barrier Reef. PLoS One 8:e58447
Torres AF, Forsman ZH, Ravago-Gotanco R (2020) Shifts in coral clonality along a gradient of disturbance: insights on reproduction and dispersal of Pocillopora acuta. Mar Biol 167:1–18
Vanwonterghem I, Webster NS (2020) Coral reef microorganisms in a changing climate. Iscience 23:100972
Veal CJ, Holmes G, Nunez M, Hoegh-Guldberg O, Osborn J (2010) A comparative study of methods for surface area and three-dimensional shape measurement of coral skeletons. Limnol Oceanogr Meth 8:241–253
Voolstra CR, Ziegler M (2020) Adapting with microbial help: microbiome flexibility facilitates rapid responses to environmental change. BioEssays 42:2000004
Wickham H, Chang W (2015) ggplot2. URL: https://github.com/tidyverse/ggplot2
Wickham H, Averick M, Bryan J, Chang W, D’Agostino McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J (2019) Welcome to the Tidyverse. J Open Source Softw 4:1686–1691
Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR (2017) Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:1–8
Ziegler M, Grupstra CG, Barreto MM, Eaton M, BaOmar J, Zubier K, Al-Sofyani A, Turki AJ, Ormond R, Voolstra CR (2019) Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun 10:1–11
Zilber-Rosenberg I, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735
Acknowledgements
The authors acknowledge the Manbarra People of the Palm Island group as the Traditional Owners of the sea country where the sampling took place. We pay our respects to their elders, past, present and emerging and we acknowledge their continuing spiritual connection to their sea country. We thank Benjamin Hume for his interest in our work and his thorough explanation of the Symbiodiniaceae phylogeny, and Jean-Baptiste Raina for fruitful comments on the manuscript. Funding for this research was provided by the Australian Institute of Marine Science appropriation funds.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflict of interest.
Data availability
The sequencing data generated and analysed in this study can be found in the NCBI Sequence Reads Archive (https://www.ncbi.nlm.nih.gov/sra) under submission PRJNA743719.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Topic Editor Steve Vollmer
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Botté, E.S., Cantin, N.E., Mocellin, V.J.L. et al. Reef location has a greater impact than coral bleaching severity on the microbiome of Pocillopora acuta. Coral Reefs 41, 63–79 (2022). https://doi.org/10.1007/s00338-021-02201-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-021-02201-y