Abstract
Coral reefs are home to the greatest diversity of marine life, and many species on reefs live in symbiotic associations. Studying the historical biogeography of symbiotic species is key to unravelling (potential) coevolutionary processes and explaining species richness patterns. Coral-dwelling gall crabs (Cryptochiridae) live in obligate symbiosis with a scleractinian host, and are ideally suited to study the evolutionary history between heterogeneous taxa involved in a symbiotic relationship. The genus Opecarcinus Kropp and Manning, 1987, like its host coral family Agariciidae, occurs in both Indo-Pacific and Caribbean seas, and is the only cryptochirid genus with a circumtropical distribution. Here, we use mitochondrial and nuclear DNA gene fragments of Opecarcinus specimens sampled from 21 Indo-Pacific localities and one Atlantic (Caribbean) locality. We applied several species delimitation tests to characterise species diversity, inferred a Bayesian molecular-clock time-calibrated phylogeny to estimate divergence times and performed an ancestral area reconstruction. Time to the most recent common ancestor (tMRCA) of Opecarcinus is estimated at 15−6 Mya (middle Miocene—late Miocene). The genus harbours ~ 15 undescribed species as well as several potential species complexes. There are indications of strict host-specificity patterns in certain Opecarcinus species in the Indo-Pacific and Atlantic, however, a robust phylogeny reconstruction of Agariciidae corals—needed to test this further—is currently lacking. The Indo-West Pacific was inferred to be the most probable ancestral area, from where the Opecarcinus lineage colonised the Western Atlantic and subsequently speciated into O. hypostegus. Opecarcinus likely invaded from the Indo-West Pacific across the East Pacific Barrier to the Atlantic, before the full closure of the Isthmus of Panama. The subsequent speciation of O. hypostegus, is possibly associated with newly available niches in the Caribbean, in combination with genetic isolation following the closure of the Panama Isthmus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coral reefs are home to the greatest diversity of marine life, and many species on reefs live in symbiotic associations. Symbiosis plays a key role in maintaining the health and balance of diversity of reef systems (Stewart et al. 2006). The biodiversity of coral reefs is dominated by invertebrates, many of which rely on hosts for food, habitat, or settlement cues (Stella et al. 2011; Hoeksema et al. 2012). While the diversity, distribution, and relationships of some reef organisms are fairly well-studied, we know relatively less about coral symbionts other than zooxanthellae. The study of the historical biogeography of symbiont taxa is important for our understanding of the evolution of symbiotic relationships and their species richness gradients (Pinto-Ledezma et al. 2017).
While reefs and reef corals exist in all four tropical marine regions, they are best developed and most diverse in the Indo-West Pacific (IWP) and the West Atlantic (WA), and occur to a more limited extent in the East Pacific (EP) and East Atlantic (EA). The origin and evolution of reef biota in the two great reef regions have been complex. Within the IWP the Coral Triangle (CT) is the centre of marine biodiversity (Renema et al. 2008), and diversity of most marine organisms declines from there with both latitude (Ukuwela et al. 2016) and longitude (Miller et al. 2018). These diversity clines have long been studied and numerous hypotheses advanced to explain them (Rosen 1988; Paulay 1997; Bellwood et al. 2005; Huang et al. 2018).
Diversity in the IWP is about an order of magnitude greater than in the WA (Paulay 1997). Part of the biota of both regions have radiated in situ, while other lineages have not diversified since their arrival. In situ radiations dominate the IWP fauna, while migrant lineages that have not diversified are more common in the WA. In situ diversification is nevertheless common in the WA, and characterises much of the biota, as exemplified by several coral clades (Fukami et al. 2004, 2008), mithracid crabs (Windsor and Felder 2014), and cone snails (Kohn 2014). Other WA species represent isolated lineages that have not diversified within the basin (e.g. O’Hara et al. 2019).
WA lineages that have IWP ancestry range broadly in age. Phylogenetic analyses reveal that some species that range across the IWP and WA show little differentiation and are recently or currently connected (Collin et al. 2020). Other species that were thought to be so wide-ranging turned out to be cryptic complexes, with divergent lineages in the IWP and WA (Michonneau 2015; Dudoit et al. 2018). Many well-characterised and older WA endemics are nestled in IWP clades (O’Hara et al. 2019).
Some clades or lineages that range across the IWP and WA have attained their wide ranges by crossing the East Pacific Barrier (EPB) prior to when the Isthmus of Panama separated the EP and WA (Glynn and Ault 2000; Lessios and Robertson 2006; Baraf et al. 2019), others have colonised the WA around the Cape of Good Hope via the Benguela Current (Rocha et al. 2005; Andrews et al. 2016), and some have done both (Bowen et al. 2001). The more species-rich IWP has typically been the source for inter-regional dispersal, with some notable exceptions (Levinton et al. 1996; Huang et al. 2018).
To what extent is the diversification and distribution of symbiotic groups coordinated? Here we investigate the evolutionary dynamics of a crab lineage that is obligately symbiotic with stony corals. The modern scleractinian faunas of both IWP and WA are dominated by locally diversified lineages, such as the endemic Faviidae, Meandrinidae, and Agaricia Lamarck, 1801 of the WA, and most coral clades in the IWP. In contrast, local radiations appear to be common in coral-symbiotic crabs in the IWP, but not in the WA.
Several crab lineages have evolved obligate or facultative symbioses with scleractinian corals (Castro 2015), and these symbionts are much more diverse in the IWP than WA. Cryptochiridae and Domeciidae (not Maldivia Borradaile, 1902, which associates with gorgonians) include representatives in both the IWP and WA, while the Tetraliidae, Trapezia Latreille, 1828 (Trapeziidae), Tanaocheles Kropp, 1984 (Tanaochelidae) and Cymo De Haan, 1833 (Xanthidae) associate with scleractinians in the Indo-Pacific (Lai et al. 2009; Castro 2015). Currently 47 cryptochirid species have been described from the IWP, and only four are known from the WA in three genera, with one of these genera endemic to the WA (Kropp and Manning 1987; Ng et al. 2008; Van der Meij 2014b; Castro 2015; WoRMS 2021). Five domeciids are known from the IWP and only one from the WA (Castro et al. 2004). Thus it appears that symbiotic crabs may not have diversified within the WA, although this needs further testing given the high diversity of undiscovered and cryptic species in these groups (as we also demonstrate below) (Van Tienderen and Van der Meij 2017).
Our goal is to explore the diversity and distributional dynamics of the cryptochirid genus Opecarcinus, obligate symbionts of the scleractinian coral family Agariciidae. These crabs are a prime example of species living in obligate symbiosis with a scleractinian coral host (Castro 1988). Van der Meij and Schubart (2014) demonstrated that the Cryptochiridae is monophyletic, and their most recent common ancestor (MRCA) is estimated at 50–23 Mya (Van der Meij and Klaus 2015). The cryptochirid MRCA was previously estimated at ~ 83 Mya in a study on the infra-order Brachyura by Tsang et al. (2014), however, the clade containing the cryptochirid specimen has poor support. The Agariciidae currently includes seven genera that range across the IWP, EP, and WA, although ongoing taxonomic revisions will likely lead to changes in generic classification (Terraneo et al. 2017). Agariciidae are mostly zooxanthellate reef corals, common in tropical shallow-waters and also well represented in mesophotic reefs (Terraneo et al. 2017). The genera Agaricia and Helioseris Milne Edwards & Haime, 1849 are restricted to the WA; Leptoseris Milne Edwards and Haime, 1849 occurs in both the Indo-Pacific and WA, and the remaining four genera are limited to the IWP, with two (Pavona Lamarck, 1801 and Gardineroseris Scheer and Pillai, 1974) extending to the EP.
We explore the diversity of the genus using a multimarker dataset to assess how much undiscovered and cryptic diversity exists and where these additional species live. With a time-calibrated, multigene phylogeny we then explore how the diversity of this group has evolved across the tropical reefscape, with special attention to how WA and IWP species are related. Do Opecarcinus in these regions represent sister lineages or are they nested? What is the timing and likely route of colonisation?
Materials and Methods
Sample collection and data collection
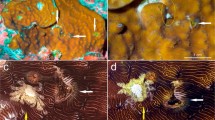
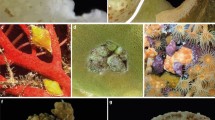
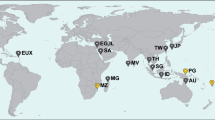
Species of Opecarcinus and Pseudohapalocarcinus ransoni Fize and Serène, 1956 (and cryptochirid outgroups) were collected from 21 localities in the IWP and WA (Fig. 1, Table S1), between 2006 and 2017. Nine species belonging to seven cryptochirid genera were chosen as outgroups according to Van der Meij and Nieman (2016). Specimens were photographed alive to document colour patterns, then fixed and stored in 80% ethanol. The material collected from the Red Sea, Maldives, Coral Triangle, Japan, New Caledonia, and Curaçao is deposited in Naturalis Biodiversity Center, Leiden, The Netherlands (RMNH), whereas specimens from the remaining localities are deposited in the Florida Museum of Natural History, University of Florida, Gainesville, USA (UF) (Table S1). Most sampled localities were extensively explored for gall crabs, with the exception of Japan, Taiwan, Hawaii and New Caledonia, for which a limited number of Opecarcinus specimens were available for analyses. DNA extractions, PCR and sequencing followed the protocol in Van der Meij (2015). Specimens were identified using Kropp (1989) and Van der Meij (2014b), using morphological characters combined with host and distribution data. Provisional names were assigned to species that did not fit established described taxa, using the prefix SET (for SET van der Meij) and a numeric designation. These names will be consistently applied to these OTUs in the future until a proper name is established for each.
Map of sampling sites, constructed in ArcGIS v10.5.1 (ESRI, Redlands, CA, USA). SAU–Saudi Arabia, Red Sea; MAD–Faafu Atoll, Maldives; LAX–Layang-Layang, Spratly Islands, Malaysia; TMP–Tun Mustapha Park, Kudat, N Borneo, Malaysia; SEM–Semporna, N Borneo, Malaysia; MEN–Manado, N Sulawesi, Indonesia; LEM–Lembeh, N Sulawesi, Indonesia; TER–Ternate, Halmahera, Indonesia; RAJ–Raja Ampat, Papua, Indonesia; RYU–Okinawa, Ryukyus, Japan; TWI–Taiwan Island; NC–New Caledonia; CAO–Curaçao. Hawaii includes Maui and Oahu; SE Polynesia includes Moorea; A, B and D are Scattered Islands, C is Nosy Be, Madagascar, and E is Réunion
Phylogenetic analyses and divergence time estimation
All analyses were performed on a concatenated Opecarcinus dataset of two mitochondrial genes (Cytochrome Oxidase I (COI) and 16S rRNA) and a nuclear gene (Histone H3). The total data set consisted of 1539 bp: 658 bp for COI, 594 bp for 16S rRNA and 287 bp for H3. The sequences of each marker were aligned separately using Clustal W 2.1 (Thompson et al. 1994) and then adjusted manually. All sequences were concatenated by Phylosuite 1.2.1 (Zhang et al. 2020); subsequently PartitionFinder 2 (Lanfear et al. 2017) was applied to find the best partition scheme for the complete dataset consisting of 230 terminals. The best-fit scheme corresponded with the markers (COI, 16S, H3) in the original dataset. PartitionFinder was also used to find the best-fit nucleotide substitution models for the respective partitions, based on the Bayesian Information Criterion (BIC; Schwarz 1978).
Bayesian Inference (BI) analyses and divergence time estimations were conducted on the concatenated data set in BEAST v1.10.4 (Suchard et al. 2018) by running the Markov chain for 100 × 106 steps iterations, sampling every 5000 iterations. The TN93 + Γ + I + X substitution model was applied to COI, while the best model for 16S and H3 was GTR + Γ + I + X. A Yule tree prior with default settings for the speciation rate and an uncorrelated relaxed clock with lognormal distribution were applied. Tracer v1.7.1 (Rambaut et al. 2018) was used to test for convergence, where Effective Sample Size (ESS) of all parameters exceeded 200. Maximum Clade Credibility (MCC) tree was obtained through TreeAnnotator v1.10.4, with the first 10% trees discarded as burn-in. The phylogeny reconstruction was visualised using Figtree v1.4.4 (Rambaut and Drummond 2018).
Calibration information for divergence time estimation can come from several sources, such as substitution rates, fossils, and geological data (Heath 2015). There are no known cryptochirid fossils (only trace fossils, see Klompmaker et al. 2016), hence substitution rates for each of the three gene fragments were used for calibration (Van der Meij and Klaus 2015). The priors for substitution rates were set as follows. Substitution rates of the COI locus in arthropods range between 0.7% and 2.0% per Myr (e.g. Schubart et al. 1998; Daniels et al. 2015). Here the mean rate of 1.17% per Myr for COI locus was used with an SD of 0.9%, and 95% highest posterior density (HPD) was from 0.20 to 2.69%. The base substitution rate of 16S rRNA was set to 1.09 ± 0.24% (mean ± SD) per Myr and 95% HPD was from 0.63 to 1.41%. Histone H3 was set to 0.19 ± 0.04% per Myr distribution and 95% HPD was from 0.12 to 0.26% (Van der Meij and Klaus 2015). Substitution rates for the latter two genes are derived from divergence time estimates of freshwater crabs from the Old World (Asia, Africa and Europe) based on three fossil calibration points (Klaus et al. 2010). All priors of gene fragments were calculated using a normal distribution.
In addition to the time-calibrated phylogenetic reconstruction, a ML analysis based on three concatenated markers (COI, 16S rRNA and H3) including Opecarcinus, Pseudohapalocarcinus ransoni and nine cryptochirid outgroups was conducted by IQ-TREE (Nguyen et al. 2015) for 10,000 ultrafast bootstraps (Minh et al. 2013). The best-fit nucleotide substitution model for each marker was GTR + I + G.
Three species delimitation tests were applied to the Opecarcinus dataset, separately for the COI and the three-marker concatenated dataset (Reid and Carstens 2012): (1) a General Mixed Yule-Coalescent (GMYC) approach (Fujisawa and Barraclough 2013) implemented with the R package ‘splits’ (Ezard et al. 2009; R Core Team 2020); (2) Automatic Barcode Gap Discovery (ABGD) (Puillandre et al. 2012); and (3) the Poisson Tree Processes (PTP) method (Zhang et al. 2013). The most conservative outcome from these three tests was used for delimiting Opecarcinus species (Table S1).
Ancestral area reconstruction
The Opecarcinus samples were collected from IWP and WA, and these two regions were applied to ancestral area reconstruction. To estimate ancestral ranges across the Opecarcinus phylogeny, a Maximum Clade Credibility (MCC) tree was implemented with BEAST using the same process as described above for the divergence time estimation. The best-fit nucleotide substitution model for 16S was GTR based on PartitionFinder 2. However, the eigenvalues did not converge, likely because the GTR model was applied to small partitions with too few taxa (Drummond and Bouckaert 2015), so HKY was used instead for 16S. Parametric methods (e.g. DEC and its extension; Yu et al. 2015) have been developed as a response to the shortcomings in event-based methods, which focus on integrating biogeographic processes and patterns (e.g. Dispersal-Vicariance Analysis, DIVA) (Ronquist 1997). Hence, ancestral range estimation was computed using the R package ‘BioGeoBEARS’ under the Dispersal-Extinction Cladogenesis model (DEC) (Ree et al. 2005; Ree and Smith 2008; R Core Team 2020). Considering the criticism of the DEC + j model (Ree and Sanmartín 2018), ‘jump’ speciation was not considered in our analyses.
Results
Phylogenetic inference and divergence time of Opecarcinus
The phylogenetic reconstruction and species delimitation tests recovered 25 species in Opecarcinus by all species delineation methods (Fig. 2, Table S1) and all with high branch support. Additional species were recovered by some, but not all, delineation methods within seven species: O. hypostegus Shaw and Hopkins, 1977, O. pholeter Kropp, 1989, O. SET7, O. SET8, O. SET12, O. SET14, and O. SET16 (Table S1). We treated each of these latter as single species.
Time-calibrated, three-marker MCC tree of Opecarcinus highlighting the diversity in the genus. Three inner circles (yellow to soft pink) in different colours are the results of species delimitation tests (GMYC, ABGD, and PTP) based on a single gene (COI), and three outer circles (blue to light green) are based on three concatenated markers. Photographs by Sancia van der Meij / Bastian Reijnen
Time to the Most Recent Common Ancestor (tMRCA) of Opecarcinus was estimated at 15–6 Mya (middle Miocene—late Miocene). Within Opecarcinus two main clades can be discerned (Fig. 2, Fig. S2). Clade I (tMRCA 12-4 Mya) contains two deeply divergent species: (1) Opecarcinus SET11 inhabiting Pavona venosa Ehrenberg, 1834 and P. varians Verrill, 1864 in the Red Sea and O. SET13 inhabiting various Pavona species and Gardineroseris planulata Dana, 1846 from the Red Sea to SE Polynesia.
Clade II (tMRCA 10-5 Mya) contains all other Opecarcinus species. Within this clade several groupings can be discerned, and several potential species complexes are revealed (Fig. 2, S2). Potential (cryptic) speciation and/or high levels of intraspecific genetic diversity is observed in O. pholeter, O. hypostegus, O. SET7, O. SET8, O. SET12, O. SET14, and O. SET16. Opecarcinus SET1, O. SET2 and O. SET8 all inhabit Leptoseris yabei Pillai & Scheer, 1976, however, there are no indications that L. yabei is a species complex (F. Benzoni, pers. comm). Opecarcinus cathyae van der Meij, 2014a, b, O. SET10, and O. SET19 inhabit Pavona minuta Wells, 1954, P. clavus Dana, 1846 and P. bipartita Nemenzo, 1979, whereas Opecarcinus lobifrons Kropp 1989 and O. pholeter inhabit Gardinoseris planulata and Pavona explanulata Lamarck, 1816, respectively. A well-supported clade containing Opecarcinus SET3, O. SET4, O. SET7, O. SET12, O. SET15, O. SET17, and O. SET18, associates with a range of Leptoseris and Pavona corals, similar to the remaining species Opecarcinus sierra Kropp, 1989, Opecarcinus peliops Kropp, 1989, O. SET9, O. SET14, and O. SET16. Opecarcinus peliops and O. SET9 are morphologically very similar and further work is needed to understand the morphological boundaries between the two species. The closely related species O. SET5 and O. SET6, inhabit various plate-forming Leptoseris and Pavona species. Both species are restricted to the Coral Triangle, and, interestingly, are sister taxa to the Atlantic species O. hypostegus inhabiting Agaricia and Helioseris (Van der Meij 2014a; Hoeksema 2017). The latter shows high levels of intraspecific divergence.
Ancestral area reconstruction
The IWP was recovered as the most probable ancestral area for Opecarcinus as a whole, as well as for all nodes within the genus (Fig. 3). Opecarcinus colonised the WA from the IWP, and speciated into O. hypostegus. Our divergence time estimation indicates that Opecarcinus colonised the Atlantic ca. 3.25 Mya (95% CI [2.07, 4.82]; Fig. S2).
Discussion
Diversity of Opecarcinus
Currently Opecarcinus contains nine described species (Van der Meij 2014b; WoRMS 2021), however, our results suggest that the genus includes at least 25 species (Figs. 2, S2, Table S1). These results will form the basis of a taxonomic revision of the genus. Moreover, substantial genetic variation in several species (e.g. O. pholeter, O. SET7, and O. SET12; see Figs. 2, S2), suggests further, potential cryptic species diversity, which warrants investigation. The only Atlantic species, O. hypostegus, is also a potential species complex (Fig. 2, S2). This cryptochirid inhabits species of Agaricia (Kropp and Manning, 1987; Van der Meij 2014a), and Helioseris cucullata Ellis and Solander, 1786 (Hoeksema et al. 2017). Our results are in line with those of Van Tienderen and Van der Meij (2017), who identified high levels of genetic divergence within this species, with significant genetic differentiation across its host species. The authors hypothesised that this differentiation may represent early signs of host speciation in O. hypostegus, but still considered this gall crab a single species.
Opecarcinus is strictly associated with the Agariciidae (Kropp 1989; Van der Meij 2014b). This coral family also hosts the monotypic gall crab genera Pseudohapalocarcinus and Luciades Kropp and Manning, 1996, neither of which occurs in the Atlantic. The position of P. ransoni is not fixed in the various reconstructions of the Cryptochiridae (e.g. Van der Meij and Klaus 2015; Van der Meij and Nieman 2016), however, our results show that it falls well within Opecarcinus with full support (Fig. S1), making Opecarcinus paraphyletic. Further study with additional markers, combined with morphological data, is needed to robustly place P. ransoni within Opecarcinus. No fresh material of Luciades agana Kropp and Manning, 1996 is currently available for genetic analyses, hence we cannot assess its phylogenetic position. However, the overall morphology of this species is similar to Opecarcinus (distinguished only by the lack of a distal expansion of pereopod 2), as is its association with the agariciid Leptoseris papyracea Dana, 1846, suggesting that L. agana may also fall within Opecarcinus (Kropp and Manning 1996; Komatsu and Takeda 2013).
The origin of Opecarcinus
Van der Meij and Klaus (2015) established the first time-calibrated phylogenetic reconstruction of Cryptochiridae, including species belonging to 17 of 21 described genera. They estimated the MRCA of Cryptochiridae at 50–23 Mya, much later than the estimated Middle Ordovician origin of Scleractinia (Seiblitz et al. 2020). Such a discrepancy in diversification between host and symbiont has been observed in other taxa, such as coral-dwelling gobies (Duchene et al. 2013). A more focussed approach studying gall crab species in a single genus, allowing for the study of biogeographic and host use patterns in more detail, has been lacking.
Our divergence time estimation indicates that the tMRCA for Opercarcinus is around 15–6 Mya (middle Miocene—late Miocene), in line with the results (11–5 Mya) of Van der Meij and Klaus (2015). Our ancestral area reconstruction based on samples collected from IWP and WA is the first analyses of the evolutionary history of Opecarcinus, and indicates the IWP as the area of origin.
Timing and route of colonisation
How did the largely endemic biotas of the major reef regions develop? Cryptochirids are most diverse in the IWP, and only four species are currently recorded from the WA. What are the origins of these WA gall crabs? Our results indicate that Opecarcinus hypostegus is a relatively recent (ca. 3.25 Mya) colonist in the WA from the IWP. Limited phylogenetic information on two other WA gall crabs (Kroppcarcinus siderastreicola Badaro, Neves, Castro and Johnsson, 2012, Troglocarcinus corallicola Verrill, 1908) suggests they also have sister taxa in the IWP (Van der Meij and Klaus 2015; Van der Meij and Nieman 2016). These studies did not aim to trace the origin and route of colonisation of the WA species. Van der Meij and Klaus (2015) estimated that K. siderastreicola and T. corallicola diverged early within their respective clades [36–15 Mya], at a time when the connection between the Atlantic/Mediterranean Sea and IWP across the Tethys seaway was still open and thus may have served as a colonisation route, in addition to the two other routes discussed below (Bialik et al. 2019).
Subsequent to the closure of the Tethys in the early Miocene, dispersal between the IWP and the Atlantic could occur around the Cape of Good Hope, or across the EPB prior to the rise of the Isthmus of Panama. Reef organisms appear to have utilised both paths. Atlantic populations that established after the closure of the Isthmus had to have dispersed around the Cape of Good Hope as has been demonstrated in brachyuran crabs (Guinot and Castro 2007; Rahayu and Ng 2014; Shih et al. 2016) and other organisms, such as Etelis Cuvier, 1828 snappers, Gnatholepis Bleeker, 1874 gobies, Stenopus Latreille, 1819 shrimp, and the sea star Valvaster Perrier, 1875, some potentially facilitated by unusual life history strategies, such as larval cloning (Rocha et al. 2005; Andrews et al. 2016; Dudoit et al. 2018; Collin et al. 2020).
The EPB is a semipermeable biogeographic barrier as evidenced by comparisons of populations across this vast expanse of open ocean. There are some examples from corals (Glynn and Ault 2000) and molluscs (Emerson and Chaney 1995). Dispersal from the IWP to the WA across the EPB prior to the closure of the Isthmus has been put forward to account for the presence of numerous marine taxa in the Atlantic (e.g. Barber and Bellwood 2005; Baraf et al. 2019), including several crabs (Harrison and Crespi 1999; Thiercelin and Schubart 2014; Magalhães et al. 2016).
Opecarcinus appears to have crossed the EPB, one of the world’s most potent marine biogeographic barriers, multiple times. Two Opecarcinus species are recorded from both the IWP and EP. Opecarcinus crescentus Edmondson, 1925 has been recorded from Vietnam, Palau and Johnston Island in the IWP (Garth 1965), and from Clipperton Island to the Gulf of California in the EP (Garth and Hopkins 1968). Opecarcinus lobifrons is known from the Red Sea to French Polynesia in the IWP, and Clipperton Atoll off the American mainland in the EP (Kropp 1989). Unfortunately, we lack samples from the EP, hence have not been able to assess the origin and diversity of Opecarcinus from this region directly. Opecarcinus SET5 and O. SET6, the likely sister taxa of O. hypostegus, are currently only known from the Pacific and not from the Indian Ocean. The estimated divergence time of O. hypostegus at ca. 3.25 Mya (Fig. S2) roughly coincides with the closure time of the Isthmus of Panama at 2.8 Mya (O’Dea et al. 2016), suggesting that this lineage could have arrived in the Atlantic by crossing the EPB, before the closure of the Isthmus of Panama.
Agariciids have a well-established fossil record in the Caribbean indicating that suitable hosts were available at the time Opecarcinus colonised the region. Pavona and Gardineroseris, currently restricted to the Indo-Pacific, have a Caribbean fossil record from the late Miocene to the Middle Pleistocene (Budd et al. 1994). The Caribbean endemic Agaricia first appeared in the Early to Middle Miocene (Budd 2000). Trace fossils (dwellings) of gall crabs are recorded from late Pliocene–Pleistocene corals from the WA, including Agaricia (Klompmaker et al. 2016). Given Opecarcinus’ high levels of host specificity, we hypothesise that gall crabs diverged over closely related coral species, and subsequently speciated through host-switching to newly available niches (i.e. Agaricia) in the Atlantic. This result is in contrast with a study on coral-associated hydrozoans of the genus Zanclea Gegenbaur, 1856, where the Caribbean harbours the same generalist hydrozoan species as the Indo-Pacific (Maggioni et al. 2020), highlighting the suitability of Cryptochiridae crabs for co-evolutionary studies.
References
Andrews KR, Williams AJ, Fernandez-Silva I, Newman SJ, Copus JM, Wakefield CB, Randall JE, Bowen BW (2016) Phylogeny of deepwater snappers (Genus Etelis) reveals a cryptic species pair in the Indo-Pacific and Pleistocene invasion of the Atlantic. Mol Phylogenet Evol 100:361-71
Baraf LM, Pratchett MS, Cowman PF (2019) Ancestral biogeography and ecology of marine angelfishes (F: Pomacanthidae). Mol Phylogenet Evol 140:106596
Barber PH, Bellwood DR (2005) Biodiversity hotspots: evolutionary origins of biodiversity in wrasses (Halichoeres: Labridae) in the Indo-Pacific and new world tropics. Mol Phylogenet Evol 35:235-253
Bellwood DR, Hughes TP, Connolly SR, Tanner J (2005) Environmental and geometric constraints on Indo-Pacific coral reef biodiversity Global coral reef biodiversity. Ecol Lett 8:643-651
Bialik OM, Frank M, Betzler C, Zammit R, Waldmann ND (2019) Two-step closure of the Miocene Indian Ocean Gateway to the Mediterranean. Sci Rep 9(1):1-10
Bowen BW, Bass AL, Rocha LA, Grant WS, Robertson DR (2001) Phylogeography of the trumpetfishes (Aulostomus): ring species complex on a global scale. Evolution (N Y) 55:1029-1039
Budd AF (2000) Diversity and extinction in the Cenozoic history of Caribbean reefs. Coral Reefs 19:25-35
Budd AF, Stemann TA, Johnson KG (1994) Stratigraphic distributions of genera and species of Neogene to recent caribbean reef corals. J Paleontol 68:951-977
Castro P (1988) Animal symbioses in coral reef communities: A review. Symbiosis 5:161-184
Castro P (2015) Symbiotic Brachyura. In: Treatise on zoology - anatomy, taxonomy, biology, Decapoda: Brachyura (P Castro, PJF Davie, D Guinot, FR Schram & JC von Vaupel Klein, eds.), Vol. 9C-I, pp. 543–581. Brill, Leiden, The Netherlands
Castro P, Ng PK, Ahyong ST (2004) Phylogeny and systematics of the Trapeziidae Miers, 1886 (Crustacea: Brachyura), with the description of a new family. Zootaxa 643(1):1-70
Collin R, Venera-Pontón DE, Paulay G, Boyle MJ (2020) World travelers: DNA barcoding unmasks the origin of cloning asteroid larvae from the Caribbean. Biol Bull 239:73-79
Cowman PF, Bellwood DR (2013) The historical biogeography of coral reef fishes: global patterns of origination and dispersal. J Biogeogr 40:209-224
Daniels SR, Phiri EE, Klaus S, Albrecht C, Cumberlidge N (2015) Multilocus phylogeny of the Afrotropical freshwater crab fauna reveals historical drainage connectivity and transoceanic dispersal since the Eocene. Syst Biol 64:549-567
Drummond AJ, Bouckaert RR (2015) Bayesian evolutionary analysis with BEAST. Cambridge University Press, Cambridge
Duchene D, Klanten SO, Munday PL, Herler J, van Herwerden L (2013) Phylogenetic evidence for recent diversification of obligate coral-dwelling gobies compared with their host corals. Mol Phylogenet Evol 69:123-132
Dudoit AA, Iacchei M, Coleman RR, Gaither MR, Browne WE, Bowen BW, Toonen RJ (2018) The little shrimp that could: phylogeography of the circumtropical Stenopus hispidus (Crustacea: Decapoda), reveals divergent Atlantic and Pacific lineages. PeerJ 6:e4409
Emerson W, Chaney H (1995) A zoogeographic review of the Cypraeidae (Mollusca: Gastropoda) occurring in the eastern Pacific Ocean. Veliger 38:8-21
Ezard T, Fujisawa T, Barraclough T (2009) Splits: species’ limits by threshold statistics. https://rdrr.io/rforge/splits/man/splits-package.html
Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the generalized mixed yule coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol 62:707-724
Fukami H, Budd A, Paulay G, Solé-Cava A, Chen C, Iwao K, Knowlton N (2004) Conventional taxonomy obscures deep divergence between Pacific and Atlantic Corals. Nature 427:832-835
Fukami H, Chen CA, Budd AF, Collins A, Wallace C, Chuang YY, Chen C, Dai CF, Iwao K, Sheppard C, Knowlton N (2008) Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (Order Scleractinia, Class Anthozoa, Phylum Cnidaria). PloS one 3:e3222
Garth JS (1965) The brachyuran decapod crustaceans of Clipperton Island. Proc Calif Acad Sci 33:1-46
Garth JS, Hopkins TS (1968) Pseudocryptochirus crescentus (Edmondson), a second crab of the corallicolous family Hapalocarcinidae (Crustacea, Decapoda) from the Eastern Pacific with remarks of phragmosis, host specificity, and distribution. Bull South Calif Acad Sci 67:40-48
Glynn PW, Ault JS (2000) A biogeographic analysis and review of the far eastern Pacific coral reef region. Coral Reefs 19:1-23
Guinot D, Castro P (2007) A new species of Goneplax Leach, 1814 (Crustacea, Decapoda, Brachyura, Goneplacidae) from the south Atlantic and the western limits of the Indo-West Pacific region, long confused with G. rhomboides (Linnaeus, 1758). Zootaxa 1577(1):17–31
Harrison MK, Crespi BJ (1999) Phylogenetics of Cancer Crabs (Crustacea: Decapoda: Brachyura). Mol Phylogenet Evol 12:186-199
Heath TA (2015) Divergence time estimation using BEAST v2. Dating species divergences with the fossilized birth-death process.
Hoeksema BW, van Beusekom M, Harry A, Ivanenko VN, van der Meij SET, van Moorsel GW (2017) Helioseris cucullata as a host coral at St. Eustatius, Dutch Caribbean. Mar Biodivers 47:71-78
Hoeksema BW, van der Meij SET, Fransen CHJM (2012) The mushroom coral as a habitat. J Mar Biol Assoc U K 92:647-663
Hoeksema JD, Bever JD, Chakraborty S, Chaudhary VB, Gardes M, Gehring CA, Hart MM, Housworth EA, Kaonongbua W, Klironomos JN, Lajeunesse MJ, Meadow J, Milligan BG, Piculell BJ, Pringle A, Rúa MA, Umbanhowar J, Viechtbauer W, Wang Y-W, Wilson GWT, Zee PC (2018) Evolutionary history of plant hosts and fungal symbionts predicts the strength of mycorrhizal mutualism. Commun Biol 1:116
Huang D, Goldberg EE, Chou LM, Roy K (2018) The origin and evolution of coral species richness in a marine biodiversity hotspot. Evolution 72:288-302
Klaus S, Schubart CD, Streit B, Pfenninger M (2010) When Indian crabs were not yet Asian - biogeographic evidence for Eocene proximity of India and Southeast Asia. BMC Evol Biol 10:287
Klompmaker AA, Portell RW, van der Meij SET (2016) Trace fossil evidence of coral-inhabiting crabs (Cryptochiridae) and its implications for growth and paleobiogeography. Sci Rep 6:23443
Kohn AJ (2014) Conus of the Southeastern United States and Caribbean. Princeton University Press, Princeton
Komatsu H, Takeda M (2013) Second record of Luciades agana Kropp and Manning, 1996 (Crustacea, Decapoda, Cryptochiridae) from Tonga, South Pacific. Bull Natl Mus Nat Sci Ser A 39:11-14
Kropp RK (1989) A revision of the Pacific species of gall crabs, genus Opecarcinus (Crustacea: Cryptochiridae). Bull Mar Sci 45:98-129
Kropp RK, Manning RB (1987) The Atlantic gall crabs, family Cryptochiridae (Crustacea: Decapoda: Brachyura). Smithson Contrib Zool:1–21
Kropp RK, Manning RB (1996) Crustacea Decapoda: two new genera and species of deep water gall crabs from the Indo-west Pacific (Cryptochiridae). Résultats des Campagnes MUSORSTOM 15:531-539
Lai JC, Ahyong ST, Jeng MS, Ng PK (2009) Are coral-dwelling crabs monophyletic? A phylogeny of the Trapezioidea (Crustacea: Decapoda: Brachyura). Invertebr Syst 23(4):402-8
LaJeunesse TC (2004) “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene Transition. Mol Biol Evol 22:570-581
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34:772-773
Leigh EG, O'Dea A, Vermeij GJ (2014) Historical biogeography of the Isthmus of Panama. Biol Rev Camb Philos Soc 89:148-172
Lessios HA (2008) The great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu Rev Ecol Evol Syst 39:63-91
Lessios HA, Robertson DR (2006) Crossing the impassable: genetic connections in 20 reef fishes across the eastern Pacific barrier. Proc Biol Sci 273:2201-2208
Levinton J, Sturmbauer C, Christy J (1996) Molecular data and biogeography: resolution of a controversy over evolutionary history of a pan-tropical group of invertebrates. J Exp Mar Biol Ecol 203:117-131
Magalhães T, Robles R, Felder DL, Mantelatto FL (2016) Integrative taxonomic study of the purse crab genus Persephona Leach, 1817 (Brachyura: Leucosiidae): combining morphology and molecular data. PLoS One11(4):e0152627
Maggioni D, Arrigoni R, Seveso D, Galli P, Berumen ML, Denis V, Hoeksema BW, Huang D, Manca F, Pica D (2020) Evolution and biogeography of the Zanclea-Scleractinia symbiosis. Coral Reefs:1–17
Michonneau F (2015) Cryptic and not-so-cryptic species in the complex “Holothuria (Thymiosycia) imaptiens” (Forsskål, 1775) (Echinodermata: Holothuroidea: Holothuriidae). Biorxiv:014225
Miller EC, Hayashi KT, Song D, Wiens JJ (2018) Explaining the ocean's richest biodiversity hotspot and global patterns of fish diversity. Proc Biol Sci 285:20181314
Minh BQ, Nguyen T, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188-1195
Ng PK, Guinot D, Davie PJ (2008) Systema Brachyurorum: Part I. An annotated checklist of extant brachyuran crabs of the world. Raffles Bull Zool 17(1):1–286
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268-274
O'Dea A, Lessios HA, Coates AG, Eytan RI, Restrepo-Moreno SA, Cione AL, Collins LS, de Queiroz A, Farris DW, Norris RD, Stallard RF, Woodburne MO, Aguilera O, Aubry M-P, Berggren WA, Budd AF, Cozzuol MA, Coppard SE, Duque-Caro H, Finnegan S, Gasparini GM, Grossman EL, Johnson KG, Keigwin LD, Knowlton N, Leigh EG, Leonard-Pingel JS, Marko PB, Pyenson ND, Rachello-Dolmen PG, Soibelzon E, Soibelzon L, Todd JA, Vermeij GJ, Jackson JBC (2016) Formation of the Isthmus of Panama. Sci Adv 2:e1600883-e1600883
O'Hara TD, Hugall AF, Cisternas PA, Boissin E, Bribiesca-Contreras G, Sellanes J, Paulay G, Byrne M (2019) Phylogenomics, life history and morphological evolution of ophiocomid brittlestars. Mol Phylogenet Evol 130:67-80
Paulay G (1997) Diversity and distribution of reef organisms. In C. Birkeland, ed., Life. and Death of Coral Reefs, pp. 298–353
Pinto-Ledezma JN, Simon LM, Diniz-Filho JAF, Villalobos F (2017) The geographical diversification of Furnariides: the role of forest versus open habitats in driving species richness gradients. J Biogeogr 44:1683-1693
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol 21:1864-1877
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Rahayu DL, Ng PK (2014) New genera and new species of Hexapodidae (Crustacea, Brachyura) from the Indo-West Pacific and east Atlantic. Raffles Bull Zool 4:62
Rambaut A (2018) FigTree v1. 4.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol 67:901-904
Ree RH, Moore BR, Webb CO, Donoghue MJ (2005) A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59:2299-2311
Ree RH, Sanmartín I (2018) Conceptual and statistical problems with the DEC+J model of founder-event speciation and its comparison with DEC via model selection. J Biogeogr 45:741-749
Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol 57:4-14
Reid NM, Carstens BC (2012) Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol Biol 12:196
Renema W, Bellwood DR, Braga JC, Bromfield K, Hall R, Johnson KG, Lunt P, Meyer CP, McMonagle LB, Morley RJ, Dea A, Todd JA, Wesselingh FP, Wilson MEJ, Pandolfi JM (2008) Hopping hotspots: global shifts in marine biodiversity. Science 321:654
Rocha LA, Robertson DR, Rocha CR, Van Tassell JL, Craig MT, Bowen BW (2005) Recent invasion of the tropical Atlantic by an Indo‐Pacific coral reef fish. Mol Ecol 14:3921-3928
Ronquist F (1997) Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst Biol 46:195-203
Rosen BR (1988) Progress, problems and patterns in the biogeography of reef corals and other tropical marine organisms. Helgoländer Meeresuntersuchungen 42:269-301
Schubart CD, Diesel R, Hedges SB (1998) Rapid evolution to terrestrial life in Jamaican crabs. Nature 393:363-365
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461-464
Seiblitz IGL, Capel KCC, Stolarski J, Quek ZBR, Huang D, Kitahara MV (2020) The earliest diverging extant scleractinian corals recovered by mitochondrial genomes. Sci Rep 10:20714
Spalding MD, Fox HE, Allen GR, Davidson N, Ferdaña ZA, Finlayson M, Halpern BS, Jorge MA, Lombana A, Lourie SA, Martin KD, McManus E, Molnar J, Recchia CA, Robertson J (2007) Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience 57:573-583
Shih HT, Ng PK, Davie PJ, Schubart CD, Türkay M, Naderloo R, Jones D, Liu MY (2016) Systematics of the family Ocypodidae Rafinesque, 1815 (Crustacea: Brachyura), based on phylogenetic relationships, with a reorganization of subfamily rankings and a review of the taxonomic status of Uca Leach, 1814, sensu lato and its subgenera. Raffles Bull Zool 64
Stella J, Pratchett M, Hutchings P, Jones G (2011) Coral-associated invertebrates: diversity, ecological importance and vulnerability to disturbance. Oceanogr. Mar. Biol 49:43-104
Stewart HL, Holbrook SJ, Schmitt RJ, Brooks AJ (2006) Symbiotic crabs maintain coral health by clearing sediments. Coral Reefs 25:609-615
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4
Terraneo TI, Arrigoni R, Benzoni F, Tietbohl MD, Berumen ML (2017) Exploring the genetic diversity of shallow-water Agariciidae (Cnidaria: Anthozoa) from the Saudi Arabian Red Sea. Mar Biodivers 47:1065-1078
Thiercelin N, Schubart CD (2014) Transisthmian differentiation in the tree-climbing mangrove crab Aratus H. Milne Edwards, 1853 (Crustacea, Brachyura, Sesarmidae), with description of a new species from the tropical eastern Pacific. Zootaxa 3793(5):545-60
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673-4680
Tsang LM, Schubart CD, Ahyong ST, Lai JC, Au EY, Chan TY, Ng PK, Chu KH (2014) Evolutionary history of true crabs (Crustacea: Decapoda: Brachyura) and the origin of freshwater crabs. Mol Biol Evol 31:1173-1187
Ukuwela K, Lee M, Rasmussen A, Silva A, Mumpuni M, Ghezellou P, Rezaie-Atagholipour M, Sanders K (2016) Evaluating the drivers of Indo-Pacific biodiversity: Speciation and dispersal of sea snakes (Elapidae: Hydrophiinae). J Biogeogr 43:243–255
van der Meij SET (2014a) Host species, range extensions, and an observation of the mating system of Atlantic shallow-water gall crabs (Decapoda: Cryptochiridae). Bull Mar Sci 90:1001-1010
van der Meij SET (2014b) A new species of Opecarcinus Kropp & Manning, 1987 (Crustacea: Brachyura: Cryptochiridae) associated with the stony corals Pavona clavus (Dana, 1846) and P. bipartita Nemenzo, 1980 (Scleractinia: Agariciidae). Zootaxa 3869:44-52
van der Meij SET (2015) Host relations and DNA reveal a cryptic gall crab species (Crustacea: Decapoda: Cryptochiridae) associated with mushroom corals (Scleractinia: Fungiidae). Contrib Zool 84:39-57
van der Meij SET, Klaus S (2015) Evolutionary diversification of coral-dwelling gall crabs (Cryptochiridae). PhD thesis, Naturalis Biodiversity Center, Faculty of Science, Leiden University, pp 79 - 86
van der Meij SET, Nieman AM (2016) Old and new DNA unweave the phylogenetic position of the eastern Atlantic gall crab Detocarcinus balssi (Monod, 1956) (Decapoda: Cryptochiridae). J Zool Syst Evol Res 54:189-196
van der Meij SET, Schubart CD (2014) Monophyly and phylogenetic origin of the gall crab family Cryptochiridae (Decapoda: Brachyura). Invertebr Syst 28:491–500
van Tienderen KM, van der Meij SET (2017) Extreme mitochondrial variation in the Atlantic gall crab Opecarcinus hypostegus (Decapoda: Cryptochiridae) reveals adaptive genetic divergence over Agaricia coral hosts. Sci Rep 7:39461
Windsor AM, Felder DL (2014) Molecular phylogenetics and taxonomic reanalysis of the family Mithracidae MacLeay (Decapoda: Brachyura: Majoidea). Invertebr Syst 28:145-173
WoRMS (2021) Cryptochiridae Paulson, 1875. Accessed at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=106753 on 2021–06–09
Yu Y, Harris AJ, Blair C, He X (2015) RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol Phylogenet Evol 87:46-49
Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour 20:348-355
Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869-2876
Acknowledgements
We thank Francesca Benzoni (KAUST) for providing background information on the systematics of the Agariciidae, and Jorn Claassen (RUG) for help with BioGeoBEARS. The fieldwork in Indonesia was organised by Naturalis and the Research Centre for Oceanography (RCO‐LIPI), under the umbrella of Ekspedisi Widya Nusantara (E‐Win). We are grateful to LIPI and RISTEK for granting research permits. The fieldwork in Malaysia was WWF‐Malaysia, Universiti Malaysia Sabah's Borneo Marine Research Institute, and Universiti Malaya's Institute of Biological Sciences, Sabah Parks and funded through WWF‐Malaysia, the Ministry of Science, Technology and Innovation (MOSTI) and USAID CTSP (Coral Triangle Support Partnership). The research permits were granted by the Economic Planning Unit, Prime Minister's Department and Sabah Biodiversity Centre. Collecting in New Caledonia was done during the mission CORALCAL 4. Provinces Sud and Nord of New Caledonia provided sampling permits. Loyalty Island samples were collected during the BIBELOT campaign in 2014 onboard RV Alis of IRD at Nouméa. Sampling permits were granted by the Loyalty Islands Province, New Caledonia. For the samples from the Maldives, the help of the University of Milano‐Bicocca Marine Research and High Education Centre in Magoodhoo, the Ministry of Fisheries and Agriculture, Republic of Maldives, and the community of Magoodhoo, Faafu Atoll is gratefully acknowledged. Fieldwork in the Red Sea was supported by the King Abdullah University of Science and Technology under the Biodiversity in the Saudi ArabianRed Sea program, award number CRG-1-BER-002 to Michael Berumen. Support for the various fieldwork trips by SETM was provided by Van Tienhoven Foundation for International Nature Protection, Stichting Fonds C. Tussenbroek (Nell Ongerboerfonds), Academy Ecology Fund (KNAW), LUF International Study Fund (Leiden University), TREUB-maatschappij, L.B. Holthuisfonds, J.‐J. ter Pelkwijkfonds, and A. M. Buitendijkfonds. We are also grateful for the scholarship of TX, provided by the China Scholarship Council (CSC), and NSF DEB 1856245 for funding GP.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Topic Editor Francesca Benzoni (francesca.benzoni@kaust.edu.sa)
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, T., Bravo, H., Paulay, G. et al. Diversification and distribution of gall crabs (Brachyura: Cryptochiridae: Opecarcinus) associated with Agariciidae corals. Coral Reefs 41, 699–709 (2022). https://doi.org/10.1007/s00338-021-02163-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-021-02163-1