Abstract
In this study, we delved into the interaction between corallivorous marine gastropods, the muricid Coralliophilinae Chenu, 1859, and their cnidarian food targets. Coralliophilinae is a subfamily of specialised corallivorous caenogastropods that feed by browsing on octocorals or hexacorals. Only sparse information is available on the phylogenetic relationships and the degree of specificity of the trophic relationships within this corallivorous lineage. To address these gaps, we generated the largest molecular dataset to date, comprising two mitochondrial (cox1 and 16S rDNA) and one nuclear gene (ITS2 rDNA) from 586 specimens collected worldwide. The coral hosts of coralliophilines were identified through an integrative approach, combining literature data with new records, employing morphological and/or molecular markers, and incorporating data from DNA barcoding of the snail stomach content. Our comprehensive approach unveiled the existence of numerous cryptic species in Coralliophilinae, while the phylogeny showed that most of the currently accepted genera are not monophyletic. The molecular dating confirmed the origin of the Coralliophilinae in Middle Eocene, with diversification of most lineages during the Miocene. Our results indicate that the subfamily’s ancestor evolved in shallow waters in association with Scleractinia. Through the evolutionary history of Coralliophilinae, multiple host shifts to other cnidarian orders were observed, not correlated with changes in the depth range. The results of diversification analyses within the subfamily further suggest that the association with the host has influenced the evolutionary patterns of Coralliophilinae, but not vice versa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Symbiotic interactions play crucial roles in marine ecosystems and are fundamental in shaping community dynamics (Pita et al. 2018). Asymmetric symbiotic interactions (such as host–parasite) may involve co-evolutionary patterns characterised by a delicate balance of adaptations and counter-adaptations between host and parasite (Van Der Laan and Hogeweg 1997) or reflect a sequential evolution mechanism (Jermy 1976), where the evolution of the host influences the evolution of the parasite, but not vice versa. Exploring the evolution of symbiosis necessitates reliable data on the associations as well as a robust systematic and phylogenetic framework of the groups involved in such associations.
Coral reefs have been recognised as exceptionally diverse ecosystems, sustaining multiple symbiotic interactions (Stella et al. 2011). The vulnerability of cnidarians to climate change implies that the threats currently affecting oceans could not only affect their global decline but also have profound repercussions on the associated fauna (Pandolfi et al. 2003). Various gastropod groups have evolved the ability to feed on corals, despite their inherent toxicity, providing intriguing models for studying the evolutionary ecology of the association with corals. In particular, the muricid subfamily Coralliophilinae Chenu, 1859, is a highly diverse lineage of Neogastropods, currently comprising 268 extant species (WoRMS 2024). Coralliophilinae are distributed worldwide, mostly in warm temperate and tropical oceans (Oliverio et al. 2009b). All species for which the ecology is known exhibit symbiotic relationships (ecto or endobiotic) with anthozoans, including sea-anemones, soft corals, and scleractinians, on which they feed (Fig. 1). While some snail species feed on solitary polyps, others are associated with colonial anthozoans, with exceptional cases of species able to swap between solitary and colonial hexacorals (Oliverio 2008; Oliverio and Mariottini 2001).
Living specimens of coralliophiline species on their hosts. a Galeropsis monodontus on Pocilloporidae, Papua New Guinea. b Coralliophila radula on Poritidae, New Caledonia. c Coralliophila rubrococcinea on Gorgoniidae, Philippines. d Coralliophila violacea on Poritidae, Kenya. e Coralliophila meyendorffii on Parazoanthidae, Italy. f Leptoconchus sp. in Merulinidae, Vanuatu. g Leptoconchus sp. in Fungiidae, Vanuatu. Photograph credits: a Laurent Charles (MNHN); b Philippe Maestrati (MNHN); c Guido Poppe (http://www.www.poppe-images.com); d–e Paolo Mariottini (University of Roma Tre); f–g Anne-Lise Fleddum (MNHN)
For some coralliophiline species, significant impact of their trophic habits on coral reef communities has been reported (Hayes and Bush 1990). However, limited information is so far available regarding the level of specificity and the adaptive mechanisms underlying coralliophiline-cnidarian interactions (e.g. Oliverio et al. 2009a, b). While common in shallow waters, Coralliophilinae are better represented in the deep-water ecosystems, particularly in the mesophotic zone, but also in bathyal and abyssal habitats. Although Oliverio et al. (2009b) suggested that coralliophilines might have repeatedly invaded deep habitats, a formal analysis of the ancestral habitat for the subfamily has not been conducted. The monophyly of Coralliophilinae is strongly supported and their most probable sister groups has been identified in the Rapaninae and Ergalataxinae (Russini et al. 2023). However, the clarification of the evolutionary relationships within the subfamily has been complicated by the remarkable interspecific variation of Coralliophilinae shell features (Richter and Luque 2002). Preliminary molecular studies of Coralliophilinae phylogenetic relationships (Oliverio and Mariottini 2001; Oliverio et al. 2002a, b, 2009a) were penalised by a limited taxon sampling.
To establish a robust phylogenetic framework for taxonomic and macroevolutionary investigations of the subfamily Coralliophilinae, we have generated the most extensive molecular dataset to date for this subfamily. Additionally, we assembled a comprehensive dataset containing association data between coralliophiline snails and their cnidarian hosts. This involved the integration of literature data with collection data pertaining to the specimens used in our analyses. Moreover, DNA barcoding of the stomach content of the snails, or of the coral host sample, when collected with the molluscs, has been incorporated in our dataset.
In this study, phylogeny and association data were integrated with data on the depth range of the examined coralliophiline species. By mapping trophic ecology and habitat data on the phylogenetic hypothesis, we carried out macroevolutionary analyses aiming to:
-
(1)
Reconstruct the evolution of the trophic ecology in Coralliophilinae, by identifying the ancestral host for the subfamily and the occurrence of host shifts across the phylogenetic tree
-
(2)
Test the hypothesis that the Coralliophilinae originated in shallow waters, with subsequent repeated colonisations of deep habitats
-
(3)
Investigate the occurrence of clade-specific rate shifts across the Coralliophilinae radiation, which might be suggestive of a diversification driven by the acquisition of key adaptive innovations.
Material and methods
Material examined
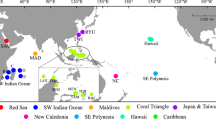
The dataset consisted of 586 specimens (plus five for the out-group), morphologically ascribed to 111 species belonging to 10 of the 13 accepted extant genera of Coralliophilinae (WoRMS 2023). Specimens from tropical, subtropical and temperate areas were included (Fig. 2). A total of 74 specimens were collected by one of the authors (M.O.) or kindly provided by Francisco Otero of Las Palmas de Gran Canaria University. Two samples were provided by the Florida Museum (UF), one by the Museum of New Zealand (NMZN) and one by KwaZulu-Natal Museum (NMSA). The majority of samples (350) belonged to the Muséum national d’Histoire Naturelle (MNHN), Paris (catalogue numbers MNHN-IM), having been collected during several scientific expeditions (www.expeditions.mnhn.fr). Sequences of 163 additional specimens were obtained from GenBank.
Global map featuring sampling locations of the Coralliophilinae dataset created using QGIS. See Table S1 for corresponding locality identifiers
Samples were collected from 0 m to approximately 1000 m depth and fixed in the field specifically for molecular analyses. The majority of shells were kept intact for identification and deposited as vouchers in the collections of MNHN, NMZN, UF, NMSA and the Department of Biology and Biotechnologies ‘Charles Darwin’ (BAU). In some cases, coralliophiline samples have been collected with their host and stored together. When the host was missing and the coralliophiline whole body was available, the stomach and foregut were dissected and processed for the amplification of cnidarian DNA directly from the gut content (Oliverio et al. 2009a) using cnidarian primer sets. Sequences from five Muricidae species were used as out-groups: Orania rosea (Houart, 1996); Pascula darrosensis (Smith, 1884); Claremontiella nodulosa (Adams, 1845); Semiricinula squamosa (Pease, 1868); Mancinella herberti (Houart, 1998). For details on samples and GenBank accession numbers, see the Supporting Information (Table S1).
Molecular analyses and gastropod species delimitation
Laboratory work was carried out in part at the Service de Systématique Moléculaire (UAR 2AD, MNHN, Paris, France) and in part at Molecular Systematic Laboratory, Department of Biology and Biotechnologies ‘Charles Darwin’, Sapienza University of Rome, Italy. For samples processed at MNHN, total genomic DNA was extracted from the foot and, when available, also from the stomach, using the Macherey–Nagel NucleoSpin 96 Tissue Kit and following the manufacturer’s protocol. For samples processed at Sapienza University of Rome, total genomic DNA was extracted using ‘salting out’ protocols (Fassio et al. 2023). Cnidarians DNA extraction was performed following Nocella et al. (2024). Polymerase chain reactions were performed following Fassio et al. (2022). In both laboratories, the same primers and amplification protocols were used. For molecular analysis and gastropod species delimitation details, see Supplementary Part 1.
Cnidarian host identification and depth range
Cnidarians samples collected with the associated coralliophilines (for a few of which field observation and/or photographs were also available) were observed under a Wild-M6 microscope. Our examination focused on the arrangement and distribution of polyps and/or the calcareous skeleton. Taxa were identified following Fabricius and Alderslade (2001), Zibrowius (1980), and Veron (2000). To gather anatomical data, alcohol-preserved specimens were observed and dissected under a Wild-M6 microscope.
All 16S and ITS2 cnidarian nucleotide sequences, either obtained from the cnidarian samples associated with the snails or from the stomach contents, were matched with sequences available in Genbank with the NCBI BLAST web interface (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Molecular taxonomic attributions were preliminarily determined using the BLAST taxonomy lineage report, taking in consideration all IDs with per cent of identity ≥ 89% and e-value ≤ 4e−04. Preliminary taxonomic identification was then cross-referenced for consistency with morphology (when the host sample was available) and geographical range. Cnidarian identification was conducted to the lowest taxonomic level possible and in all cases at least to the family level.
Recognising the considerable challenges in morphologically identifying coralliophiline and cnidarian species, literature records of coralliophiline-coral associations were considered reliable only when supported by morphological (on actual specimens or field photographs) or genetic identification pertaining to both gastropods and cnidarians.
The habitat of each species (deep v. shallow) was determined based on literature data (e.g. Oliverio 2009; Marshall and Oliverio 2009), on collection data of the assayed specimens, and on M.O. pers. observations. Species have been assigned to a shallow habitat (“S”, in Table S1) if living specimens are consistently collected in the euphotic zone (including cases with occasional collections of living specimens in the upper mesophotic, or empty shells from deeper habitats). Species have been assigned to a deep habitat (“D”, in Table S1) if living specimens are collected in the mesophotic or aphotic zone.
Phylogenetic reconstruction and temporal calibration
Phylogenetic analyses were performed on the three single-gene datasets (cox1 partitioned by codon, 16S and ITS2) and on the combined dataset (cox1 partitioned by codon + 16S + ITS2) with both maximum likelihood (ML) and Bayesian inference (BI). For the combined dataset, only the specimens with at least the cox1 data were kept. Sequences alignment, maximum likelihood and Bayesian analyses were performed following Nocella et al. (2024).
We identified four reliable fossil records of Coralliophilinae in the literature suitable for our phylogeny (Table 1). The first appearance of this neogastropod lineage is reported for the Middle Eocene of Mississippi and Louisiana in the Claiborne formation (42–44 mya; Dockery 1986) with the earliest known species attributed to the subfamily, Coralliophila (Timothia) aldrichi (Cossmann, 1903). The oldest known record of the genus Galeropsis Hupé, 1860, is from the Aquitanian, Early Miocene (20.44–23.03 mya) with Galeropsis lavenayanus Hupé, 1860 (Lozouet and Renard 1988). Coralliophila richardi P. Fischer, 1882, and Hirtomurex squamosus Bivona & Bernardi, 1838, are witnessed by the early Pleistocene in the Mediterranean Sea (0.77–2.58 mya; Vazzana 1996).
We selected a single specimen (indicated by an asterisk in Table S1) for each SSH retrieved with the integrative approach, to create a new combined dataset comprising 128 samples (including the out-group). Temporal calibration analysis was performed following Nocella et al. (2024).
Diversification rates through time and ancestral state analysis
Macroevolutionary dynamics of diversification and ancestral state reconstruction were modelled across the phylogeny using the Bayesian analysis of macroevolutionary mixtures (BAMM v.2.5.0: Rabosky 2014) on the maximum clade credibility tree obtained in BEAST, after out-group removal, following Nocella et al. (2024). We reconstructed the ancestral states for the depth range, using Deep v. Shallow as character states; either the families or the orders (Scleractinia, Actiniaria, Malacalcyonacea, Antipatharia and Zoantharia) of cnidarian hosts exploited by each coralliophiline species were used as character in two distinct analyses of the trophic ecology. Ancestral state reconstructions were carried out using the Bayesian binary Markov chain Monte Carlo (BBM; Ali et al., 2012) in Reconstruct Ancestral State in Phylogenies (RASP, v. 4.2: Yu et al. 2015) Analyses priors were set following Nocella et al. (2024).
Results
Integrative taxonomy
The molecular dataset included 972 gastropod sequences (542 cox1, 344 16S, 87 ITS2) from 586 in-group and 5 out-group specimens. The combined dataset included sequences from 542 samples (after the removal of samples for which no cox1 sequences were available). Preliminary identification of the 586 in-group specimens retrieved 116 PSH (primary species hypothesis). We excluded the amplification of nuclear pseudogene of the cox1, by checking the absence of indels and of internal stop codons, and the vast prevalence of 3rd position substitutions. All the 481–658 bp long cox1 sequences (516, thus excluding 21 shorter minibarcodes) were included in the ASAP analysis, which divided the dataset into 67–118 hypothetical species (in the ten best partitions: Supporting Information, Fig. S1).
The first and second ASAP best partitions (threshold distance = 11, 12%, number of species 67, 71, respectively) evidently overlumped multiple clearly distinct entities. For instance, these partitions merged into a single species hypothesis samples showing substantial differences in shell morphology (i.e. the smooth-shelled C. violacea, C. galea, C. erosa, C. salebrosa, C. radula, almost all Leptoconchus spp., along with the spiny-shelled H. squamosus A and B, B. mansfieldi, B. bernardi, H. filiaregis A), making a largely polyphyletic assemblage. The same holds for the seventh and ninth partitions, which were not considered due to their high threshold distance (10%) again resulting in inconsistent overlumping. The partitions 4–5, 6, 8, and 10 in the rank (threshold distance = 4–5.5%; number of species 104–117) were only slightly less splitting than partition 3; the latter (threshold distance = 3.9%; number of species 118) mostly aligned with our shell morphology results, was not contradicted by the phylogenetic ones, and was thus retained as the most reliable.
All the hypothetical species, except four, identified by ASAP in the third-best partition corresponded to monophyletic groups highly supported in the ML and BI phylogenetic reconstructions: Ufb (Ultrafast bootstrap) = 99–100, PP (Posterior Probability) = 0.98–1. The exceptions were Hirtomurex oyamai (Ufb = 75, PP = 1) not supported in the ML combined tree, and Mipus vicdani (Ufb = 100, PP = 0.86) in the BI, whereas Coralliophila fearnleyi and Coralliophila violacea A were not supported in any of the combined trees.
Almost all specimens that were not included in the ASAP analysis because their cox1 sequence was too short, did not represent new independent lineages in the phylogenetic analyses, but ended up into clades corresponding to hypothetical species already identified by ASAP, except five: MNHN-IM-2009-5423 Babelomurex nagahorii, MNHN-IM-2013-11444 Leptoconchus sp. H, MNHN-IM-2013-12027 Coralliophila bulbiformis, BAU-2892 Babelomurex benoiti, BAU-2968 Coralliophila basileus. These five samples formed five lineages clearly distinct from all other clades in the combined tree and we consider that they represent five additional hypothetical species.
In four cases, two or more PSH were retrieved in all the ASAP partitions as a single hypothetical species (confirmed by the phylogenetic analyses): (1) Leptoconchus ingranulosa, L. incycloseris, L. inpleuractis, L. ingravis (Ufb = 100, PP = 0.99). Among the species identified by Gittenberger and Gittenberger (2011), L. inpleuractis and L. ingravis were not retrieved as reciprocally monophyletic, but rather intertwined within a single clade, which also includes a sequence (MNHN-IM-2009-6385 Leptoconchus sp.) generated in this study. Conversely, the sequences assigned to L. ingranulosa and L. incycloseris were reciprocally monophyletic. However, the genetic distance calculated among the four morphospecies ranged 1.2–3.8%. The inter-/intraspecific threshold corresponding to the partition chosen in this work is 3.9%, which resulted in consolidating this clade of four morphospecies into a single SSH (secondary species hypothesis). (2) Leptoconchus inpileus and L. infungites (Ufb = 100, PP = 1). These sequences were reciprocally monophyletic; however, their rather low genetic distance (1.6%) is suggestive of another case of synonymy; (3) Babelomurex cariniferoides and Latiaxis nippoleifera (Ufb = 100, PP = 1). The single specimen of B. cariniferoides was collected in New Caledonia, in the same locality of one specimen ascribed to L. nippoleifera. (4) Coralliophila richardi and Emozamia licina (Ufb = 100, PP = 1). The specimens ascribed to these two nominal species, from the Caribbean (Harasewych et al. 2022), New Zealand and Mediterranean (Russini et al. 2023) were shown to represent a single genetic species, seemingly cosmopolitan.
In ten cases, one PSH was split by ASAP into two or more hypothetical species (each confirmed as monophyletic by the phylogenetic analyses): (1) Galeropsis monodontus into eight SSH (Ufb = 99–100, PP = 0.99–1). This is a remarkable case of cryptic diversity within Coralliophilinae. Our 27 samples morphologically ascribed to this nominal species, actually belong to eight different molecular species. Samples of different SSH displayed no differences in shell shape, were frequently collected in the same locality and occasionally from the same station, and we retrieved no evidence of host differences; (2) Coralliophila fimbriata into five SSH (Ufb = 100, PP = 1). It emerged as the second most splitted nominal species in our dataset. Samples collected in the same locality belonged to different SSH; (3) Coralliophila pulchella into two SSH (Ufb = 97–100, PP = 1). Samples are found to belong to two distinct sister species, one corresponding to samples collected in the Philippines, South Madagascar and Papua New Guinea, and the other from the Maldives; (4) Rapa rapa into three SSH (Ufb = 100, PP = 1). The three specimens ascribed to this coralliophiline nominal species, actually correspond to three distinct species. R. rapa B and C were collected in the same locality; (5) Hirtomurex filiaregis into four SSH (Ufb = 100, PP = 1). The four samples morphologically ascribed to this morphospecies actually correspond to as many different species. Notably, H. filiaregis A is phylogenetically quite unrelated to all the other species in all phylogenetic trees, despite having been collected in the same area (New Caledonia) as H. filiaregis C; (6) Coralliophila violacea (the type species of the genus Coralliophila) into three SSH (A not supported, B represented by a single samples and C Ufb = 99, PP = 0.97). As already noted by Simmonds et al. (2018), it actually represents a complex of cryptic species. Of the three putative species detected herein, C. violacea B is represented by only one sample, and C. violacea A lacks consistent phylogenetic support in the combined trees. Consequently, the species delimitation for this complex remains uncertain; (7) Babelomurex japonicus into two SSH (Ufb = 100, PP = 1). These two clades (the first comprising one sample collected in the China Sea and the second comprising four samples from the New Caledonia) do not emerge as sister species in any combined trees; (8) Babelomurex spinosus into two SSH (Ufb = 100, PP = 1). These two specimens (collected one in the China Sea and one in New Caledonia) correspond to two distinct sister species; (9) Coralliophila costularis into two SSH (Ufb = 100, PP = 1). These samples are identified as two sister species one corresponding to samples collected in Papua New Guinea and the other from various eastern Indian Ocean localities (Madagascar, Mozambique and Yemen); (10) Hirtomurex squamosus into two SSH (Ufb = 100, PP = 1). The cox1 sequence of H. squamosus A is shorter than those representing the putative species B, which may have biased the ASAP analysis. However, considering that the specimens of the two hypothetical species feed on different cnidarian families, we have preferred to keep them into two separate SSH, which has not affected the subsequent analyses.
In the integrative taxonomic process, we identified a total of 123 SSH as depicted in the collapsed tree (Fig. 3). The PSHs that have undergone further subdivision into multiple SSHs were named with alphabetical suffixes from A to H.
Phylogenetic relationships of the subfamily Coralliophilinae (maximum likelihood tree on combined dataset), with clades collapsed by species. Numbers at nodes indicate branch support values [ultrafast bootstrap (Ufb) values and posterior probability (PP), respectively]; support values are shown only when at least one of them is ≥ 95%; black circles at nodes indicate maximum support (Ufb = 100, PP = 1). Photograph credits: Mélanie Van Weddingen (MNHN)
Phylogenetic reconstruction
The final alignment comprises 658 bp of cox1, 469 of 16S and 785 of ITS2. Comparison of the major nodes among the ML and BI combined trees revealed no inconsistencies (Supporting Information, Figs S2–S9): the monophyly of the subfamily Coralliophilinae was strongly supported (Ufb = 100, PP = 1), while all the genera as traditionally conceived, except Galeropsis and Rapa, did not prove monophyletic.
Leptoconchus was split into two lineages, one corresponding to Leptoconchus lamarckii (Ufb = 100, PP = 1) (the type of Magilopsis G. B. Sowerby III, 1919), and one including all the other Leptoconchus species (Ufb = 99, PP = 1).
The three species traditionally ascribed to the genus Latiaxis (the type species Latiaxis mawae, L. pilsbryi, and L. hayashii) represented three distinct lineages.
The species of the genus Mipus were split into three lineages: two corresponded respectively to M. sugitani (Ufb = 100, PP = 1) and M. eugeniae (Ufb = 100, PP = 1), the latter included M. vicdani and M. alis (Ufb = 100, PP = 1).
The species morphologically ascribed to the genus Hirtomurex were split into ten lineages, with a high number of cryptic species: (i) Hirtomurex sp. A, (ii) H. tangaroa, (iii) Hirtomurex sp. B, (iv) H. marshalli, (v) H. oyamai, (vi) H. filiaregis A; (vii) H. kawamurai (Ufb = 100, PP = 1); (viii) H. filiaregis B and Hirtomurex sp. C-D-E (Ufb = 98, PP = 0.96); similarly, (ix) H. filiaregis C and D (Ufb = 100, PP = 1); (x) the type species H. squamosus that was split into two cryptic species, H. squamosus A and B (Ufb = 100, PP = 1).
The genus Babelomurex included 9 lineages: (i) B. lischkeanus (Ufb = 100, PP = 1); (ii) B. bernardi, B. mansfieldi, B. oldroydi and the type species B. cariniferus (Ufb = 100, PP = 1); (iii) B. tectumsinensis (Ufb = 100, PP = 1); (iv) B. nakamigawai (Ufb = 100, PP = 1); (v) B. tosanus (Ufb = 100, PP = 1); (vi) B. nagahorii; (vii) B. pruvosti, B. benoiti and B. japonicus A and B (Ufb = 100, PP = 1); (viii) B. cariniferoides (Ufb = 100, PP = 1); (ix) B. nakayasui, B. diadema, B. armatus, B. pallox, B. yumimarumai, B. cf spinosus, B. cristatus and B. spinosus A and B (not supported).
The species traditionally ascribed to the genus Coralliophila included twenty lineages. C. trigoi, C. kaofitorum, C. curacaoensis, C. brevis, C. mitraeforma, C. cf ovoidea, C. galea, C. ahuiri, C. fearnley and C. curta represented 10 distinct lineages The remaining 10 clades included: (1) C. clathrata, sister to C. salebrosa (Ufb = 100, PP = 1); (2) C. robillardi and the five species identified as C. fimbriata A-E (Ufb = 100, PP = 1); (3) C. profundicola, sister to C. meyendorffii (Ufb = 100, PP = 1); (4) C. rubrococcinea, C. carnosa and the two species identified as C. pulchella A and B (Ufb = 100, PP = 1); (5) C. caroleae, sister to C. roseocephala (Ufb = 100, PP = 1); (6) C. basileus, sister to C. richardi (Ufb = 95, PP = 0.97); (7) C. ovoidea, sister to C. xenophila (Ufb = 100, PP = 1); (8) C. australis, C. mira and the type species C. violacea splitted into three species A-C (not supported); (9) C. nodosa, C. radula and C. erosa (not supported); (10) C. bulbiformis, C. amirantium and the two species identified as C. costularis A and B (not supported).
Hereafter we will use Leptoconchus to indicate the clade including all Leptoconchus species except L. lamarckii, and Babelomurex sensu stricto for the clade comprising B. bernardi, B. mansfieldi, B. oldroydi and the type species B. cariniferus.
Dating major lineages
All our calibration nodes were dated in the BEAST output congruently with the corresponding fossil data. The time-calibrated phylogeny (Fig. 4; see Table 1 for 95% HPD) estimated the origin of the subfamily Coralliophilinae at 42.34 mya (95% HPD 41.65–43.03) during the Lutetian (Middle Eocene). The origin of the genus Galeropsis was dated at 21.22 mya (95% HPD 18.53–23.91) from the Burdigalian to the Aquitanian (Early Miocene). The Hirtomurex squamosus complex was estimated as having originated 1.3 mya (95% HPD 0.77–2.39) during the Meghalayan (Holocene). Similarly, Coralliophila richardi diverged from C. basileus approximately 1.54 mya (95% HPD 0.77–2.77). The origin of Leptoconchus was dated at 12.4 mya (95% HPD 9.5–25.37) during the Miocene. Babelomurex sensu stricto was estimated as having originated during the Miocene, with the node dated at 10.9 mya (95% HPD 3.78–22.32). Rapa was estimated to have originated around 9.73 mya (95% HPD 4.18–17.79) in the Late Miocene. The clade including C. rubrococcinea, C. carnosa, C. pulchella A and B, C. brevis, C. mitraeforma, Hirtomurex sp. A, M. sugitanii, C. roseocephala, C. caroleae, H. tangaroa, C. richardi, C. basileus, Hirtomurex sp. B, H. marshalli, C. cf ovoidea, H. oyamai, C. xenophila, C. ovoidea, H. filiaregis A, Hirtomurex sp. C-D-E, was estimated as having originated 17.64 mya (95% HPD 12.04–24.42) during the Late Miocene.
Diversification rate of Coralliophilinae
We modelled the diversification rates within the Coralliophilinae as a function of time. The best model supported by BAMM analysis indicated a steady rate of diversification over time, with a posterior probability of 1. The analysis consistently upheld this model, demonstrating alignment with every significant shift in the rate of speciation considered in our prior assessments. The credible shifts plot depicted a non-core shift across all lineages (Fig. 5a), with a 100% posterior probability. The rate-through-time BAMM plot supported a scenario with an initial higher rate of diversification (speciation rate ∼0.15) that decreased gradually over time (to ∼ 0.12) until the present day (Fig. 5b).
Ancestral state reconstruction
Of the 123 species recognised by our integrative taxonomy approach, 75 (61%) come from shallow-water habitats, while 48 (39%) inhabit deep waters. The results of the ancestral state reconstruction for habitat types (shallow vs. deep) are shown in Fig. 6. (Nodes are numbered to ease following the description). The ancestral Coralliophilinae (node 245) were estimated to have evolved in shallow waters (99.9% marginal probability), which were retrieved as the ancestral habitat for most clades. The first shift to deep waters took place at node 237 (89.5% deep), followed by a reversal to shallow water at node 231 (93.6%) that gave rise to several common shallow-water species (e.g. C. radula, C. erosa, C. bulbiformis). Finally, another shift to deep waters is estimated to have occurred at node 227 (93%), subtending one of the spiny clades. A second shift to deep waters likely occurred at node 187 (37% deep, 35% both shallow and deep), in the ancestor of 19 species of which only Coralliophila brevis is estimated to have secondarily migrated back into shallow waters. Several additional shifts to deep waters were retrieved scattered across the tree: Babelomurex lischkeanus, Coralliophila profundicola, the pair Hirtomurex squamosus A–B, and Mipus eugeniae. Instances of reversals to shallow waters were also observed: the pair C. mira—C. australis, C. violacea A–C, C. fearnleyi, and the pair B. pruvosti—B. benoiti.
Graphical representation of the ancestral state reconstruction at each node of the phylogeny of the subfamily Coralliophilinae obtained from RASP by BBM analysis using depth range as a prior. Pie charts at each node (from 124 to 222) show the probabilities of alternative ancestral states; numbers inside the pie charts identify each node. The legend shows the colour key to the depth range. X-axis represents time in millions of years
A total of 82 distinct coralliophiline-cnidarian associations were identified (Table S3). We have identified the cnidarian hosts (26 families) of 51% (63) of the tested gastropod species. For 36 coralliophiline species the host was retrieved exclusively from literature, whereas for the remaining 46 host identification was based on our molecular and/or morphological analyses. Among them, 15 resulted from DNA barcoding of coralliophiline stomach content (17 sequences, out of the 72 stomach samples), 6 from coral tissue amplification, and 25 were exclusively identified by morphology due to the unavailability of coral tissue or unsuccessful DNA amplification. When multiple association records were available for a species (in 26 instances), usually they congruently indicated its association with a same cnidarian family, with six exceptions (B. tectumsinensis, B. cariniferus, C. meyendorffii, C. richardi, C. salebrosa and C. galea). Congruence was even higher in analysing the association with cnidarian orders, with only two exceptions (Coralliophila salebrosa and C. meyendorffii).
The results of the ancestral state reconstruction using cnidarian families as prior for the states (Fig. 7) show that for the Coralliophilinae (node 245) the scleractinian family Pocilloporidae was the most likely inferred ancestral host (65.8% marginal probability), followed by a multiple association with Caryophyllidae and Cladocoridae (13%) and by Agariciidae (11.1%). Pocilloporidae resulted as the most likely ancestral host also for the genus Galeropsis (node 130, 98.9%). At node 151, Faviidae was estimated as the ancestral host of the clade comprising all the species belonging to the genus Leptoconchus (52.7%), followed by Merulinidae (26%). At node 160 Sarcophytidae was the most likely ancestral host of the genus Rapa (99%). Sarcophytidae resulted as the ancestral host also for a clade including Rapa and Rhizochilus (node 161, 61.2%). The gastropod-coral associations among other coralliophiline major clades did not display a phylogenetically consistent pattern: different species within the same clade in our tree feed frequently on different cnidarian families. However, the clade comprising Coralliophila robillardi and the five species identified as C. fimbriata A-E (node 157), exhibited a clear pattern, with the ancestor of this clade estimated to have had Agariciidae as the most likely host (97.7%).
Graphical representation of the ancestral state reconstruction at each node of the phylogeny of the subfamily Coralliophilinae obtained from RASP by BBM analysis using cnidarian families as a prior. Pie charts at each node (from 124 to 222) show the probabilities of alternative ancestral states; numbers inside the pie charts identify each node. The legend shows the colour key to the hosts; black represents other unknown ancestral states. White barred circles represent no host information. X-axis represents time in millions of years
In the ancestral state reconstruction using cnidarian orders as prior for the states (Fig. 8), the ancestral Coralliophilinae (node 245) were estimated to be associated with scleractinian Hexacorallia (99.4% marginal probability). Scleractinia were retrieved as the ancestral host for most clades. The main shifts occurred in the ancestral host of the genera Rhizochilus and Rapa (node 161, shift to Octocorallia Malacalcyonacea 43.9%) similarly to the ancestor of the clade comprising 23 species (node 188, shift to Octocorallia Malacalcyonacea, 79%). Within the latter clade, three species feed on hexacorals (one on Scleractinia, one on Zoantharia, one on Actiniaria), while the host of the remaining species, when known, is an Octocorallia Malacalcyonacea.
Graphical representation of ancestral state reconstruction at each node of the phylogeny of the subfamily Coralliophilinae obtained from RASP by BBM analysis cnidarian orders as a prior. Pie charts at each node (from 124 to 222) show the probabilities of alternative ancestral states; numbers inside the pie charts identify each node. The legend shows the colour key to the hosts; black represents other unknown ancestral states. White barred circles represent no host information. X-axis represents time in millions of years
Discussion
The assessment of Coralliophilinae diversity
The integrative taxonomy analysis was imperative to correctly estimate the alpha-diversity of the Coralliophilinae and robustly infer the species tree, crucial for the subsequent macroevolutionary analyses.
We have significantly increased the taxonomic span of Coralliophilinae diversity, with molecular data for 46% of the accepted species and 77% of the accepted genera, compared to 12% of species and 54% of genera in previous works (Oliverio et al. 2009b).
The 116 morphospecies initially identified actually comprise 123 distinct species, with some exceptions discussed in detail below. Our analysis revealed only four instances of potential oversplitting of actual species diversity. Among them, particularly interesting is the case of Leptoconchus ingranulosa/L. incycloseris/L. inpleuractis/L. ingravis, since these four species were defined on the basis of their host preference, assuming a process of speciation driven by adaptation to the host (Gittenberger and Gittenberger 2011). While for L. inpleuractis and L. ingravis we are prone to consider the assayed specimens as quite evidently conspecific (low genetic distance, and non-reciprocally monophyletic), the two other nominal species (L. ingranulosa and L. incycloseris) may either represent two distinct species with a clear ecological distinction (being associated to different genera of Fungiidae), or a single species, distinct from L. inpleuractis/L. ingravis. Additional data would be needed to make a robust taxonomic decision.
Conversely, most instances of morphological-molecular incongruence indicated an underestimation of the actual species diversity, leading to the splitting of ten nominal species. This is a common situation in molluscs where cryptic species complexes are particularly abundant (e.g. Barco et al., 2008). Among them, particularly interesting for the high number of cryptic species detected, are the cases of Galeropsis monodontus and Coralliophila fimbriata, split into eight and five molecularly supported species, respectively.
Out of the 10 nominal extant genera represented in our dataset, a total of six (Babelomurex, Coralliophila, Mipus, Leptoconchus, Latiaxis, Hirtomurex) were not monophyletic as traditionally conceived. Although the present work is not aimed at reassessing the coralliophiline systematics, the following general taxonomic considerations may prove useful in a future work on the systematics of this subfamily.
Galeropsis proved monophyletic, comprising at least 8 cryptic species within the G. monodontus complex; it would be interesting to test whether Purpura porphyroleuca Crosse, 1870 (currently included in Coralliophila s.l.; Cernohorsky 1980: 114, figs. 4, 5) actually belongs in this genus as morphology suggests.
Rapa also proved monophyletic, with at least three cryptic species within Rapa rapa.
Rhizochilus was based on specimens of a single species, and both its relationships with Rapa (topological, but not supported by PP or Ufb), and its actual diversity need to be further tested.
The actual magnitude of the radiation of Leptoconchus needs to be re-evaluated; however, it proved monophyletic only if excluding L. lamarcki, for which Magilopsis G. B. Sowerby III, 1919 could be reinstated.
Coralliophila robillardi and the complex of C. fimbriata A-E may deserve a genus on their own, which might be the available Coralliobia H. Adams & A. Adams, 1853.
Five species are included in a clade that could be defined as Babelomurex s.s.: the type species B. cariniferus, along with B. oldroydi, B. bayeri, B. mansfieldi, B. bernardi. All the other species sharing a morphologically similar spiny shell are interspersed across the tree among smooth-shelled species: spiny and smooth shells have probably been acquired and lost multiple times during the coralliophiline evolution, implying that an increased number of species should be analysed before drawing any conclusions on the evolution of shell morphology in this subfamily.
The type species of Coralliophila (C. violacea, which is actually a complex of at least three cryptic species) is retrieved in our analyses as topologically related (albeit with a weak support) to C. australis and C. mira; however, the relationships with C. bulbiformis, C. costularis and C. radula, previously obtained in a molecular phylogenetic analysis of a reduced dataset (Oliverio et al. 2009b), has not been retrieved herein. While it is evident that the use of the genus Coralliophila (the type genus of the subfamily) should be restricted to a much smaller clade than currently done, the extent of this lineage needs to be better defined.
Overall, the phylogenetic patterns emerging from this work are more in contrast than in agreement with shell morphology-based traditional taxonomy, casting serious doubts on the validity of the current classification. The alternate hypothesis would weaken the reliability of molecular phylogenetic approaches based on mitochondrial genes. Indeed, the so-called ‘Davison-effect’, i.e. the accelerated accumulation of mutations in mitochondrial genes of protandrous hermaphrodites (Davison 2006), such as Coralliophilinae, may have obscured the phylogenetic signal of our dataset. However, preliminary data from a phylogenetic analysis on an Exon-Capture dataset (N.P. unpublished), which should be less affected by this issue (Abdelkrim et al. 2018), broadly confirmed the patterns retrieved herein, with Galeropsis as the most basal lineage among extant coralliophilines, Leptoconchus as sister to the remaining lineages, Coralliophila as strongly polyphyletic and the smooth-shelled C. violacea falling within a clade of spiny-shelled species. For this reason, we are more inclined towards considering shell morphology highly plastic probably in response to adaptive pressures, and therefore not suitable as an uncritical source of diagnostic features for supraspecific taxonomy, a pattern not unusual in gastropods (e.g. Fassio et al. 2021). Probably, only an in-depth reassessment of characters from the anatomy might provide morphological features suitable to delimit major lineages worthy of genus-level recognition in Coralliophilinae (Richter and Luque 2002).
Macroevolutionary patterns
Robust molecular evidence indicates that the origin and early diversification of coralliophilines could have occurred between 41 and 43 mya (congruent with the age of the fossil Coralliophila aldrichi). Despite Coralliophilinae are known to be more diverse in deep water habitats, from the mesophotic zone down to bathyal and abyssal bottoms (75% of deep-water species: Oliverio 2008), our dataset is biased towards shallow-water taxa (61% of shallow-water species), as expected due to the difficulties in collecting in deep-water rocky bottoms. However, given the good taxonomic coverage obtained, we do not expect that this bias affects the results of the ancestral state reconstructions. Our analysis strongly supported the hypothesis that the Coralliophilinae originated in shallow waters with multiple subsequent instances of colonisations of deep waters (Oliverio et al. 2009b). At least three major shifts to deep habitats were detected (two shifts concern unsupported nodes, allowing, in alternative topologies, additional shifts to deep habitats).
Coralliophilinae feed on five cnidarian orders belonging to the anthozoan subclasses Hexacorallia and Octocorallia. The present study, that includes 41 new coralliophiline-cnidarian associations, demonstrates that the vast majority of coralliophiline species is rather specialised in its parasitic behaviour. Although, our dataset was based on a majority of single records per coralliophiline species (which may bias the perception towards a “one coralliophiline-one cnidarian” pattern), many of these specialised associations are confirmed by authors’ observation, reliable anecdotal reports or photographic material in scuba divers’ websites (e.g. http://www.www.poppe-images.com). In most cases, individuals of the same coralliophiline species feed on cnidarians belonging to a single family, with a few exceptions such as C. galea, which feeds on corals of ten different families, the broadest diet so far reported for the subfamily, and C. meyendorffii and C. salebrosa, each of them feeding on five families belonging to two different orders (see Table S3).
The BBM analysis indicated that shallow-water scleractinians were the most likely ancestral host for the Coralliophilinae. The Pocilloporidae served as host for the ancestral Coralliophilinae since the beginning of coralliophiline history (41–43 mya) and has been maintained until the present day in the genus Galeropsis. The association with scleractinians persisted in all major ancestral lineages except two. The first shift occurred 18–26 mya in the common ancestor of the genera Rhizochilus and Rapa, which is supposed to have moved to the octocoral Malacalyonacea; this association has persisted up to the present day in the genus Rapa (with Sarcophytidae), while the sister lineage Rhizochilus might have secondarily shifted back to hexacorals, evolving a new association with the family Anthipatidae. The second and major dietary shift occurred in the ancestor of the “node 188 clade”, which also moved to the octocoral Malacalcyonacea 18–20 mya. Within this clade, at least three reversals to hexacorals have occurred in deep waters: one to deep-water Scleractinia carried out by C. richardi (2–5 mya); another in C. cf ovoidea to Zoantharia (3–8 mya); the third in the ancestor of the clade comprising C. xenophila, C. ovoidea, H. filiaregis A and the three species of Hirtomurex (sp. A-C), that moved to Actiniaria approximately 6–7 mya. Among the remaining clades, some species are not associated with Scleractinia (C. kaofitorum and B. oldroydi), or are clearly polyphagous (C. meyendorffii), but these instances did not bear any influence on the ancestral nodes’ host probability. Both shifts to octocorals actually concern nodes with weak or no support in our phylogeny (Fig. 3); alternative topologies may require more shifts, and we have discussed only the most conservative scenario.
We did not retrieve a consistent pattern for the major coralliophiline clades, some of which display a clear trend of specialisation, whereas others exhibit a higher degree of dietary variance. The Galeropsis monodontus complex represents an extreme case of specialisation, including at least six species that feed on Pocilloporidae, probably the original coralliophiline heritage. Similarly, the ancestor of the clade comprising C. robillardi and the five species ascribed to C. fimbriata, has been associated with the shallow-water Agariciidae since approximately 12–33 mya. Conversely, the shallow-water Leptoconchus spp. are estimated to have first evolved in association with Faviidae (13–23 mya), then underwent a broad host diversification in which one major lineage transitioned to Fungiidae (6–10 mya), while other lineages switched to Merulinidae or maintained their association with Faviidae (5–16 mya). The clade subtended by node 188, after an initial association with shallow-water Poritidae (around 18–20 mya), underwent a broad diversification in host associations, with extant species feeding on five different families.
The results obtained by the BAMM analysis of coralliophiline diversification rates across time did not highlight any core shift in diversification rates across the phylogeny of the subfamily (Fig. 5a). The shape of the rate-through-time plot suggests that the diversification rate of the family started decreasing at the beginning of the coralliophiline evolutionary history and continued to steadily slow down until the present day (Fig. 5b). The same pattern has been observed in another corallivorous gastropod lineage, the family Ovulidae (Nocella et al. 2024). As in the Ovulidae, we suggest that only during the initial stages of the evolution of the Coralliophilinae, a high diversification rate (∼0.15) existed; subsequently, this rate gradually declined in all lineages as time progressed. Following an initial burst of diversification, the progressive occupation of all available niches may result in a deceleration of the speciation rate within a density-dependent model (Moen and Morlon 2014). In our case, the initial high diversification rate could be linked to the emergence, in the ancestral coralliophilines, of the capability to exploit cnidarians as a trophic resource. The enduring association of Coralliophilinae with cnidarians has forged such robust relationships that have effectively constrained diversification rates for over 40 million years. In this context, in the only study to date available on macroevolutionary patterns of corallivorous gastropods (Nocella et al. 2024), the lack of distinct shifts in diversification rates within specific clades despite variations in species richness was instead associated with a pattern of slow, steady increase of diversification across the entire subfamily, linked to environmental variables. In Coralliophilinae this association started in shallow water with scleractinian hexacorals, with repeated instances of colonisations of deep-water habitats in several lineages. The origin of Scleractinia can be located from the Carboniferous to Silurian (324–447 mya, McFadden et al. 2021) followed by a rapid expansion and diversification across shallow marine environments during the Ladinian, about 240 mya (Frankowiak et al. 2023), thus providing a broad range of trophic niches available to corallivorous species. An ancestral exploitation of shallow-water hexacorals, with subsequent colonisations of deeper habitats, follows an onshore-offshore trend widely documented in marine fauna (Jacobs et al. 1998). However, we did not detect co-occurring shifts in depth and host.
Our results do not substantiate the existence of global co-evolutionary processes between coralliophiline gastropods and their cnidarian hosts. Instead, we found multiple unrelated switches from one cnidarian family to another. The exploited families of Scleractinia, Antipatharia, Zoantharia, Actiniaria and Malacalcyonacea, originated significantly earlier than Coralliophilinae (Lindner et al. 2008; McFadden et al. 2021). Therefore, neither the origin nor the diversification of the Coralliophilinae seems to have been strictly coupled with cnidarian evolution, suggesting the absence of a pervasive host–parasite co-evolutionary pattern. Similarly to what has been observed in the family Ovulidae (Nocella et al. 2024), the evolutionary trajectories of Coralliophilinae are better defined as a pattern of sequential evolution, where the evolution of the host may have influenced the evolution of the parasite, but not vice versa, as proposed in some association of phytophagous insects (Hardy and Otto 2014). The exploitation of the new niche offered by the corals represented a remarkable ecological opportunity, resulting in an ecological release that might have driven the evolution and diversification of corallivorous snails (Yoder et al. 2010), yet without a major bearing on cnidarian evolution. Nevertheless, we cannot exclude patterns of co-evolutionary processes within specific lineages, but their detection would require a denser sampling and a finer taxonomic identification of the cnidarian hosts at the genus or species level.
References
Abdelkrim J, Aznar-Cormano L, Fedosov AE, Kantor YI et al (2018) Exon-Capture-Based Phylogeny and Diversification of the Venomous Gastropods (Neogastropoda, Conoidea). Mol Biol Evol 35:2355–2374
Cernohorsky WO (1980) The Taxonomy of Some Indo-Pacific Mollusca: PART 7. Rec Auckland Inst Mus 16:171–187
Davison A (2006) The ovotestis: an underdeveloped organ of evolution. BioEssays 28:642–650
Dockery DT III (1986) Punctuated succession of Paleogene mollusks in the northern Gulf Coastal Plain. Palaios 1(6):582–589
Fabricius K, Alderslade P (2001) Soft corals and sea fans: a comprehensive guide to the tropical 731 shallow-water genera of the Central-West Pacific, the Indian Ocean and the Red Sea. Australian Institute of Marine Science, Townsville, Queensland, Australia, 204 pp
Fassio G, Bouchet P, Lozouet P, Modica MV et al (2021) Becoming a limpet: An ‘intermittent limpetization’ process driven by host features in the kleptoparasitic gastropod family Capulidae. Mol Phylogenet Evol 155:107014
Fassio G, Russo P, Bonomolo G, Fedosov AE, Modica MV et al (2022) A molecular framework for the systematics of the Mediterranean spindle-shells (Gastropoda, Neogastropoda, Fasciolariidae, Fusininae). Mediterr Mar Sci 23:623–636
Fassio G, Stefani M, Russini V, Buge B et al (2023) Neither slugs nor snails: a molecular reappraisal of the gastropod family Velutinidae. Zool J Linn Soc 197:924–964
Frankowiak K, Wang XT, Sigman DM, Gothmann AM et al (2023) Photosymbiosis and the expansion of shallow-water corals. Sci Adv 2:e1601122
Gittenberger A, Gittenberger E (2011) Cryptic, adaptive radiation of endoparasitic snails: sibling species of Leptoconchus (Gastropoda: Coralliophilidae) in corals. Org Divers Evol 11:21–41
Harasewych MG, Sei M, Uribe JE (2022) The complete mitochondrial genome of Coralliophila richardi (P. Fischer, 1882) (Neogastropoda: Muricidae: Coralliophilinae). The Nautilus 136(1-2), 1–11
Hardy NB, Otto SP (2014) Specialization and generalization in the diversification of phytophagous insects: tests of the musical chairs and oscillation hypotheses. Proc Roy Soc B: Biological Sciences 281:20132960
Hayes RL, Bush PG (1990) Microscopic observations of recovery in the reef-building 799 scleractinian coral, Montastrea annularis, after bleaching on a Cayman reef. Coral Reefs 8(203–800):209
Jacobs DK, Lindberg DR (1998) Oxygen and evolutionary patterns in the sea: Onshore/offshore trends and recent recruitment of deep-sea faunas. Proc Natl Acad Sci 95(16):9396–9401. https://doi.org/10.1073/pnas.95.16.9396
Jermy T (1976) Insect—Host-plant Relationship—Co-evolution or Sequential Evolution? In: Jermy T (ed) The Host-Plant in Relation to Insect Behaviour and Reproduction. Springer, US, Boston, MA, pp 109–113
Lindner A, Cairns SD, Cunningham CW (2008) From offshore to onshore: Multiple origins of shallow-water corals from deep-dea ancestors. PLoS ONE 3(6):e2429. https://doi.org/10.1371/journal.pone.0002429
Lozouet P, Renard P (1988) Les Coralliophilidae, Gastropoda de l’Oligocène et du Miocène inferieur d’Aquitaine (sud-ouest de la France): systématique et coraux hôtes. Geobios 31(2):171–185
McFadden CS, Quattrini AM, Brugler MR, Cowman PF et al (2021) Phylogenomics, Origin, and Diversification of Anthozoans (Phylum Cnidaria). Syst Biol 70:635–647
Moen D, Morlon H (2014) Why does diversification slow down? Trends Ecol Evol 29:190–197
Nocella E, Zvonareva SS, Fassio G, Pica D et al (2024) Spicy food for the egg-cowries: the evolution of corallivory in the Ovulidae (Gastropoda: Cypraeoidea). Front Mar Sci 10:1323156
Oliverio M (2009) Diversity of Coralliophilinae (Mollusca, Neogastropoda, Muricidae) at Austral Islands (South Pacific). Zoosystema 31:759–789
Oliverio M, Mariottini P (2001) Contrasting morphological and molecular variation in Coralliophila meyendorffii (Muricidae, Coralliophilinae). J Mollus Stud 67(2):243–246
Oliverio M, Cervelli M, Mariottini P (2002b) ITS2 rRNA evolution and its congruence with the phylogeny of muricid neogastropods (Caenogastropoda, Muricoidea). Mol Phylogenet Evol 25:63–69
Oliverio M, Barco A, Modica MV, Richter A et al (2009a) Ecological barcoding of corallivory by second internal transcribed spacer sequences: hosts of coralliophiline gastropods detected by the cnidarian DNA in their stomach. Mol Ecol Resour 9:94–103
Oliverio M, Barco A, Richter A, Modica MV (2009b) The coralliophiline (Gastropoda: Muricidae) radiation: repeated colonizations of the deep sea? The Nautilus 123(3):113–120
Oliverio M, Cervelli M, Mariottini P (2002a) ITS2 rRNA evolution and its congruence with the phylogeny of muricid neogastropods (Caenogastropoda). Muricoidea. Mol Phylogenet Evol 25(1):63–69. https://doi.org/10.1016/S1055-7903(02)00227-0
Oliverio, M. (2008). Coralliophilinae (Neogastropoda: Muricidae) from the southwest Pacific. In: Tropical Deep-Sea Benthos 25 (Héros V., Cowie R, Bouchet P. eds). Mém Mus Na Hist Nat 196: 451–555. Paris
Pandolfi JM, Bradbury RH, Sala E, Hughes TP et al (2003) Global Trajectories of the Long-Term Decline of Coral Reef Ecosystems. Science 301:955–958
Pita L, Rix L, Slaby BM, Franke A et al (2018) The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46
Rabosky DL (2014) Automatic Detection of Key Innovations, Rate Shifts, and Diversity-Dependence on Phylogenetic Trees. PLoS ONE 9:e89543. https://doi.org/10.1371/journal.pone.0089543
Richter A, Luque ÁA (2002) Current knowledge on Coralliophilidae (Gastropoda) and phylogenetic implication of anatomical and reproductive. Boll Malacol 38(4):5–18
Russini V, Fassio G, Nocella E, Houart R et al (2023) Whelks, rock-snails, and allied: a new phylogenetic framework for the family Muricidae (Mollusca: Gastropoda). Eur Zool J 90:856–868
Simmonds, S.E., Chou, V., Cheng, S.H. et al. (2018) Evidence of host-associated divergence from coral-eating snails (genus Coralliophila) in the Coral Triangle. Coral Reefs 37:355–371. https://doi.org/10.1007/s00338-018-1661-6
Stella J, Pratchett M, Hutchings P, Jones G (2011) Coral-associated invertebrates: diversity, ecological importance and vulnerability to disturbance. Oceanogr Mar Biol Annu Rev 49:43–104. https://doi.org/10.1201/b11009-3
Van Der Laan JD, Hogeweg P (1997) Predator—prey coevolution: interactions across different timescales. Proc R Soc Lond B Biol Sci 259:35–42
Vazzana A (1996) Malacofauna batiale del Pleistocene inferiore del Vallone Catrica (Reggio Calabria, Italia). Bollettino Malacologico 31:143–162
Veron JEN (2000). Corals of the world. Townsville: Australian Institute of Marine Science. Volumes 1–3. 1410 pp
WoRMS Editorial Board (2024). World Register of Marine Species. Available from https://www.marinespecies.org/. Accessed 2024-07-29. https://doi.org/10.14284/170
Yoder JB, Clancey E, Des Roches S, Eastman JM et al (2010) Ecological opportunity and the origin of adaptive radiations. J Evol Biol 23:1581–1596. https://doi.org/10.1111/j.1420-9101.2010.02029.x
Yu Y, Harris AJ, Blair C, He X (2015) RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol Phylogenet Evol 87:46–49. https://doi.org/10.1016/j.ympev.2015.03.008
Zibrowius H (1980) Les Scléractiniaires de la Méditerranée et de l’Atlantique nord-oriental. Mém Inst Océanogr Monaco 11:1–284
Acknowledgements
Many of the specimens sequenced herein were collected under the auspices of the Our Planet Reviewed Tropical Deep Sea Benthos programmes at the MNHN (expeditions ATIMO VATAE, DOI: https://doi.org/10.17600/10110040; BIOMAGLO, DOI: https://doi.org/10.17600/17004000; CONCALIS, DOI: https://doi.org/10.17600/8100010; CORSICABENTHOS 2; CORSICABENTHOS 3; EBISCO, DOI: https://doi.org/10.17600/5100080; EXBODI, DOI: https://doi.org/10.17600/11100080; GUYANE 2014; INHACA 2011; KARUBENTHOS 2, DOI: https://doi.org/10.17600/15005400; KAVIENG 2014, DOI: https://doi.org/10.17600/14004400; LIFOU 2000; MAINBAZA ;MD208 (Walters Shoal), DOI: https://doi.org/10.17600/17002700; MIRIKI; NanHai 2014; MADIBENTHOS; NORFOLK 2, DOI: https://doi.org/10.17600/3100030; PAKAIHI I TE MOANA; PANGLAO 2004; PANGLAO 2005; PAPUA NIUGINI, DOI: https://doi.org/10.17600/18000841; TARASOC; TERRASSES, DOI: https://doi.org/10.17600/8100100; Tuhaa Pae 2013, https://doi.org/10.17600/13100030; SW Australia 2013; SANTO 2006; ZhongSha 2015). These expeditions operated under the regulations then in force in the countries in question and satisfy the conditions set by the Nagoya Protocol for access to genetic resources. We extend our acknowledgements to Paolo Mariottini (University of Roma Tre), Carlo Smriglio (University of Roma Tre) and Francisco Otero (Las Palmas de Gran Canaria University) for providing molecular grade materials. We are grateful to, Laurent Charles (Muséum de Bordeaux), Anne-Lise Fleddum (MNHN) and Mélanie Van Weddingen (MNHN) for providing precious photographic material. We are thankful to Barbara Buge (MNHN), Philippe Maestrati (MNHN) and Virginie Héros (MNHN) for their help with MNHN samples and to Daniela Pica (SZN) for cnidarian identification. The research was partially funded by the Doctorate School in Evolutionary and Environmental Biology of Sapienza University of Rome, and by the 2023 Early-Career Research Award from the Malacological Society of London (to E.N.); by the Sapienza 2020 INV-EVO/2 grant (RM12017276B846BB) and Sapienza 2021 SEED-3840974 grant (to M.O.); and by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme [grant agreement no. 865101] (to N.P).
Funding
Open access funding provided by Università degli Studi di Roma La Sapienza within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors certify that they have NO affiliations with or involvement in any organisation or entity with any financial interest, or non-financial interest in the subject matter or materials discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nocella, E., Fassio, G., Zuccon, D. et al. From coral reefs into the abyss: the evolution of corallivory in the Coralliophilinae (Neogastropoda, Muricidae). Coral Reefs (2024). https://doi.org/10.1007/s00338-024-02537-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00338-024-02537-1