Abstract
Pathogenic variants in SCN1A result in a spectrum of phenotypes ranging from mild febrile seizures to Dravet syndrome, a severe infant-onset epileptic encephalopathy. Individuals with Dravet syndrome have developmental delays, elevated risk for sudden unexpected death in epilepsy (SUDEP), and have multiple seizure types that are often refractory to treatment. Although most Dravet syndrome variants arise de novo, there are cases where an SCN1A variant was inherited from mildly affected parents, as well as some individuals with de novo loss-of-function or truncation mutations that presented with milder phenotypes. This suggests that disease severity is influenced by other factors that modify expressivity of the primary mutation, which likely includes genetic modifiers. Consistent with this, the Scn1a+/− mouse model of Dravet syndrome exhibits strain-dependent variable phenotype severity. Scn1a+/− mice on the 129S6/SvEvTac (129) strain have no overt phenotype and a normal lifespan, while [C57BL/6Jx129]F1.Scn1a+/− mice have severe epilepsy with high rates of premature death. Low resolution genetic mapping identified several Dravet syndrome modifier (Dsm) loci responsible for the strain-dependent difference in survival of Scn1a+/− mice. To confirm the Dsm5 locus and refine its position, we generated interval-specific congenic strains carrying 129-derived chromosome 11 alleles on the C57BL/6J strain and localized Dsm5 to a 5.9 Mb minimal region. We then performed candidate gene analysis in the modifier region. Consideration of brain-expressed genes with expression or coding sequence differences between strains along with gene function suggested numerous strong candidates, including several protein coding genes and two miRNAs that may regulate Scn1a transcript.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dravet syndrome is an infant-onset epileptic encephalopathy caused by haploinsufficiency for SCN1A. Seizure onset usually occurs between 6 and 18 months of age with generalized tonic–clonic or hemiclonic seizures, often precipitated by fever. Individuals subsequently develop pleiomorphic afebrile seizure types that often respond poorly to conventional therapies (Dravet 2011; Dravet and Oguni 2013; Wirrell et al. 2017). Development is normal prior to onset, but disease progression is frequently accompanied by stagnation or decline of psychomotor and cognitive development (Battaglia et al. 2021; Dravet 2011). Individuals with Dravet syndrome have a significantly increased risk of mortality, mainly attributed to prolonged status epilepticus in early childhood and sudden unexplained death in epilepsy (SUDEP) in adolescents and adults (Dravet 2011). In most cases, the SCN1A variants arise de novo and result in heterozygous loss-of-function (Li et al. 2021). However, there have been reports of Dravet syndrome caused by SCN1A variants inherited from mildly affected parents, as well as some individuals with de novo loss-of-function or premature truncation variants that presented with milder phenotypes, like generalized epilepsy with febrile seizures plus (GEFS+) (Depienne et al. 2010, 2009; Goldberg-Stern et al. 2014; Guerrini et al. 2010; Nabbout et al. 2003; Osaka et al. 2007; Yordanova et al. 2011; Yu et al. 2010). This variable expressivity suggests that disease severity is influenced by additional factors, which may include genetic modifiers (de Lange et al. 2020; Hammer et al. 2017).
Mice with heterozygous targeted deletion of Scn1a (Scn1a+/−) are a well-validated model and recapitulate core features of Dravet syndrome, including epilepsy and sudden unexpected death following a seizure in otherwise healthy animals (SUDEP-like) (Kalume et al. 2013; Miller et al. 2014; Yu et al. 2006). The highest incidence of SUDEP-like deaths occurs in the 4th postnatal week and survival stabilizes after 6 weeks of age (Kalume et al. 2013; Miller et al. 2014). Genetic background dramatically influences phenotype severity and survival of Scn1a+/− mice (Miller et al. 2014; Rubinstein et al. 2015; Yu et al. 2006). On the 129S6/SvEvTac (129) strain, there is no overt phenotype and 129.Scn1a+/− mice live a normal lifespan. However, when 129.Scn1a+/− mice are crossed with C57BL/6J (B6), the resulting [129xB6]F1.Scn1a+/− mice (F1.Scn1a+/−) have a severe phenotype with spontaneous seizures and premature lethality (Miller et al. 2014). We previously performed genetic mapping and identified several Dravet syndrome modifier (Dsm) loci that influence the strain-dependent difference in survival (Miller et al. 2014).
In this study, we used interval-specific congenic (ISC) strains to fine map the Dsm5 locus on chromosome 11 and identified a minimal interval that influenced survival, particularly in female Scn1a+/− mice. Within the mapped interval, we performed an initial survey of potential candidate modifier genes and identified several high-priority candidate modifier genes.
Methods
Mice (NCBI Taxon ID 10090)
Scn1atm1Kea mice (MMRRC 037107-JAX) were generated by homologous recombination in TL1 ES cells (129S6/SvEvTac) as previously described (Miller et al. 2014). The line has been maintained as isogenic on 129S6/SvEvTac (129) by continuous backcrossing of 129.Scn1a+/− heterozygotes to 129 inbred mice (129SVE, Taconic Biosciences, Germantown, NY, USA). Trpv1 knockout mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA; RRID:IMSR_JAX:003,770). C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA; RRID:IMSR_JAX:000664).
Mice were maintained in a Specific Pathogen Free (SPF) barrier facility with a 14:10 light:dark cycle and ad libitum access to food and water. All animal care and experimental procedures were approved by the Northwestern University Animal Care and Use Committee in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The principles outlined in the ARRIVE (Animal Research: Reporting of in vivo Experiments) guideline were considered when planning experiments (Percie du Sert et al. 2020).
Interval specific congenic (ISC) lines
We generated nine ISC lines carrying 129-derived alleles on chromosome 11 on a C57BL/6 J (B6) background, designated as B6.129-Dsm5A through B6.129-Dsm5F. F1 progeny were generated by crossing 129 males with B6 females, and then successively crossed to B6 to generate congenic lines. Genotyping for chromosome 11 markers was performed at each generation and mice retaining 129 alleles were propagated. Whole genome and selective genotyping was performed at generations N2 and N5 to select breeders with low percentages of 129 in the rest of the genome. All lines were crossed to B6 for ≥ N9 generations prior to any experiments.
Genotyping
DNA was prepared from tail biopsies (Gentra Puregene Mouse Tail Kit, Qiagen, Valencia, CA, USA). Scn1a genotype was determined by multiplex PCR using a common primer (5’- AGTCTGTACCAGGCAGAACTTG) and two allele specific primers (WT: 5’-CCCTGAGATGTGGGTGAATAG; KO: 5’‐AGACTGCCTTGGGAAAAGCG). Amplicons include a 357 bp WT product and a 200 bp KO product. Genotyping of microsatellite markers was performed by analysis of PCR products on 7% denaturing polyacrylamide gels stained with ethidium bromide. ISC breakpoints were refined using the mini Mouse Universal Genotyping Array (miniMUGA) (Neogen, Lincoln, NE, USA). Strain background was surveyed for Trpv1 mice by miniMUGA genotyping (Neogen).
Phenotyping
B6.129-Dsm5 females were bred with heterozygous 129.Scn1a+/− males to generate F1 offspring carrying homozygous 129/129 alleles or heterozygous 129/B6 alleles in Dsm5. Offspring were ear-tagged and genotyped at P12-14. At P19-21, mice were weaned into standard vivarium cages containing 4–5 mice of the same sex and age. Wild-type littermates were included in holding cages. Survival was monitored to 8 weeks of age. Over the 8-week period, mice were monitored daily for general health and any visibly unhealthy mouse (e.g., underweight, dehydrated, poorly groomed, or immobile) was euthanized and excluded from the study; this occurred rarely. The focus of the study was sudden and unexpected death in otherwise healthy appearing Scn1a+/− mice. Survival was compared between groups using Kaplan–Meier analysis with p-values determined by LogRank Mantel–Cox tests. Group sizes were based on data simulations using data from our prior survival studies.
Candidate gene analysis
We defined the Dsm5 gene set using the Mouse GRCm38.p6 reference genome (B6) in Ensembl BioMart, which included classification by gene type. To define a subset of genes with CNS expression, we used the MGI database and EMBL-EBI Expression Atlas. Differential expression of Dsm5 genes between 129 and B6 or F1 was assessed using two RNA-Seq datasets that we previously reported: (1) B6 and 129 forebrain (Hawkins et al. 2016); and (2) F1 and 129 hippocampus (Hawkins et al. 2019) (NCBI GEO GSE112627). Differential expression of miRNAs was assessed using a previously reported dataset (Trontti et al. 2018). To identify consequential coding sequence changes between the strains, we performed whole genome re-sequencing of 129S6/SvEvTac and compared with the C57BL/6J reference sequence and mouse strains in Ensembl (Mouse GRCm38.p6) (NCBI SRA PRJNA817075). The effect of single-nucleotide variants (SNVs) was assessed using the Ensembl Variant Effect Predictor tool (VEP) (McLaren et al. 2016), and SNVs classified as deleterious were retained. TargetScan and LncRRIsearch were used to predict targets of Dsm5 high confidence miRNAs and lncRNAs, respectively (Agarwal et al. 2015; Fukunaga et al. 2019).
Results
Fine mapping of Dsm5 to central chromosome 11
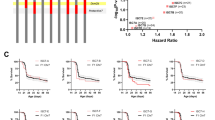
Low resolution mapping of Dsm5 on chromosome 11 localized a 1.5-LOD support interval to 4.7–39.7 cM (6.99–63.9 Mb; GRCm38.p6) (Miller et al. 2014). To refine the map interval, we used ISC lines carrying varying 129-derived chromosome 11 segments on a congenic B6 background (Fig. 1A; Table 1). Each B6.129-Dsm5 strain was crossed to 129.Scn1a+/− to generate offspring with either homozygous 129/129 or heterozygous 129/B6 alleles in Dsm5. Survival of Scn1a+/− offspring was monitored to 8 weeks of age and compared between those carrying homozygous (129/129) alleles at Dsm5 or F1 controls with heterozygous (B6/129) alleles at Dsm5. First, we compared survival between males and females with control heterozygous alleles and observed a sex-difference (p < 0.01 LogRank Mantel-Cox), with females having worse survival than males (Hazard Ratio (HR): 1.561; 95% confidence interval: 1.088–2.240) (Fig. 1B), consistent with other reports (Gerbatin et al. 2021; Niibori et al. 2020). Therefore, all subsequent ISC comparisons were performed separately by sex. Next, we compared survival between mice that were homozygous 129/129 in each ISC interval with their sex-matched F1 (129/B6) controls. Improvement in survival was observed with some ISC lines (Fig. 1C, D). Improved survival of females was observed with 129/129 alleles from lines ISC-D (65–88 Mb), ISC-E (65–76 Mb) and ISC-F (70–88 Mb) (Fig. 1C), while males only showed improvement in survival with ISC-D. Furthermore, the magnitude of the effect of 129/129 alleles from lines ISC-D was greater in females (HR: 3.183; Table 1) relative to males (HR: 2.851; Table 1) (Fig. 1). This suggests that the effect of 129 alleles in Dsm5 is more pronounced in females. Under a single modifier gene model, the smallest 129-derived region to harbor a modifier gene would be the ~ 7 Mb region of overlap between strains ISC-E and ISC-F.
Fine mapping of Dsm5 with ISC strains. A Dsm5 ISC lines carry varying 129-derived chromosome 11 segments (colors) on a congenic B6 background (black). B6.129-Dsm5 ISC lines were crossed with 129.Scn1a+/− mice and survival of resulting Scn1a+/− offspring was monitored to 8 weeks of age. B Comparison of female and male F1 controls with heterozygous 129/B6 alleles in Dsm5 showed a sex-dependent difference in survival, with females (n = 80) having worse survival than males (n = 84) (p < 0.01, LogRank Mantel-Cox). C–D Hazard ratios for all ISC lines relative to F1 controls are plotted against –log10(p-values) as determined by LogRank Mantel–Cox test, and Kaplan–Meier survival plots are shown for lines ISC-D, ISC-E and ISC-F. Survival was significantly improved in female mice with homozygous 129/129 alleles in ISC-D (p < 0.001), ISC-E (p < 0.05), and ISC-F (p < 0.05) compared to F1.KO heterozygous controls (C), while only males with homozygous 129/129 alleles in ISC-D (p < 0.05) had improved survival compared to F1 controls (D). Shaded areas on Kaplan–Meier plots are 95% confidence intervals for F1 controls with heterozygous 129/B6 alleles
To further refine the ISC intervals, breakpoints were defined by SNP genotyping using the mini MUGA platform that includes over 11,000 SNP probes. For ISC-D and ISC-E, the proximal breakpoint was localized between rs216826621 (65.018062 Mb) and rs48061329 (65.423779 Mb), while the distal breakpoint for ISC-E was between rs26888826 (74.871795 Mb) and rs29407735 (76.169847 Mb). For ISC-D and ISC-F, the distal breakpoint was localized between UNC20075505 (88.402458 Mb) and S3J113539515 (88.487894 Mb), while the proximal breakpoint for ISC-F was between gUNC19880825 (70.808812 Mb) and rs26890640 (71.870339 Mb). Thus, the area of overlap between ISC-E and ISC-F is defined as the 5.4 Mb region between 70.808812 Mb and 76.169847 Mb. Bold indicates P < 0.05.
Candidate gene analysis
Under a single modifier gene model, the Dsm5 locus was narrowed to a 5.4 Mb interval (70.808812–76.169847 Mb, GRCm38.p6) that contains 126 known genes, including 12 RNA genes, and 114 protein coding genes (including 30 olfactory receptors genes). Of those, 28 protein coding and 10 RNA genes have confirmed brain expression.
Examination of candidate genes within the refined Dsm5 interval in existing mRNA-seq datasets revealed two differentially expressed genes (DEGs) between wild-type 129 and B6 females (Sgsm2, Smg6) (Fig. 2A, B); and three DEGs between P14 or P24 wild-type 129 and F1 mice in pooled samples containing both sexes (Pafah1b1, Ywhae, 6330403K07Rik) (Fig. 2C-E). There were three DEGs (Rtn4rl1, Serpinf1, Smyd4) when comparing F1.Scn1a+/− mice without seizures and F1.Scn1a+/− mice with recent seizures (Fig. 2F-H). From whole genome sequencing, we identified 24 genes in the refined Dsm5 interval with predicted deleterious variants, including missense, splice site, and indels (4930563E22Rik, 4933427D14Rik, 6330403K07Rik, Aspa, Cluh, E130309D14Rik, Pimreg, Haspin, Hic1, Itgae, Nlrp1a, Nlrp1b, Ovca2, Pitpnm3, Rpa1, Rtn4rl1, Smg6, Spns3, Tax1bp3, Tekt1, Trpv1, Trpv3, Xaf1, Zzef1) (Table 2). Of the 30 protein coding genes with identified coding sequence or expression differences, ten had a prior association with seizure or epilepsy based on literature and database searches, including Aspa, Hic1, Nlrp1a, Nlrp1b, Pafah1b1, Smg6, Trpv1, Trpv3, Ywhae and 6330403K07Rik (Table 2). Pathway analysis of these Dsm5 candidate genes showed that Trpv1 is the only first neighbor to Scn1a, while Trpv3 and Aspa are included in this network through their interactions with Trpv1 (Fig. 3).
Differential expression of positional candidate modifier genes from our published RNA-seq datasets (Hawkins et al. 2019, 2016). A–B Two genes, Sgsm2 (A) and Smg6 (B) had differential expression between 129 and B6 females (Hawkins et al. 2016). C–E Three genes were differentially expressed between wild-type 129 and F1 mice in pooled samples containing both sexes, with Pafah1b1 (C) and Ywhae (D) differentially expressed at P24, and 6330403K07Rik differentially expressed at P14 (E) (Hawkins et al. 2019). F–H Comparison of F1.Scn1a+/− mice with or without seizures in the antecedent 24 h showed differential expression of three genes, Rtn4rl1 (F), Serpinf1 (G) and Smyd4 (H) (Hawkins et al. 2019). Floating bars represent the range of values for each group and significance is denoted as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
String protein association network for candidate protein coding modifier genes and Scn1a. Only Trpv1 had a direct association with Scn1a, while Aspa and Trpv3 had indirect associations. The 30 protein coding candidate genes in Table 2 and Scn1a were used as input in String (v. 11.0) (Szklarczyk et al. 2019). At least medium confidence associations are displayed (interaction score ≥ 0.4). Edges represent protein associations, with line color indicating the type of evidence as follows: yellow, text-mining; black, co-expression; pink, experimentally determined interactions; blue, known interactions from curated databases; purple, protein homology
Three of the brain-expressed miRNAs (miRs-22, -132, -212) had prior association as seizure-responsive genes according to EpimiRBase, a database cataloging published reports of miRNA up- and downregulation following seizures (Jimenez-Mateos et al. 2011; Mooney et al. 2016). It is notable that two of these miRNAs (miRs-132, and -212) are shown to be upregulated following seizures and are predicted to target voltage-gated sodium channel genes, including Scn1a. Furthermore, a recent study demonstrated that miR-132 regulates Nav1.1 expression in a chronic cerebral hypoperfusion model (Hu et al. 2019). To assess whether seizures affected expression of miR-132-3p or miR-212-3p, we performed RT-ddPCR and compared expression between F1.Scn1a+/− mice with and without recent seizures. Consistent with prior reports, we found that F1.Scn1a+/− mice with recent seizures had elevated levels of miR-132-3p and a trend toward elevated levels of miR-212-3p relative to those that were seizure-free in the antecedent 24 h (Fig. 4). This suggests the possibility that seizure-mediated elevation of miR-132-3p and/or miR-212-3p could downregulate expression of NaV1.1 and exacerbate the effect of Scn1a heterozygous deletion.
Seizure responsive positional candidate miRNA genes with seed match sites in the Scn1a 3’UTR. F1.Scn1a+/− mice with recent seizures had elevated levels of miR-132-3p (A) and a trend toward elevated levels of miR-212-3p (B) relative to those that were seizure-free in the antecedent 24 h. Each group had three pooled samples consisting of 4 mice each, with either no seizures or 3–7 seizures per mouse in the 24 h prior to collection. Floating bars represent the range of values for each group. p-values are from Welch’s t-tests
Discussion
In the present study, we constructed a set of ISCs to fine map the Dsm5 locus on mouse chromosome 11. Under a single gene modifier model, our results narrow the modifier interval to a 5.7 Mb region of overlap between ISC-E and ISC-F. Within this interval, we identified a number of potential candidate modifier genes with coding sequence variation and/or expression differences between the strains. However, future studies will be needed to validate candidate modifier genes.
Among the Dsm5 potential modifier genes, Trpv1 is an intriguing candidate as a proposed target of cannabidiol, an FDA/EMA-approved Dravet syndrome therapeutic (Gray and Whalley 2020). Recently, we reported differential expression of Trpv1 transcript in cortex, with higher expression in F1.Scn1a+/− mice relative to seizure resistant 129.Scn1a+/− mice (Satpute Janve et al. 2021). However, deletion of Trpv1 had both pro- and anti-convulsant effects when combined with the Scn1a+/− allele. Double mutant F1.Scn1a+/−;Trpv1+/− mice had lower temperature thresholds for hyperthermia-induced seizure compared to F1.Scn1a+/− mice, suggesting a pro-convulsant effect. Conversely, Trpv1 deletion resulted in reduced severity of spontaneous GTCS in double mutant F1.Scn1a+/−;Trpv1+/− mice, although it did not affect seizure frequency or survival compared to F1.Scn1a+/− mice (Satpute Janve et al. 2021). Although Trpv1 deletion alters seizure phenotypes in Scn1a+/− mice, it is not possible to separate an effect of Trpv1 deletion from residual 129 alleles in the chromosome 11 region remaining from the homologous recombination in JM-1 ES cells (129X1/SvJ) (Caterina et al. 2000). Lack of any effect of a Trpv1-selective antagonist on F1.Scn1a+/− phenotypes lends support for an effect of residual 129 alleles rather than Trpv1 deletion itself on Scn1a+/− seizure phenotypes (Satpute Janve et al. 2021). Additional intriguing candidates include the miRNA genes for miR-132-3p and miR-212-3p that are predicted to target Scn1a and are seizure-responsive genes. However, to date, we have no evidence for altered expression of Scn1a transcript between strains or in response to seizures. It is possible that miRNA regulation could be post-transcriptional and alter protein levels,
The Dsm5 region overlaps previously identified seizure susceptibility QTL, including Szs10 and Gss4 (Ferraro et al. 2001; Gu et al. 2020). In addition, this region of mouse chromosome 11 is syntenic with recurrent CNVs implicated in epilepsy and neurodevelopmental disorders (Coppola et al. 2019; Kolishovski et al. 2019). This includes synteny with Miller-Dieker lissencephaly 17p13.3 microdeletion syndrome, which includes epilepsy as part of the neurological features. The 17p13.3 microdeletion includes deletion of PAFAH1B1 and YWHAE, as well as deletion of the intervening genes. The lissencephaly phenotype is attributed to deletion of PAFAH1B1, formerly known as LIS1, while disruption of YWHAE, encoding 14–3-3ε, is associated with variable structure brain abnormalities, cognitive impairment and seizures (Cardoso et al. 2000; Noor et al. 2018; Romano et al. 2020). Rare microduplications at 17q12 are associated with epilepsy, including familial FS/GEFS + that is also often caused by SCN1A variants (Hardies et al. 2013; Mefford et al. 2016, 2007). Genes associated with the 17q12 microduplication are AATF, ACACA, C17orf78, DDX52, DHRS11, DUSP14, GGNBP2, HNF1B, LHX1, MRM1, MYO19, PIGW, SYNRG, TADA2A, and ZNHIT3 (Mefford et al. 2016). LHX1, encoding LIM homeobox protein 1, and ACACA, encoding acetyl-CoA carboxylase, have been proposed as potential candidate genes responsible for neurodevelopmental and epilepsy phenotypes, although there is not sufficient evidence to confirm causality to date (Hardies et al. 2013; Mefford et al. 2007). The 17p13.3 deletion overlaps with the region of overlap between ISC-E and ISC-F, while the 17q12 microduplication overlaps with the non-overlapping segment of ISC-F.
One caveat of our study is that the fine mapping data could also support an alternative interpretation of additive modifier genes from the non-overlapping portions of ISC-E and ISC-F. This seems less likely when examining the Kaplan–Meier survival curves (Fig. 1C, D), where the net effect of ISC-E, ISC-F or ISC-D appear similar and are not significantly different from one another (p > 0.3, LogRank Mantel-Cox). However, examination of the female hazard ratios shows a modest trend toward smaller magnitude of effects for ISC-E (2.258 HR) and ISC-F (2.162 HR) relative to ISC-D (3.183 HR) (Fig. 1C), which could support the possibility of each interval harboring modifiers with additive effects. Additional support for this hypothesis comes from males that showed robust improvement in survival with ISC-D, whereas ISC-E or ISC-F alone had no protective effect. Ideally, these two possibilities could be tested empirically by generation of ISC lines separating the E–F overlapping and non-overlapping regions; however, such recombination events have been elusive in our ISC colony. It may be possible in the future to use genome engineering to further dissect the interval.
Haploinsufficiency for SCN1A is a major cause of Dravet syndrome, and yet there is variable expressivity among patients with this shared genetic basis. This suggests that clinical presentation is influenced by other factors, which may include genetic modifiers. Identification of modifier genes that influence disease course will provide insights into understanding the molecular basis of Dravet syndrome and identify potential pathways for intervention.
Data availability
Genomic and transcriptomic datasets generated during and/or analyzed during the current study are available in the NCBI GEO repository [GSE112627] and SRA repository [PRJNA817075]. Other datasets are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife. https://doi.org/10.7554/eLife.05005
Battaglia D, Chieffo D, Lucibello S, Marini C, Sibilia V, Mei D, Darra F, Offredi F, Fontana E, Specchio N, Cappelletti S, Granata T, Ragona F, Patrini M, Baglietto MG, Prato G, Ferrari A, Vigevano F, Mercuri E, Bernardina BD, Guerrini R, Dravet C, Guzzetta F (2021) Multicenter prospective longitudinal study in 34 patients with Dravet syndrome: Neuropsychological development in the first six years of life. Brain Dev 43:419–430. https://doi.org/10.1016/j.braindev.2020.10.004
Cardoso C, Leventer RJ, Matsumoto N, Kuc JA, Ramocki MB, Mewborn SK, Dudlicek LL, May LF, Mills PL, Das S, Pilz DT, Dobyns WB, Ledbetter DH (2000) The location and type of mutation predict malformation severity in isolated lissencephaly caused by abnormalities within the LIS1 gene. Hum Mol Genet 9:3019–3028. https://doi.org/10.1093/hmg/9.20.3019
Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D (2000) Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 288:306–313. https://doi.org/10.1126/science.288.5464.306
Coppola A, Cellini E, Stamberger H, Saarentaus E, Cetica V, Lal D, Djémié T, Bartnik-Glaska M, Ceulemans B, Helen Cross J, Deconinck T, Masi S, Dorn T, Guerrini R, Hoffman-Zacharska D, Kooy F, Lagae L, Lench N, Lemke JR, Lucenteforte E, Madia F, Mefford HC, Morrogh D, Nuernberg P, Palotie A, Schoonjans AS, Striano P, Szczepanik E, Tostevin A, Vermeesch JR, Van Esch H, Van Paesschen W, Waters JJ, Weckhuysen S, Zara F, De Jonghe P, Sisodiya SM, Marini C (2019) Diagnostic implications of genetic copy number variation in epilepsy plus. Epilepsia 60:689–706. https://doi.org/10.1111/epi.14683
de Lange IM, Mulder F, van ’t Slot R, Sonsma ACM, van Kempen MJA, Nijman IJ, Ernst RF, Knoers N, Brilstra EH, Koeleman BPC, (2020) Modifier genes in SCN1A-related epilepsy syndromes. Mol Genet Genomic Med 8:e1103. https://doi.org/10.1002/mgg3.1103
Depienne C, Trouillard O, Saint-Martin C, Gourfinkel-An I, Bouteiller D, Carpentier W, Keren B, Abert B, Gautier A, Baulac S, Arzimanoglou A, Cazeneuve C, Nabbout R, LeGuern E (2009) Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. J Med Genet 46:183–191. https://doi.org/10.1136/jmg.2008.062323
Depienne C, Trouillard O, Gourfinkel-An I, Saint-Martin C, Bouteiller D, Graber D, Barthez-Carpentier MA, Gautier A, Villeneuve N, Dravet C, Livet MO, Rivier-Ringenbach C, Adam C, Dupont S, Baulac S, Héron D, Nabbout R, Leguern E (2010) Mechanisms for variable expressivity of inherited SCN1A mutations causing Dravet syndrome. J Med Genet 47:404–410. https://doi.org/10.1136/jmg.2009.074328
Dravet C (2011) The core Dravet syndrome phenotype. Epilepsia 52(Suppl 2):3–9. https://doi.org/10.1111/j.1528-1167.2011.02994.x
Dravet C, Oguni H (2013) Dravet syndrome (severe myoclonic epilepsy in infancy). Handb Clin Neurol 111:627–633. https://doi.org/10.1016/b978-0-444-52891-9.00065-8
Ferraro TN, Golden GT, Smith GG, Longman RL, Snyder RL, DeMuth D, Szpilzak I, Mulholland N, Eng E, Lohoff FW, Buono RJ, Berrettini WH (2001) Quantitative genetic study of maximal electroshock seizure threshold in mice: evidence for a major seizure susceptibility locus on distal chromosome 1. Genomics 75:35–42. https://doi.org/10.1006/geno.2001.6577
Fukunaga T, Iwakiri J, Ono Y, Hamada M (2019) LncRRIsearch: A Web Server for lncRNA-RNA interaction prediction integrated with tissue-specific expression and subcellular localization data. Front Genet 10:462. https://doi.org/10.3389/fgene.2019.00462
Gerbatin RR, Augusto J, Boutouil H, Reschke CR, Henshall DC (2021) Sexual dimorphism in epilepsy and comorbidities in Dravet syndrome mice carrying a targeted deletion of exon 1 of the Scn1a gene. bioRxiv. https://doi.org/10.1101/2021.08.27.457904
Goldberg-Stern H, Aharoni S, Afawi Z, Bennett O, Appenzeller S, Pendziwiat M, Kuhlenbäumer G, Basel-Vanagaite L, Shuper A, Korczyn AD, Helbig I (2014) Broad phenotypic heterogeneity due to a novel SCN1A mutation in a family with genetic epilepsy with febrile seizures plus. J Child Neurol 29:221–226. https://doi.org/10.1177/0883073813509016
Gray RA, Whalley BJ (2020) The proposed mechanisms of action of CBD in epilepsy. Epileptic Disord 22:10–15. https://doi.org/10.1684/epd.2020.1135
Gu B, Shorter JR, Williams LH, Bell TA, Hock P, Dalton KA, Pan Y, Miller DR, Shaw GD, Philpot BD, Pardo-Manuel de Villena F (2020) Collaborative Cross mice reveal extreme epilepsy phenotypes and genetic loci for seizure susceptibility. Epilepsia 61:2010–2021. https://doi.org/10.1111/epi.16617
Guerrini R, Cellini E, Mei D, Metitieri T, Petrelli C, Pucatti D, Marini C, Zamponi N (2010) Variable epilepsy phenotypes associated with a familial intragenic deletion of the SCN1A gene. Epilepsia 51:2474–2477. https://doi.org/10.1111/j.1528-1167.2010.02790.x
Hammer MF, Ishii A, Johnstone L, Tchourbanov A, Lau B, Sprissler R, Hallmark B, Zhang M, Zhou J, Watkins J, Hirose S (2017) Rare variants of small effect size in neuronal excitability genes influence clinical outcome in Japanese cases of SCN1A truncation-positive Dravet syndrome. PLoS ONE 12:e0180485. https://doi.org/10.1371/journal.pone.0180485
Hardies K, Weckhuysen S, Peeters E, Holmgren P, Van Esch H, De Jonghe P, Van Paesschen W, Suls A (2013) Duplications of 17q12 can cause familial fever-related epilepsy syndromes. Neurology 81:1434–1440. https://doi.org/10.1212/WNL.0b013e3182a84163
Hawkins NA, Zachwieja NJ, Miller AR, Anderson LL, Kearney JA (2016) Fine mapping of a dravet syndrome modifier locus on mouse chromosome 5 and candidate gene analysis by RNA-Seq. PLoS Genet 12:e1006398. https://doi.org/10.1371/journal.pgen.1006398
Hawkins NA, Calhoun JD, Huffman AM, Kearney JA (2019) Gene expression profiling in a mouse model of Dravet syndrome. Exp Neurol 311:247–256. https://doi.org/10.1016/j.expneurol.2018.10.010
Hu XL, Wang XX, Zhu YM, Xuan LN, Peng LW, Liu YQ, Yang H, Yang C, Jiao L, Hang PZ, Sun LH (2019) MicroRNA-132 regulates total protein of Nav1.1 and Nav1.2 in the hippocampus and cortex of rat with chronic cerebral hypoperfusion. Behav Brain Res 366:118–125. https://doi.org/10.1016/j.bbr.2019.03.026
Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, Stallings RL, Henshall DC (2011) miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol 179:2519–2532. https://doi.org/10.1016/j.ajpath.2011.07.036
Kalume F, Westenbroek RE, Cheah CS, Yu FH, Oakley JC, Scheuer T, Catterall WA (2013) Sudden unexpected death in a mouse model of Dravet syndrome. J Clin Invest 123:1798–1808. https://doi.org/10.1172/jci66220
Kolishovski G, Lamoureux A, Hale P, Richardson JE, Recla JM, Adesanya O, Simons A, Kunde-Ramamoorthy G, Bult CJ (2019) The JAX Synteny Browser for mouse-human comparative genomics. Mamm Genome 30:353–361. https://doi.org/10.1007/s00335-019-09821-4
Li W, Schneider AL, Scheffer IE (2021) Defining Dravet syndrome: An essential pre-requisite for precision medicine trials. Epilepsia 62:2205–2217. https://doi.org/10.1111/epi.17015
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F (2016) The ensembl variant effect predictor. Genome Biol 17:122. https://doi.org/10.1186/s13059-016-0974-4
Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, Kapur R, Pinkel D, Cooper GM, Ventura M, Ropers HH, Tommerup N, Eichler EE, Bellanne-Chantelot C (2007) Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet 81:1057–1069. https://doi.org/10.1086/522591
Miller AR, Hawkins NA, McCollom CE, Kearney JA (2014) Mapping genetic modifiers of survival in a mouse model of Dravet syndrome. Genes Brain Behav 13:163–172. https://doi.org/10.1111/gbb.12099
Mooney C, Becker BA, Raoof R, Henshall DC (2016) EpimiRBase: a comprehensive database of microRNA-epilepsy associations. Bioinformatics 32:1436–1438. https://doi.org/10.1093/bioinformatics/btw008
Nabbout R, Gennaro E, Dalla Bernardina B, Dulac O, Madia F, Bertini E, Capovilla G, Chiron C, Cristofori G, Elia M, Fontana E, Gaggero R, Granata T, Guerrini R, Loi M, La Selva L, Lispi ML, Matricardi A, Romeo A, Tzolas V, Valseriati D, Veggiotti P, Vigevano F, Vallée L, Dagna Bricarelli F, Bianchi A, Zara F (2003) Spectrum of SCN1A mutations in severe myoclonic epilepsy of infancy. Neurology 60:1961–1967. https://doi.org/10.1212/01.wnl.0000069463.41870.2f
Niibori Y, Lee SJ, Minassian BA, Hampson DR (2020) Sexually divergent mortality and partial phenotypic rescue after gene therapy in a mouse model of dravet syndrome. Hum Gene Ther 31:339–351. https://doi.org/10.1089/hum.2019.225
Noor A, Bogatan S, Watkins N, Meschino WS, Stavropoulos DJ (2018) Disruption of YWHAE gene at 17p13.3 causes learning disabilities and brain abnormalities. Clin Genet 93:365–367. https://doi.org/10.1111/cge.13056
Osaka H, Ogiwara I, Mazaki E, Okamura N, Yamashita S, Iai M, Yamada M, Kurosawa K, Iwamoto H, Yasui-Furukori N, Kaneko S, Fujiwara T, Inoue Y, Yamakawa K (2007) Patients with a sodium channel alpha 1 gene mutation show wide phenotypic variation. Epilepsy Res 75:46–51. https://doi.org/10.1016/j.eplepsyres.2007.03.018
Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, Emerson M, Garner P, Holgate ST, Howells DW, Karp NA, Lazic SE, Lidster K, MacCallum CJ, Macleod M, Pearl EJ, Petersen OH, Rawle F, Reynolds P, Rooney K, Sena ES, Silberberg SD, Steckler T, Würbel H (2020) The ARRIVE guidelines 20: updated guidelines for reporting animal research. BMJ Open Sci 4:e100115. https://doi.org/10.1136/bmjos-2020-100115
Romano C, Ferranti S, Mencarelli MA, Longo I, Renieri A, Grosso S (2020) 17p13.3 microdeletion including YWHAE and CRK genes: towards a clinical characterization. Neurol Sci 41:2259–2262. https://doi.org/10.1007/s10072-020-04424-3
Rubinstein M, Westenbroek RE, Yu FH, Jones CJ, Scheuer T, Catterall WA (2015) Genetic background modulates impaired excitability of inhibitory neurons in a mouse model of Dravet syndrome. Neurobiol Dis 73:106–117. https://doi.org/10.1016/j.nbd.2014.09.017
Satpute Janve V, Anderson LL, Bahceci D, Hawkins NA, Kearney JA, Arnold JC (2021) The Heat Sensing Trpv1 Receptor Is Not a Viable Anticonvulsant Drug Target in the Scn1a (+/-) Mouse Model of Dravet Syndrome. Front Pharmacol 12:675128. https://doi.org/10.3389/fphar.2021.675128
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607-d613. https://doi.org/10.1093/nar/gky1131
Trontti K, Väänänen J, Sipilä T, Greco D, Hovatta I (2018) Strong conservation of inbred mouse strain microRNA loci but broad variation in brain microRNAs due to RNA editing and isomiR expression. RNA 24:643–655. https://doi.org/10.1261/rna.064881.117
Wirrell EC, Laux L, Donner E, Jette N, Knupp K, Meskis MA, Miller I, Sullivan J, Welborn M, Berg AT (2017) Optimizing the diagnosis and management of dravet syndrome: recommendations from a North American consensus panel. Pediatr Neurol 68:18-34.e13. https://doi.org/10.1016/j.pediatrneurol.2017.01.025
Yordanova I, Todorov T, Dimova P, Hristova D, Tincheva R, Litvinenko I, Yotovska O, Kremensky I, Todorova A (2011) One novel Dravet syndrome causing mutation and one recurrent MAE causing mutation in SCN1A gene. Neurosci Lett 494:180–183. https://doi.org/10.1016/j.neulet.2011.03.008
Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, Catterall WA (2006) Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci 9:1142–1149. https://doi.org/10.1038/nn1754
Yu MJ, Shi YW, Gao MM, Deng WY, Liu XR, Chen L, Long YS, Yi YH, Liao WP (2010) Milder phenotype with SCN1A truncation mutation other than SMEI. Seizure 19:443–445. https://doi.org/10.1016/j.seizure.2010.06.010
Mefford H, Mitchell E, Hodge J (2016) 17q12 Recurrent Duplication. In Gene Reviews [Internet], M.P. Adam, H.H. Ardinger, R.A. Pagon, e. al, eds. Seattle, WA: University of Washington Seattle
Acknowledgements
We thank Nathan Speakes for technical assistance. This study was supported by National Institutes of Neurological Disorders and Stroke grant R01 NS084959 (JAK).
Funding
This work was supported by NIH/NINDS Grant NS084959 (JAK).
Author information
Authors and Affiliations
Contributions
Conceptualization: JAK; Formal analysis: JAK; Investigation: JAK, LDCH, NAH, SD, NAZ, IKE; Methodology: NAH, LDCH, JAK; Project administration: JAK; Supervision: JAK; Visualization: JAK; Writing – original draft: JAK; Writing – review & editing: LDCH, SD, NAZ, IKEF, NAH, JAK; Funding acquisition: JAK.
Corresponding author
Ethics declarations
Conflict of interest
JAK serves as a consultant to Encoded Genomics, Praxis Precision Medicines and NeuroCycle Therapeutics, and serves on the Scientific Advisory Boards of the Dravet Syndrome Foundation and FamilieSCN2A Foundation. JAK receives research funding from Praxis Precision Medicines and Pfizer. NAH serves as a consultant to NeuroCycle Therapeutics. All other authors have declared that no competing interests exist.
Ethical approval
All animal care and experimental procedures were approved by the Northwestern University Animal Care and Use Committee in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kearney, J.A., Copeland-Hardin, L.D., Duarte, S. et al. Fine mapping and candidate gene analysis of a dravet syndrome modifier locus on mouse chromosome 11. Mamm Genome 33, 565–574 (2022). https://doi.org/10.1007/s00335-022-09955-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-022-09955-y