Abstract
Objectives
Existing brain extraction models should be further optimized to provide more information for oncological analysis. We aimed to develop an nnU-Net–based deep learning model for automated brain extraction on contrast-enhanced T1-weighted (T1CE) images in presence of brain tumors.
Methods
This is a multi-center, retrospective study involving 920 patients. A total of 720 cases with four types of intracranial tumors from private institutions were collected and set as the training group and the internal test group. Mann–Whitney U test (U test) was used to investigate if the model performance was associated with pathological types and tumor characteristics. Then, the generalization of model was independently tested on public datasets consisting of 100 glioma and 100 vestibular schwannoma cases.
Results
In the internal test, the model achieved promising performance with median Dice similarity coefficient (DSC) of 0.989 (interquartile range (IQR), 0.988–0.991), and Hausdorff distance (HD) of 6.403 mm (IQR, 5.099–8.426 mm). U test suggested a slightly descending performance in meningioma and vestibular schwannoma group. The results of U test also suggested that there was a significant difference in peritumoral edema group, with median DSC of 0.990 (IQR, 0.989–0.991, p = 0.002), and median HD of 5.916 mm (IQR, 5.000–8.000 mm, p = 0.049). In the external test, our model also showed to be robust performance, with median DSC of 0.991 (IQR, 0.983–0.998) and HD of 8.972 mm (IQR, 6.164–13.710 mm).
Conclusions
For automated processing of MRI neuroimaging data presence of brain tumors, the proposed model can perform brain extraction including important superficial structures for oncological analysis.
Clinical relevance statement
The proposed model serves as a radiological tool for image preprocessing in tumor cases, focusing on superficial brain structures, which could streamline the workflow and enhance the efficiency of subsequent radiological assessments.
Key Points
• The nnU-Net–based model is capable of segmenting significant superficial structures in brain extraction.
• The proposed model showed feasible performance, regardless of pathological types or tumor characteristics.
• The model showed generalization in the public datasets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brain extraction, or skull stripping, refers to the process of removing skull and non-brain tissue in medical images. It is considered as a preliminary but important pre-processing step as its accuracy has a direct influence on the quality of subsequent image processing and the reliability of statistical analysis [1,2,3,4,5,6]. Manual segmentation of the brain is laborious, tedious, and time-consuming, commonly leading to significant inter- and intra-reader variations that may lead to analysis deviation [7]. With the development of deep learning methods in recent years, particularly in convolutional neural network (CNN) algorithms, automated methods have obtained state-of-the-art results in medical image segmentation [8,9,10,11]. However, the brain extraction models for cases present with brain tumors on contrast-enhanced T1-weighted (T1CE) magnetic resonance images (MRIs) should be further optimized to meet the demands from both clinicians and neuroimaging researchers.
Nowadays, the criteria defined by Eskildsen et al were widely used in previous studies. It is cited as follows: (a) inclusion of cerebrum, cerebellum, brainstem, and internal vessels and arteries, along with cerebrospinal fluid in ventricles, internal cisterns, and deep sulci; (b) exclusion of the skin, skull, eyes, dura mater, external blood vessels, and nerves [12], whereas, as the most important radiological examination for brain tumor evaluation, more information should be included in region of interest (ROIs) to provide a clear depiction of lesion location, boundary, and the relationship between tumor and adjacent structures.
Generally, the first concern is that the superficial structures on brain surface are not included in the mask (mainly referring to venous sinus and superficial vessels), while exclusion of these structures can significantly affect subsequent therapeutic decisions or oncological analysis [6, 13,14,15,16]. For example, in preoperative simulation that uses extracted brain for 3D reconstruction, tumoral invasion of structures on the brain surface is one of the major concerns for neurosurgeons that can significantly determine the surgical strategy [15, 17]. Exclusion of these structures may be inadequate and not appropriate. The second one is that all previous CNN models are trained and validated on datasets with only glioma cases [18,19,20,21]. As acknowledged by previous studies, the performance of supervised CNN models that were exclusively trained on scans of glioma subjects may be limited in cases with different types of brain tumors [18, 19]. Laborious manual correction and delineation may be still needed in these studies. The third one is that whether or to what extent can tumor characteristics influence model performance has not been answered yet. Obtaining the overall highest performance has become the primary objective in all previous studies, undermining other clinical concerns such as reliability, generalization, and convenience of a new method on tumor entities [18,19,20,21,22]. Previous methodological studies showed advanced performance with Dice similarity coefficients (DSCs) of more than 0.950, while the features of different types of brain tumors showed interspecific and intraspecific differences that may also lead to performance deviation [23, 24].
Therefore, to achieve the objective of clinical translation and widespread usage, a CNN model was developed with the latest state-of-art CNN architecture to perform automated brain extraction on T1CE MRIs in presence of brain tumors. Our model was optimized by including more brain surface structures in training, and by involving a multi-center dataset covering diversified tumor entities. Moreover, a series of intra-group analyses were performed to investigate if our model could feasibly segment brain images regardless of tumor characteristics.
Materials and methods
Datasets
This was a retrospective, multi-center research. Figure 1 shows the flowchart of patient selection. In private center A and center B, 532 cases and 368 cases were initially selected from the radiological department between January 2016 and December 2021. All of the cases underwent standard pre-treatment magnetic resonance scans and received surgical resection in our institutions. In total, 180 patients were excluded based on the following exclusion criteria: (1) MRIs with severe motion artifacts (N = 85); 92) intervention history before MR scans, such as biopsy and radiotherapy (N = 52); 93) recorded history of other brain diseases, such as hypertensive intracerebral hemorrhage (N = 43). Eventually, 429 cases from center A and 291 cases from center B were included in the current study.
The T1CE images closest to clinical intervention were collected from the Picture Archiving and Communication System (PACS), including three orientations of axial, sagittal, and coronal views. Clinical features and radiological features were also collected and interpreted, including age, gender, pathological diagnosis, and imaging findings. Detailed MR scanning parameters are shown in Supplemental Material 1.
As for public datasets, a total number of 200 patients with complete, high-quality MR scans were randomly selected from three The Cancer Imaging Archive (TCIA) datasets, including The Cancer Genome Atlas Glioblastoma (TCGA-GBM) [25], The Cancer Genome Atlas Lower Grade Glioma (TCGA-LGG) [26], American College of Radiology Imaging Network (ACRIN) 6684 [27], and Vestibular-Schwannoma-SEG [28].
Definition of brain mask and ground truth manual segmentation
We defined the following criteria for ground truth mask: (a) including all cerebral and cerebellar gray and white matter, brainstem, cerebrospinal fluid in the ventricles, and the cerebellar cistern, lesion sites in the brain, superficial venous sinuses (sagittal sinus and transverse sinus), and (b) excluding the skin, skull, eyes, dura mater, cavernous sinus area, and exterior blood vessels and nerves (such as carotid arteries and optic chiasm).
Manual segmentation was performed using 3D Slicer software [29]. The brain masks were segmented by five experienced neuroradiologists with more than 10 years of experience in image reading. Following the instructions of software, the mask was delineated on three orientations in consensus reading, followed by reviewing and correction by two senior neuroradiologists (F.M.Z. and J.G.X., with more than 20 years of experience in image reading). To examine intra-observer repeatability, thirty cases were randomly selected and segmented again with at least 15-day interval.
Deep learning model for automated brain extraction
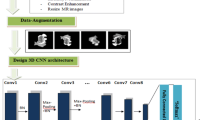
The latest state-of-art segmentation CNN network, nnU-Net, was used for modeling. It is an advanced CNN network containing a deep learning–based self-configuration module that can automatically configure image pre-processing, network architecting, cross-validation training, and post-processing [30]. The 3D full-resolution U-Net model was determined as the optimal model architecting, and the configuration of the model is provided in Supplemental Material 2. As shown in Fig. 1, the cases from private datasets were randomly divided into the training group and the internal test group in a ratio of 4:1. A fivefold cross-validation strategy was used in model training.
Public datasets were used as the independent test group, where the generalization of trained model was tested. We also validated the performance of existing methods on these TCIA datasets, including Robust Learning-Based Brain Extraction (ROBEX), HD-BET, and Brain Mask Generator (BrainMaGe) [18, 19, 31]. All experiments were written in Python language, and were performed in the machine equipped with four NVIDIA 3090 data center accelerator.
Evaluation metrics and statistical analysis
The imaging findings were interpreted by two senior radiologists (F.M.Z. and J.G.X.). The CNN models’ performance was evaluated by comparing the ground truth and the prediction. A series of commonly used metrics were calculated, including median DSC, false-negative rate (FNR), and false-positive rate (FPR). Two metrics were also introduced to make a better demonstration of model performance in segmenting brain surface, including Hausdorff distance (HD), for measuring the maximal contour distance (mm) between the ground truth masks and the predictions, and mean surface distance (MSD), for measuring the average distance (mm) between two masks’ boundaries.
Categorical variables were presented with frequencies and percentages, and continuous variables were presented with medians and interquartile range (IQR). Kolmogorov–Smirnov test, Mann–Whitney U test (U test), and Wilcoxon signed rank test (Wilcoxon test) were carried out in intra-group analysis, as appropriate. Statistical analysis was performed with GraphPad Prism. p < 0.05 implicated statistical significance. 3DMeshMetric was used to visualize the spatial distribution of errors between the ground truth mask and the prediction. Volume rendering of 3D brain image was performed by using composite with shading technology without surface smoothing.
Results
Patient characteristics
A total number of 720 patients (720 exams) were collected from private institution A and institution B. The mean age of patients was 53 years old, and the sex ratio was male:female = 163:197. As for the pathological distribution, 505 cases were diagnosed with meningioma, 50 cases with low-grade glioma, 78 cases with high-grade glioma, and 87 cases with vestibular schwannoma. For the 200 cases collected from the public dataset, twenty-one cases were present with low-grade glioma, 79 cases were with high-grade glioma, and 100 cases were with vestibular schwannoma. The clinical characteristics of the internal and external datasets are represented in Table 1.
Model performance in internal test
Overall, the automated model performed well in the internal test, with median DSC of 0.989 (IQR, 0.988–0.991), FNR of 0.012 (IQR, 0.009–0.015), FPR of 0.008 (IQR, 0.006–0.012), HD of 6.403 mm (IQR, 5.099–8.426 mm), and MSD of 0.013 mm (IQR, 0.011–0.015 mm). Model segmentation performance of cases from the internal test group is shown in Fig. 2 and Supplemental Material 3a.
The models’ performance regarding pathological type is presented in Table 2. In particular, the model exhibited slightly inferior but statistically significant performance in the meningioma dataset, with a median DSC of 0.989 (IQR, 0.987–0.990, p < 0.001) and a median HD of 7.000 mm (IQR, 5.477–9.165 mm, p = 0.001). Similar outcomes were observed in the vestibular schwannoma cohort, which represents another type of extra-axial tumor, with a median DSC of 0.990 (IQR, 0.990–0.991, p = 0.027) and a median HD of 5.385 mm (IQR, 4.243–6.708 mm, p = 0.010). However, there was no significant difference in the low-grade and high-grade glioma groups. The box chart and heat map of this intra-group analysis are shown in Supplemental Material 4a and 4b.
Model performance regarding radiological characteristics
Among the cases evaluated, 35.4% presented with peritumoral edema (PE). As demonstrated in Table 3, results from the U test revealed a significant disparity in the PE group, with a median DSC of 0.990 (IQR, 0.989–0.991, p = 0.002), and a median HD of 5.916 mm (IQR, 5.000–8.000 mm, p = 0.049). Invaded venous sinus (IVS) was detected in 240 cases (33.3%), and 51.5% of tumors were located in the skull base. However, intra-group analysis indicated non-significant results in cases with IVS (DSC = 0.990 (IQR, 0.988–0.991, p = 0.124), HD = 6.000 mm (IQR, 5.000–8.062 mm, p = 0.155)), or in cases with skull base tumors (DSC = 0.989 (IQR, 0.987–0.990, p = 0.553); HD = 7.211 mm (IQR, 5.385–8.775 mm, p = 0.398)). Supplemental Material 4c shows the box plot that indicates the intra-group analysis to investigate whether model performance is related to tumor characteristics.
Model generalization
The model achieved robust generalization in the independent datasets with median DSC of 0.991 (IQR, 0.983–0.998), FNR of 0.003 (IQR, 0.000–0.011), FPR of 0.008 (IQR, 0.004–0.020), HD of 8.972 mm (IQR, 6.164–13.710 mm), and MSD of 0.013 mm (IQR, 0.006–0.022 mm). Predictions of good and poor examples from the external test group are shown in Fig. 3 and Supplemental Material 3. However, the model exhibited significantly lower performance in the glioma dataset compared to the vestibular schwannoma group in terms of Dice similarity coefficient (DSC) (0.983 vs 0.998, p < 0.001) and Hausdorff distance (HD) (7.000 mm vs 12.860 mm, p < 0.001).
Performance of the existing models
In the internal test, the DSCs of HD-BET, BrainMaGe, Robex were 0.989 (IQR, 0.980–0.993), 0.961 (IQR, 0.945–0.970), and 0.957 (IQR, 0.948–0.963), respectively. The HDs were 9.165 mm (IQR, 7.681–11.550 mm), 27.330 mm (IQR, 12.240–53.920 mm, p < 0.001), and 12.530 mm (IQR, 11.000–14.20 mm, p < 0.001), respectively. Similar results were also suggested in the external test. The existing models’ performance is summarized in Table 4, and more detailed evaluation metrics regarding tumor pathological types are shown in Supplemental Material 5.
Automated 3D brain surface rendering
Figure 4 shows the samples of 3D brain images using the mask generated by the nnU-Net model. The overall review of brain was shown. Important structures were well displayed, including superior sagittal sinus, transverse sinus, superficial vein, anterior skull base, brain stem, vertebral artery, and basilar artery. The nnU-Net model showed relatively low error rates in the segmenting skull base, superior sagittal sinus, transverse sinus, and brain stem (Fig. 5).
Error distribution map was formed by comparison between ground truth mask and prediction of each model. Qualitative and quantitative were assessed by point value, and the points range was set from 0 to 5.00. The color in the figure corresponds to the point error (the red area represents a large error, while the green area represents a small error)
Intra-observer reproducibility, and comparison of radiologists and models
For each brain mask, it took approximately 1 h and 20 min for the manual procedure of delineation, check, and correction, while the nnU-Net model took 4 min to perform inference on both CPU and GPU. As for the randomly selected 30 patients, Bland–Altman plots suggested that there was good intra-observer reproducibility of manual segmentation (Supplemental Material 6A). Bland–Altman plots also indicated that the automated method performed as well as expert manual segmentation despite lesion characteristics, as presented in Supplemental Material 6B and Supplemental Material 6C.
Discussion
In the present study, an automated model utilizing deep learning technology was developed to achieve rapid and robust brain extraction on T1CE MRIs in the presence of intracranial tumors. Our model confers an advantage in that it incorporates superficial structures into the brain extraction mask, a crucial factor in oncological analysis that had been previously unaddressed in other studies. The proposed method is applicable to a wide range of MRI hardware and acquisition parameters commonly encountered in both clinical and research practice. The model was trained on a large multi-center MRI dataset and subsequently tested for generalizability on three independent public datasets, resulting in DSCs of over 0.980.
Compared to previous methodological studies (summarized in the Supplemental Material 7), the present study was more clinically relevant, addressed several shortcomings concerning clinicians, and improved the model performances with the latest CNN network. First, we improved the brain extraction criterion by incorporating superficial brain structures and fine-tuned model performance on these regions, which were used during model training. Unlike other imaging sequences, for neurosurgeons and neuro-radiologists, who might primarily rely on a CNN model for brain extraction on T1CE MRIs, discerning the anatomical correlation between tumors and adjacent structures is a crucial aspect of clinical decision-making [15]. Previous studies failed to address this issue, resulting in poor model performance in these areas. Secondly, we evaluated the model’s performance on four different types of tumors. Although previous studies have reported promising results, none has tested the trained model on a dataset that covers multiple types of contrast-enhanced images of tumors. In contrast, our model was trained and tested on both intra-axial and extra-axial tumors, and the good performance indicated its good generalizability. Thirdly, we conducted an intra-group analysis to examine whether the model could be generalized despite the diverse image patterns of brain tumors. Finally, we assessed the generalizability of our model on four external datasets that were independent and publicly available. All of these improvements have enhanced the practicality of our model and expanded the potential for its clinical translation and widespread usage.
Shape fidelity of the automated segmentation outline to the true brain mask is very important. While the DSC is commonly used as a metric for evaluating segmentation performance, it is insensitive to differences in edges that have a small volumetric effect relative to the total volume. Therefore, we combined the DSC with the HD in our intra-group analysis. In this analysis, we found some intriguing results that require further clarification. Overall, the model demonstrated promising performance with median DSCs exceeding 0.980 in each group, despite the distinct appearances of the four tumor entities. However, in the internal dataset, we observed slightly decreasing DSCs in the meningioma and vestibular schwannoma groups, whereas in the external dataset, the model exhibited an increase in HD in the vestibular schwannoma group. These two extra-axial tumors originate in the meninges and cranial nerves, leading to severe structural abnormalities that may account for the decreased model performance. Additionally, the results suggested that the model performed better in tumors presenting with PE, potentially because the swollen cortex displayed a darkening intensity, increasing the contrast between the brain and enhanced meninges and thereby facilitating segmentation. Although the statistical performance remained feasible, these findings are significant, as manual correction may be necessary in such cases.
Our research has several limitations. First, it was a multicenter, retrospective study with inherent selection bias. Second, compared with previous studies, only contrast-enhanced images were used, and other sequences, including T1WI, T2WI, and FLAIR, were not involved in our research. Image co-registration may be required if researchers want to perform skull stripping in these sequences. Third, our study lacked methodological novelty in terms of the used CNN structure. Although the network architecture used in this study was based on the classic nnU-net framework, its performance was remarkable, and we did not feel it necessary to optimize the network further. Fourth, due to ethical constraints, the model was solely trained and evaluated on cases with tumors. Therefore, additional studies are required to confirm the generalizability of our results.
In conclusion, we have presented a novel fully automatic deep learning model for brain extraction on T1CE MR scans. The proposed model enables extraction of the brain with tumor and provides more detailed information about the brain surface. Our study demonstrates that the model has a high level of performance and generalization in the segmentation task, which could potentially alleviate the workload of radiologists and offer a valuable tool for future neuroimaging research and oncological studies.
Data availability
The MR images or related data were not allowed to be shared publicly as privacy-protection policy, whereas we would like to share the brain-extraction model in the Git-hub (https://github.com/XU-NET/BEIT).
Abbreviations
- ACRIN:
-
American College of Radiology Imaging Network
- BrainMaGe:
-
Brain mask generator
- CE:
-
Cross-entropy
- CNN:
-
Convolutional neural network
- DCS:
-
Dice similarity coefficient
- FNR:
-
False-negative rate
- FPR:
-
False-positive rate
- HD:
-
Hausdorff distance
- IQR:
-
Interquartile range
- IVS:
-
Invaded venous sinus
- MRI:
-
Magnetic resonance image
- MSD:
-
Mean surface distance
- PACS:
-
Picture Archiving and Communication System
- PE:
-
Peritumoral edema
- ROBEX:
-
Robust Learning-Based Brain Extraction
- ROIs:
-
Region of interest
- T1CE:
-
Contrast-enhanced T1-weighted
- TCGA-GBM:
-
The Cancer Genome Atlas Glioblastoma
- TCGA-LGG:
-
The Cancer Genome Atlas Lower Grade Glioma
- TCIA:
-
The Cancer Imaging Archive
- U test:
-
Mann-Whitney U test
- Wilcoxon test:
-
Wilcoxon signed rank test
References
Cepeda S, Pérez-Nuñez A, García-García S et al (2021) Predicting short-term survival after gross total or near total resection in glioblastomas by machine learning-based radiomic analysis of preoperative MRI. Cancers (Basel) 13:5047
Haubold J, Hosch R, Parmar V et al (2021) Fully automated MR based virtual biopsy of cerebral gliomas. Cancers (Basel) 13:6186
Hsu DG, Ballangrud Å, Shamseddine A et al (2021) Automatic segmentation of brain metastases using T1 magnetic resonance and computed tomography images. Phys Med Biol 66:175014
Mert A, Buehler K, Sutherland GR et al (2012) Brain tumor surgery with 3-dimensional surface navigation. Neurosurgery 71:ons286-294 (discussion ons294-285)
Theyers AE, Zamyadi M, O’Reilly M et al (2021) Multisite comparison of MRI defacing software across multiple cohorts. Front Psychiatry 12:617997
Yang Z, Liu H, Liu Y et al (2020) A Web-based brain metastases segmentation and labeling platform for stereotactic radiosurgery. Med Phys 47:3263–3276
Souza R, Lucena O, Garrafa J et al (2018) An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. Neuroimage 170:482–494
Chan HP, Hadjiiski LM, Samala RK (2020) Computer-aided diagnosis in the era of deep learning. Med Phys 47:e218–e227
Bernal J, Kushibar K, Asfaw DS et al (2019) Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review. Artif Intell Med 95:64–81
Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E (2019) Convolutional neural networks for radiologic images: a radiologist’s guide. Radiology 290:590–606
Pratt R, Deprest J, Vercauteren T, Ourselin S, David AL (2015) Computer-assisted surgical planning and intraoperative guidance in fetal surgery: a systematic review. Prenat Diagn 35:1159–1166
Eskildsen SF, Coupé P, Fonov V et al (2012) BEaST: brain extraction based on nonlocal segmentation technique. Neuroimage 59:2362–2373
Khanna O, Fathi Kazerooni A, Farrell CJ et al (2021) Machine learning using multiparametric magnetic resonance imaging radiomic feature analysis to predict Ki-67 in World Health Organization grade I meningiomas. Neurosurgery 89:928–936
Laukamp KR, Thiele F, Shakirin G et al (2019) Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol 29:124–132
Gosal JS, Tiwari S, Sharma T et al (2021) Simulation of surgery for supratentorial gliomas in virtual reality using a 3D volume rendering technique: a poor man’s neuronavigation. Neurosurg Focus 51:E23
de Oliveira F, Santos B, Silva da Costa MD, Centeno RS et al (2018) Clinical application of an open-source 3D volume rendering software to neurosurgical approaches. World Neurosurg 110:e864–e872
Magill ST, Theodosopoulos PV, McDermott MW (2016) Resection of falx and parasagittal meningioma: complication avoidance. J Neurooncol 130:253–262
Thakur S, Doshi J, Pati S et al (2020) Brain extraction on MRI scans in presence of diffuse glioma: multi-institutional performance evaluation of deep learning methods and robust modality-agnostic training. Neuroimage 220:117081
Isensee F, Schell M, Pflueger I et al (2019) Automated brain extraction of multisequence MRI using artificial neural networks. Hum Brain Mapp 40:4952–4964
Kleesiek J, Urban G, Hubert A et al (2016) Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. Neuroimage 129:460–469
Pei L, Ak M, Tahon NHM et al (2022) A general skull stripping of multiparametric brain MRIs using 3D convolutional neural network. Sci Rep 12:10826
Moldovanu S, Moraru L, Biswas A (2015) Robust skull-stripping segmentation based on irrational mask for magnetic resonance brain images. J Digit Imaging 28:738–747
Rapalino O, Smirniotopoulos JG (2016) Extra-axial brain tumors. Handb Clin Neurol 135:275–291
Rapalino O, Batchelor T, González RG (2016) Intra-axial brain tumors. Handb Clin Neurol 135:253–274
Scarpace L, Mikkelsen T, Cha S et al (2016) The Cancer Genome Atlas Glioblastoma Multiforme Collection (TCGA-GBM) (Version 4). Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.RNYFUYE9
Pedano N, Flanders AE, Scarpace L et al (2016) The Cancer Genome Atlas Low Grade Glioma Collection (TCGA-LGG) (Version 3). Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.L4LTD3TK
Ratai EM, Zhang Z, Fink J et al (2018) ACRIN 6684: multicenter, phase II assessment of tumor hypoxia in newly diagnosed glioblastoma using magnetic resonance spectroscopy. PLoS ONE 13:e0198548
Shapey J, Kujawa A, Dorent R et al (2021) Segmentation of vestibular schwannoma from MRI, an open annotated dataset and baseline algorithm. Sci Data 8:286
Fedorov A, Beichel R, Kalpathy-Cramer J et al (2012) 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30:1323–1341
Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211
Iglesias JE, Liu CY, Thompson PM, Tu Z (2011) Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 30:1617–1634
Acknowledgements
We would like to thank Jianfeng Xu from the Third People’s Hospital of Mianyang for data curation. We would also like to thank Yang Zhang, Junkai Zheng, and Yanjie Zhao for assisting us in ground truth mask delineation.
Funding
This work was supported by 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (2020HXFH036); project for clinical research, Chinese Academy of Medical Sciences (JH2022007).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jianguo Xu.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
This multicenter study was conducted at West China Hospital, Sichuan University, and the Third Peoples’ Hospital of Mianyang. Institutional Review Board approved this multi-center, retrospective study, and waived the requirement of informed consent (ID: 2021–851).
Study subjects or cohorts overlap
No.
Methodology
• retrospective
• experimental
• multicenter study
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Teng, Y., Chen, C., Shu, X. et al. Automated, fast, robust brain extraction on contrast-enhanced T1-weighted MRI in presence of brain tumors: an optimized model based on multi-center datasets. Eur Radiol 34, 1190–1199 (2024). https://doi.org/10.1007/s00330-023-10078-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-10078-4