Abstract
Climate change is a major concern for the future of marine Arctic food webs. Diet shifts of seabirds can be used as indicators of environmental changes such as species compositions of food webs. However, studies on diets are often laborious and costly, while research in vulnerable Arctic environments benefits from short visits for data collection that minimize disturbance to Arctic wildlife and the environment. DNA-metabarcoding techniques are rapidly developing and could be used as an effective method of monitoring diet choice of seabirds. We tested DNA-metabarcoding on seabird faeces collected during short visits of typically around 30 min at breeding colonies of black-legged kittiwake Rissa tridactyla (3 colonies), Brünnich’s guillemot Uria lomvia (2 colonies) and little auk Alle alle (3 colonies). DNA metabarcoding based on COI and 18S of a limited number of faeces samples revealed a wide spectrum of fish species and crustaceans in the diets of these species, comparable with or even exceeding diversity in diet composition found in conventional, more invasive techniques where birds are shot or caught and handled to obtain samples. While previous studies on diet choice of little auk, a crustacean specialist, mainly report small fractions of unidentified fish remains, DNA metabarcoding of faeces revealed a large variety of pelagic and benthic fish species supplementing its diet. We conclude that DNA metabarcoding of seabird faeces can be an effective attribute to diet studies supporting our understanding of changes in numbers and distribution of Arctic seabirds and their marine environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arctic seabirds depend on marine fish, macrofaunal or planktonic resources and have been shown to be sensible indicators of ecosystem changes (e.g. Barrett and Krasnov 1996; Vihtakari et al. 2018; Wojczulanis-Jakubas et al. 2022). Around Svalbard, seabird diets, breeding performance, and contamination with toxic substances are well-studied and have shown significant changes in recent decades (e.g. Wold et al. 2011; Blévin et al. 2017; Vihtakari et al. 2018; Descamps and Strøm 2021). Climate change and its impacts on food webs, via for example changes in sea surface temperature and Arctic and Atlantic currents, as well as invasive species and environmental pollution constitute a complex interplay of possible stressors in the High Arctic (Ware et al. 2014; Griffith et al. 2019; Descamps and Strøm 2021; Stempniewicz et al. 2021). Food web alterations owing to increasingly northward species distributions (known as Atlantification or borealization) are of particular interest. Gradual replacement of Arctic fish and crustaceans, with high lipid and energy contents, with boreal species, that may be less nutritional for seabirds, may impact breeding success of seabirds (Vihtakari et al. 2018; Amélineau et al. 2019; Griffith et al. 2019). Therefore, information on diet composition of seabirds is key to understanding alteration in breeding ecology of seabirds and functioning of Arctic food webs.
There are many suitable methods to assess diet composition in seabirds, ranging from visual observations of food delivered at nesting sites, dissection of corpses, analyses of regurgitated material, to molecular techniques (e.g. stable isotopes, fatty acids, DNA metabarcoding) all with pro’s and con’s as to accuracy, bias or degree of disturbance to the birds (Barrett et al. 2007; Harding et al. 2008; Wold et al. 2011; Ceia et al. 2022). DNA metabarcoding in faeces is a non-invasive, relatively new method that causes little or no disturbance to birds and is more time effective in comparison to other methods to collect in the field. In recent years this method has been tested for various species and generally shows that a broad spectrum of prey items can be identified relative to conventional methods (e.g. Deagle et al. 2007; Pompanon et al. 2012; Bowser et al. 2013; Jarman et al. 2013; McInnes et al. 2016b; Oehm et al. 2011, 2017; Ceia et al. 2022; Penning et al. 2022). In addition, DNA analyses have become more and more cost-effective.
The aim of this study was to test the use of DNA metabarcoding in faeces of Arctic seabird species to identify prey species that may indicate shifts in seabird diets and the marine ecosystem of the High Arctic. Diet diversity was determined using COI and 18S DNA metabarcoding from faeces collected at breeding colonies of three focal species representing different parts of the food web: black-legged kittiwake (Rissa tridactyla), a surface-feeding piscivore, Brünnich’s guillemot (Uria lomvia), a diving piscivore, and little auk (Alle alle), a small diving planktivore and crustacean specialist (Mehlum and Gabrielsen 1993; Wold et al. 2011; Boehnke et al. 2017; Vihtakari et al. 2018; Descamps and Strøm 2021). These species are widespread on Svalbard and breed in large numbers. Seabird faeces were collected during short visits at large, or well-studied breeding colonies for a qualitative assessment of prey species allowing comparisons with earlier and ongoing diet studies.
Methods
Studied species
Black-legged kittiwake is a colonial breeding bird, building its nest against steep cliffs. It is a pelagic and coastal surface feeder on different species of fish, amphipods, cephalopods, copepods, euphausiids and decapods (Mehlum and Gabrielsen 1993). Brünnich’s guillemot breeds on steep cliff edges, often in mixed colonies with black-legged kittiwake. It is a pursuit diver mainly feeding on fish, amphipods and euphausiids (Mehlum and Gabrielsen 1993). Little auk typically breeds on mountain slopes with nests hidden under unvegetated screes. Its planktivorous diet mainly consists of copepods supplemented with young stages of amphipods, euphausiids and decapods (Mehlum and Gabrielsen 1993). Population sizes on Svalbard were estimated at 109 000 breeding pairs of black-legged kittiwake, 520 000 of Brünnich’s guillemot and over one million breeding pairs of little auk in 2014 (Anker-Nilssen et al. 2015), but populations are declining with a moderate to poor breeding season in 2022 (Decamps et al. 2013; Hanssen et al. 2023).
Field sampling
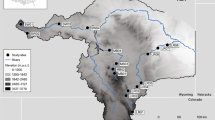
Seabird colonies were visited as part of ongoing research activities in Kongsfjorden (research groups of G.W. Gabrielsen and M.J.J.E. Loonen) and as part of SEES2022 expedition to southern Svalbard in July/August 2022 (Fig. 1, Table 1). For each of the three focal species representing different parts of marine food webs, we collected faeces at two (Brünnich’s guillemot) or three (black-legged kittiwake, little auk) breeding colonies. Methods to collect faeces varied depending on different breeding habitats of the species.
Black-legged kittiwake
Fresh faeces were mostly obtained at overhanging breeding cliffs (Blomstrandhalvøya, Kongsfjorden; Sofiekammen (Gnålberget), Hornsund) where a plastic sheet of ca 4 × 15 m was spread out some meters below occupied nests. Breeding birds showed no signs of disturbance and continued breeding and nest building while tens of faeces were collected in about 20–30 min. In addition, samples were collected from hand-held birds during nest checks as part of studies on breeding ecology and toxicology of black-legged kittiwakes at Krykkjefjellet, Kongsfjorden (Stampe 2022).
Brünnich’s guillemot
Collecting faeces was relatively difficult since guillemots use to breed at steep, non-overhanging cliffs. Successful attempts included collecting fresh faeces from hand-held birds by a mountaineer working on breeding biology of Brünnich’s guillemot at Ossian Sars, Kongsfjorden, as well as by a tourist guide on a ship approaching a breeding colony at Alkefjellet, Hinlopenstretet, collecting fresh samples from defecating birds flying to and from the colony from the shipdeck.
Little auk
Mostly fresh and some older faeces were obtained at rocks where several little auks used to gather in breeding colonies (Bjørndalen, Isfjorden; Ingeborgfjellet, Bellsundet). Disturbance in breeding colonies was limited to ca 15–30 min while carefully approaching some rocks where birds were observed defecating. In addition, some faeces samples were collected during visits of ongoing research programmes of Norsk Polarinstitutt at Feiringfjellet, Kongsfjorden, and at Bjørndalen, Isfjorden.
All seabird faeces were collected using simple equipment like disposable spoons (to avoid contamination) and small plastic jars to store the samples. Specific care was taken during sampling and sample processing to avoid any contamination between samples. Pooled samples of no more than ten faeces were preserved in a solution of DNA/RNA shield and stored at ambient temperature (maximum 1 week) or cooled (ca 7 °C).
Laboratory and data analysis
For reasons of cost effectiveness, the DNA metabarcoding analyses were performed on 1–5 pooled faeces samples of 1–10 faeces per sample per bird species per breeding colony. A total number of 22 pooled samples were analysed through a DNA metabarcoding approach at the laboratory (Wageningen University, Marine Animal Ecology group) using DNA markers COI and 18S. These markers were chosen based on the expected prey diversity and to have a wide taxonomic reach. COI is likely to give the best resolution for larger metazoans and 18S provides a wider image albeit with a lower resolution (van der Loos and Nijland 2021).
DNA extraction
DNA was extracted using the Invitrogen PureLink Microbiome Purification extraction kit (Invitrogen, Thermofisher Scientific, USA). When the PureLink extracted DNA did not yield an amplified band of the expected size in the subsequent PCRs, DNA extractions of these samples was repeated using the DNeasy Blood and Tissue extraction kit (QIAgen, Germany). For both DNA extraction methods, the faecal samples were first homogenized, and a subsample of 100–200 mg was taken. For the PureLink kit the subsample was directly added to the provided bead tube and 700 µL S1 lysis buffer was added. Bead-beating for 5 × 2 min at a frequency of 30 times per second followed. Subsequently the manufacture’s protocol was followed. DNA was eluted in 50 µL S6 elution buffer with an incubation time of 5 min. To increase the yield, the eluted volume was reapplied to the filter and incubated for another 2 min before final elution. For the DNeasy Blood and Tissue kit, the subsample was transferred to a 2 ml Eppendorf tube. DNA extraction was performed following the manufacture’s protocol for tissue samples. DNA was eluted in 50 µL AE buffer with an incubation time of 5 min. To increase the yield the eluted volume was reapplied to the filter and incubated for another 2 min before final elution.
PCR amplification
For PCR amplification 2 × Phire Tissue Direct PCR Master Mix (ThermoFisher Scientific, USA) was used. When necessary, template DNA was diluted to a concentration of < 80 ng/ µL. The mitochondrial COI gene and V4-V5 18S rRNA gene were targeted using metabarcoding primers mlCOIintF and jgHCO2198 (amplicon length of 313 bp) for COI (Leray et al. 2013) and F-566 and R-1200 (amplicon length of ~ 650) for 18S V4-V5 (Hadziavdic et al. 2014). Both primer pairs were extended with an ONT tag at the 5’ end of each primer, to allow further PCR-based sample barcoding in preparation for Oxford Nanopore sequencing. PCR reaction volume was 10 µL, containing 5 µL Phire Master Mix, 0.1 µL of each primer (10 µM), 3.8 µL nuclease-free water (NFW) and 1 µL (un)diluted DNA template. Each sample was amplified using two replicates. The COI and 18S amplicons were amplified using PCR thermoprofile and cycling conditions as follows: 98 °C for 3 min, followed by 35 cycles of 98 °C for 10 s, 55 °C for 10 s for COI / 20 s for 18S, 72 °C for 10 s, and a final extension at 72 °C for 3 min. Negative controls for DNA extraction and negative PCR controls were taken along. Gel electrophoresis using a 1.5% agarose gel was performed to assess success of all PCR reactions before pooling the successful PCR replicates.
Library preparation and Oxford Nanopore sequencing
Amplicons were barcoded using the 96 PCR barcoding kit (EXP-PCB096) (Oxford Nanopore Technologies, UK). Barcoding PCR reaction volume was 15 µL including 7.5 µL LongAmp Taq 2 × master mix (New England BioLabs (NEB) Inc), 0.3 µL barcode, 5.2 or 6.2 µL NFW and 1 or 2 µL PCR product of the first round of PCR, depending on the estimated concentration of the amplicon assessed by gel electrophoresis. Barcoding PCR was performed using the following thermoprofile and cycling conditions: 95 °C for 3 min, followed by 13 cycles of 95 °C for 15 s, 62 °C for 15 s, 65 °C for 30 s, with a final extension at 65 °C for 3 min. Another gel electrophoresis was performed assessing yield of the barcoding PCR. All barcoded amplicons were pooled equimolar based on this estimated concentration, considering the difference in amplicon length, so COI and 18S amplicons were pooled in a ratio of 1:2. Clean-up of the barcoded amplicon pool was performed using AMPure XP beads, following ONT’s clean-up protocol of the end-prep. Some adaptations to the protocol during first clean-up were made: amplicon pool and AMPure XP beads were mixed in a 1:2 ratio, pellet was resuspended in 51 µL NFW instead of 61 µL and thus 51 µL of eluate was retained in a clean 1.5 ml Eppendorf DNA LoBind tube. The purified barcoded DNA pool was quantified using a Qubit 3.0 fluorometer. End-prep was performed without DNA CS, using 1000 ng of cleaned-up barcoded pool in a volume of 50 µL. The rest of the library prep was executed according to manufacturer’s instructions (Ligation sequencing amplicons SQK-LSK114, version: ACDE_9163_v114_revH_29Jun2022-minion; ONT). After final clean-up, 200 ng of the prepared library was loaded on a primed R10.4.1 flowcell. Sequencing was performed using an Oxford Nanopore Mk1C MinION.
Bioinformatics, taxonomic assignment and further data processing
Sequence data basecalling, processing, filtering and analysis was performed using the Decona pipeline as described in Doorenspleet et al. (2023). Commands that were used for COI amplicon to run Decona were as follows: decona -T 18 -f -q 10 -l 300 -m 320 -g “GGWACWGGWTGAACWGTWTAYCCYCC;max_error_rate = 0.1;min_overlap = 20…TGRTTYTTYGGNCAYCCNGARGTNTA;max_error_rate = 0.1;min_overlap = 20” -c 0.85 -n 10 -r -o 0.99 -R 500 -k 6 -M -b path/nt_euk. For the 18S amplicon the Decona commands were: decona -T 18 -f -q 10 -l 375 -m 875 -g “CAGCAGCCGCGGTAATTCCA;max_error_rate = 0.1;min_overlap = 14…GCTTAATTTGACNCAACACGGG;max_error_rate = 0.1;min_overlap = 16” -c 0.85 -n 10 -r -o 0.99 -R 500 -k 6 -M -b path/nt_euk.
The reference database used for classification through BLAST (integrated in the Decona pipeline) was complete eukaryotic nucleotide database downloaded from NCBI (downloaded 07-06-2023). Percentage identity for species identification was set to > 99% in order to be taken for final data analysis. Taxonomically assigned output data from Decona were further analyzed using R studio (2022.12.0).
Since tag leakage is observed using the above-described protocol and the LSK114 R10.4.1 protocol, a tag leakage read correction was performed as described in Doorenspleet et al. (2023), using a 0.3% correction for both COI and 18S data. Classified reads not belonging to the potential diet of our host species were removed, such as nematodes and parasites, as well as species of terrestrial origin that probably were associated to the nesting locations of the seabirds. For COI, 10.7% of total reads was not classified at species level (identity less than 99%), of which the majority was from terrestrial mites (Sarcoptiformes) in little auk faeces. For 18S, 10.6% was not classified, representing different taxa, including intestinal parasites like Eimeria sp. Species identifications with accuracy larger than 99% but originating from non-food items (algae, protozoa, fungi, etc.) were also removed from the species list. The final species lists per sample (Online Resource 1) contain 98% of the classified COI read data and 23% of classified 18S read data. Only samples containing a single host species (either of the three focal seabird species) were accepted for diet analysis.
Results
DNA metabarcoding analysis
Out of 22 seabird faeces samples, 19 samples yielded DNA information on species level using COI as marker. Three samples were not included in further analysis. One sample did amplify in the PCR, but did not yield any data concerning the diet, and all reads were classified as contamination. One sample was identified as originating from snow bunting Plectrophenax nivalis, which is not a species of interest in this study. One sample was identified as a blend of black-legged kittiwake and Brünnich’s guillemot faeces. All other samples confirmed a single seabird host species (data in Online Resource 1, summarized data in Table 2). Using the 18S V4-V5 metabarcoding approach revealed fewer specific results. Ten pooled faeces samples yielded information on diets (Online Resource 1, Table 2). In most cases, fish and invertebrate DNA could not be identified to species level, except for calanoid copepods Calanus glacialis and C. finmarchicus in some little auk samples (Table 2).
COI revealed DNA of fish species in all diet samples. Polar cod Boreogadus saida, capelin Mallotus villosus and snakeblenny Lumpenus lampretaeformis were found in all three seabird species. Sandeel Ammodytes marinus was observed in black-legged kittiwake and Brünnich’s guillemot. American plaice Hippoglossoides platessoides and shorthorn sculpin Myoxoxephalus scorpius were found in Brünnich’s guillemot and little auk, and herring Clupea harengus in black-legged kittiwake and little auk. In little auk faeces, several other fish species were observed including both pelagic species like coalfish Pollachius virens and haddock Melanogrammus aeglefinus, and benthic species like sculpins (Myoxocephalus scorpius and Gymnocanthus tricupsis), stout blenny Anisarchus medius, and snailfish Liparis sp.
In addition, krill Thysanoessa inermis was observed in all three seabird species. Other crustaceans like calanoid copepods (Calanus finmarchicus, Calanus glacialis, Pseudocalanus sp.), shrimp Pandalus borealis, and hermit crab Pagurus pubescens were found in several little auk faeces, and a polychaete worm Glycera capitata was found in black-legged kittiwake faeces.
18S revealed some invertebrate taxa that were not identified by COI, namely amphipod Gammarus sp. in black-legged kittiwake, and amphipod Themisto sp., as well as Hippolytidae decapods in little auk diets (Table 2). We assumed that higher order taxa identified by 18S matched with species identified by COI in the same sample, e.g. Osmeriformes (18S) with Mallotus villosus (COI) and Euphausiidae (18S) with Thysanoessa inermis (COI).
Species compositions in the faeces for each seabird species showed consistent patterns across breeding colonies and pooled faeces samples. In black-legged kittiwake and Brünnich’s guillemot samples contained some fish species and a limited number of invertebrate taxa was identified. In little auk, in contrast, a high diversity in both fish species and other food taxa was observed.
Discussion
DNA metabarcoding of faeces revealed a large variety of fish and invertebrate taxa in the diet of all three seabird species in this study. It needs to be emphasized, however, that care should be taken as to what extent the observed taxa represent diet choice. The limited number of faeces collected, in most cases as single events of short visits to a breeding colony, must be considered as snapshots of prey choice, which may vary depending on for example the phase in the breeding cycle (Mehlum and Gabrielsen 1993; Deagle et al. 2007; McInnes et al. 2016a; Stampe 2022), individual foraging strategies (Karnovsky et al. 2003; Steen et al. 2007), or weather conditions (Jakubas et al. 2022). Pooling of faeces samples is cost-effective but may hide individual differences in prey choice. The diversity in prey choice we observed, however, suggested rather consistent patterns across colonies, not strongly biased by single individuals, although this aspect deserves further study.
The choice of DNA marker may affect the likelihood to identify certain species (Van der Loos and Nijland 2021). Degradation of DNA from soft tissue during digestion can influence the relative abundance of prey items discovered in the faeces when amplified by PCR metabarcoding. Furthermore, the method used to isolate the DNA might have further biased the relative abundance of amplifiable fragments of prey DNA. We chose to use two DNA markers for metabarcoding to allow for a wide spectrum of potential prey species. COI yielded satisfying results for fish species, but the resolution of both COI and 18S seemed limited to identify invertebrate species (see below). An additional DNA marker aimed at crustaceans would have likely yielded more insight in especially little auk diets where the presence of copepods was obvious from visual inspection but copepods were underrepresented in the metabarcoding data. An additional limitation of this metabarcoding study is that the DNA reference databases are still incomplete for potential prey species of Artic seabirds. A fraction of unidentified reads may therefore refer to prey species that comprised a seabird’s diet but could not be recognized as such.
For both primer sets used, large amounts of bird host DNA was amplified, which implied sequencing for a longer time to obtain sufficient sequencing depth. However, this is largely unavoidable when using more universal primer pairs, and additionally also serves as a verification of the host species. This is particularly useful when faeces are collected in large, mixed colonies of for example kittiwakes and guillemots with regularly visiting glacous gulls Larus hyperboreus.
A final consideration regarding qualitative species compositions using DNA metabarcoding in seabird diets concerns so-called secondary diet, i.e. identification of food items of the prey rather than of the seabird under study. This might have been the case for zooplankton species, e.g. Acartia hudsonica in a sample of piscivorous black-legged kittiwake. However, as discussed in the next section, for most samples we found no unexpected small taxa or other indications for this.
Diet composition
A further appraisal of our DNA metabarcoding observations is a comparison with earlier and recent diet studies of seabirds on Svalbard.
Black-legged kittiwake. Six fish species were identified in the faeces samples, mainly polar cod in the northern colonies in Kongsfjorden, and Atlantic species like capelin and sand eel, whereas Atlantic herring was only observed in the southernmost colony at Hornsund. In addition, snake blenny and lanternfish were found. In the 1980s and 1990s, polar cod was the most abundant fish species in the diet of black-legged kittiwakes supplemented with other gadoids (Pollachius virens, Melanogrammus aeglefinus), capelin, blennies (Lumpenidae, Zoarcidae) and lanternfish (Mehlum and Gabrielsen 1993), while after 2000 a higher share of Atlantic species such as capelin and herring were found (Vihtakari et al. 2018). Polar cod was also the main food item found in stomach and oesophagus samples collected in 1985 and 1986 and regurgitate samples collected between 2012 and 2020 in black-legged kittiwake colonies around Kongsfjorden (Lønne and Gabrielsen 1992; Stampe 2022; G.W. Gabrielsen, unpublished). Some snake blennies were also found, but sand eels had not been identified in those samples until 2020, although 50% of prey items could not be identified in the regurgitated material. Pelagic amphipods Themisto sp. and euphausiid Thysanoessa inermis were also identified as important food items of black-legged kittiwakes in earlier diet studies (Mehlum and Gabrielsen 1993; Stampe 2022; G.W. Gabrielsen, unpublished), but only occasionally observed in our study, possibly because of limited resolution of the DNA markers for invertebrates. Although increasing proportions of Atlantic species are found in the diets of black-legged kittiwakes, Stempniewicz et al. (2021) and Stampe (2022) observed large differences in diet composition between years.
Brünnich’s guillemot. In Brünnich’s guillemot faeces, we observed six fish species, both pelagic species like in the black-legged kittiwake samples such as polar cod, capelin, and sand eel and bottom-dwelling species like snake blenny, shorthorn sculpin, and American plaice. This coincides well with diet choice observed in colonies of Brünnich’s guillemots around the Barents Sea and around Svalbard where polar cod is the dominant prey in northern colonies and capelin, blennies, sand eel and sculpins are more often observed further south (Lønne and Gabrielsen 1992; Mehlum and Gabrielsen 1993; Barrett et al. 1997). In addition, invertebrates such as euphasiids like Thysanoessa inermis and amphipods like Themisto sp. and Gammarus sp. were often observed, but could have been missed in our study.
Little auk. In little auk, DNA metabarcoding revealed, surprisingly, a large number of fish species that could be identified reliably, although little auks are known to feed predominantly on crustaceans, especially calanoid copepods (e.g. Karnovsky et al. 2003; Boehnke et al. 2015; Jakubas et al. 2022). Indeed, microscopic visual inspection of little auk faeces suggested considerable amounts of calanoid copepods and krill in the diet, but only rather limited records of these crustaceans (C. finmarchicus, C. glacialis, Pseudocalanus sp., Acartia sp., T. inermis, P. pubescens) in COI and 18S V4-V5 metabarcoding. As mentioned above, the relatively large number of fish (larvae) DNA and limited amount of Calanus DNA we observed in little auk faeces in our study might be a bias resulting from the DNA metabarcoding method used here.
The large variety of fish species, however, is remarkable, since diets of little auks usually contain minor fractions of fish (< 1%) although frequency of occurrence (FOO) tends to be higher. Węsławski et al. (1999), for example, found unidentified fish larvae (15–20 mm) in little auk diets from Bjørnøya with FOO of 6–11% but only 0.1% of numerical diet composition. Similarly, Karnovsky et al. (2003), report that fish larvae were found in 32% of little auk diets (FOO), although the relative abundance was only 0.1% (gular pouches of adults feeding chicks) and comparably low fractions of less than 0.1% fish larvae were reported for chick-feeding little auks at Bjørndalen (Steen et al. 2007), as well as at Hornsund and Magdalenefjorden (Boehnke et al. 2015) and 0.3% fish larvae by Harding et al. (2008) based on isotope analysis. Mehlum and Gabrielsen (1993) report also fish larvae and 1-year polar cod in the diet of little auks, but it is unclear to what extent this refers to food for the adults themselves (analysis of stomach and esophagus of shot birds) or food collected for their chicks (analysis of contents of the gular pouch of the adults).
Although most diet studies on chick-feeding little auks report some fish larvae, species composition often remains unknown or unreported. The variety of both pelagic and benthic fish species in little auk faeces as revealed in our study is therefore highly interesting. It is most likely that most fish species were consumed as pelagic larvae or post-larval or small juvenile fish which frequent the water columns as is known for example for benthic species like daubed shanny Leptoclinus maculatus and snake blenny Lumpenus lampretaeformis (e.g. Meyer Ottesen et al. 2011, 2014). The nutritional value of the pelagic stages of Leptoclinus maculatus feeding on calanoid copepods can be considerable, since these post-larval stages of fish are known to store high-energy lipid components from zooplankton in a lipid sac and transfer them up the food chain to higher-order consumers (Falk-Petersen et al. 2007; Pekkoeva et al. 2017).
Although DNA metabarcoding does not allow a quantitative assessment of the importance of fish relative to invertebrate prey, the occurrence and diversity of fish species in all samples of little auks could also be related to specific ecological conditions in 2022. Little auk breeding numbers declined in recent years, while prey conditions have become worse owing to Calanus finmarchicus replacing the more nutritional C. glacialis (Descamps and Strøm 2021; Sauser et al. 2023), although Balazy et al. (2023) demonstrated that little auks are selective feeders on C. glacialis. Also, in 2022 the breeding season started late, while pack-ice conditions disappeared early in the season (https://arctic-rcc.org/climate-summary-jja-22) which could further have diminished feeding conditions for little auks in the breeding season (Amélineau et al. 2019; Hanssen et al. 2023; Sauser et al. 2023). A possible consequence could be that little auks shifted to a diet including more (post-)larval fish species as compared to earlier diet studies. Little auks are known to adjust foraging strategies to food conditions preferring, for example, areas with high visibility of favored prey items (Stempniewicz et al. 2013) and including long-distance excursions to areas with high-energy prey when food conditions nearer to the breeding colony appear poor (Steen et al. 2007). Also, Jakubas et al. (2022) showed that windy conditions may incur diets of little auk at the Hornsund breeding colony to shift to a larger fraction of the amphipod Apherusa glacialis or other food items instead of preferred calanoid copepods (mainly Calanus glacialis).
For all three seabird species, differences in species composition of the diet in this study, as compared to most earlier studies, could partly depend on differences in food choice of adults between food for the adult bird itself or food to provision to their chicks. Food items observed in DNA metabarcoding of faeces reflect the diet of adult seabirds, while most food items observed in diet studies in breeding seabirds (such as scooping gular pouches in little auks, visual observations of delivered food or regurgitated fish in other seabirds) reflect diets of chicks.
Concluding remarks
DNA metabarcoding based on COI and 18S of a limited number of seabird faeces samples in our study revealed a large variety of fish species and crustaceans in the diets of these species, comparable with or even exceeding diversity in diet composition found using more invasive techniques where birds are shot or caught and handled to obtain samples. Collecting faeces can often be done with little disturbance to seabirds and other wildlife, if direct contact with the birds can be avoided and visiting time at breeding colonies can be short. Faeces samples can also easily be stored in small plastic or glass jars at ambient temperatures and transported for later analyses in specialised laboratories (McInnes 2016; McInnes et al. 2016a, b). This makes DNA metabarcoding in most cases a convenient monitoring method, allowing data collection at remote sites and easily applied in combination with other Arctic monitoring programs. The use of plastic sheets at some distance to breeding birds can further reduce disturbance or it can be applied when breeding colonies are difficult to access, for example at steep cliffs, if birds defecate frequently in flight to and from their breeding sites. A further advantage of the use of plastic sheets might be that it reduces contamination with soil material that might impact DNA quality and amplification success (McInnes et al. 2016a).
DNA reference databases are still incomplete for potential prey species of Artic seabirds. Therefore, it is recommended to expand the amount of sequences in DNA databases. DNA metabarcoding methodology could be further improved by increasing the amount of primers used in diet studies, especially when a wide range of taxa is expected. More primers, however, increases costs and labour. Also, the number of pooled or individual samples collected and analyzed needs careful consideration with respect to the seasonal behaviour and feeding ecology of the study animal. Thus, a careful trade-off should be made for each diet study as to costs, number of samples, and taxa being able to be detected and identified to address specific research questions accordingly. With laboratory routines for molecular techniques still becoming more efficient, expansion of reference databases, and increasing knowledge on the effectiveness of different primers for identification of certain taxonomic groups, DNA metabarcoding methods become more and more cost-effective for assessment of presence or absence of taxonomic groups that may indicate changes in food conditions or the marine environment (https://geans.eu/faq; Deagle et al. 2007; Penning et al. 2022).
In conclusion, DNA metabarcoding of seabird faeces can be an effective attribute to diet studies supporting our understanding of changes in numbers and distribution of Arctic seabirds and their marine environment.
Data availability
The molecular data used in this study are available from the Integrated Marine Information System (https://doi.org/10.14284/671).
References
Amélineau F, Grémillet D, Harding AMA et al (2019) Arctic climate change and pollution impact little auk foraging and fitness across a decade. Sci Rep 9:1014. https://doi.org/10.1038/s41598-018-38042-z
Anker-Nilssen T, Barrett RT, Lorentsen S-H et al (2015) SEAPOP. De ti første årene. Nøkkeldokument 2005–2014. SEAPOP, Norsk institutt for naturforskning, Norsk Polarinstitutt & Tromsø Museum – Universitetsmuseet. Trondheim, Tromsø
Balazy K, Trudnowska E, Wojczulanis-Jakubas K et al (2023) Molecular tools prove little auks from Svalbard are extremely selective for Calanus glacialis even when exposed to Atlantification. Sci Rep 13:13647. https://doi.org/10.1038/s41598-023-40131-7
Barrett RT, Krasnov YV (1996) Recent responses to changes in stocks of prey species by seabirds breeding in the southern Barents Sea. ICES J Mar Sci 53:713–722
Barrett RT, Bakken V, Krasnov JV (1997) The diets of common and Brünnich’s guillemots Uria aalge and U. lomvia in the Barents Sea region. Polar Res 16:73–84. https://doi.org/10.3402/polar.v16i2.6626
Barrett RT, Camphuysen CJ, Anker-Nilssen T et al (2007) Diet studies of seabirds: a review and recommendations. ICES J Mar Sci 64:1675–1691. https://doi.org/10.1093/icesjms/fsm152
Blévin P, Tartu S, Ellis HI et al (2017) Contaminants and energy expenditure in an Arctic seabird: organochlorine pesticides and perfluoroalkyl substances are associated with metabolic rate in a contrasted manner. Environ Res 157:118–126. https://doi.org/10.1016/j.envres.2017.05.022
Boehnke R, Gluchowska M, Wojczulanis-Jakubas K et al (2015) Supplementary diet components of little auk chicks in two contrasting regions on the West Spitsbergen coast. Polar Biol 38:261–267. https://doi.org/10.1007/s00300-014-1568-9
Boehnke R, Balazy K, Jakubas D et al (2017) Meso-scale variations in diet composition of little auk chicks in north-west Spitsbergen. Polar Res. https://doi.org/10.1080/17518369.2017.1409585
Bowser AK, Diamond AW, Addison JA (2013) From Puffins to Plankton: a DNA-based analysis of a seabird food chain in the northern Gulf of Maine. PLoS ONE 8:e83152. https://doi.org/10.1371/journal.pone.0083152
Ceia FR, Xavier JC, Carreiro AR et al (2022) Conventional and modern approaches to study seabird trophic ecology and diet. In: Ramos JA, Pereira L (eds) Seabird biodiversity and human activities. CRC Press, Boca Raton, pp 19–35. https://doi.org/10.1201/9781003047520
Deagle BE, Gales NJ, Evans K, Jarman SN et al (2007) Studying seabird diet through genetic analysis of faeces: a case study on macaroni penguins (Eudyptes chrysolophus). PLoS ONE 2(9):e831. https://doi.org/10.1371/journal.pone.0000831
Descamps S, Strøm H (2021) As the Arctic becomes boreal: ongoing shifts in a high-Arctic seabird community. Ecol 102(11):e03485. https://doi.org/10.1002/ecy.3485
Descamps S, Strøm H, Steen H (2013) Decline of an arctic top predator: synchrony in colony size fluctuations, risk of extinction and the subpolar gyre. Oecologia 173:1271–1282. https://doi.org/10.1007/s00442-013-2701-0
Doorenspleet K, Jansen L, Oosterbroek S et al (2023) The long and the short of it: Nanopore based eDNA metabarcoding of marine vertebrates works; sensitivity and specificity depend on amplicon lengths. BioRxiv. https://doi.org/10.1101/2021.11.26.470087
Falk-Petersen S, Timofeev S, Pavlov V et al (2007) Climate variability and the effect on Arctic food chains. The role of Calanus. In: Ørbæk JB, Kallenborn R, Tombre I, Hegseth EN, Falk-Petersen S, Hoel AH (eds) Arctic-alpine ecosystems and people in a changing environment. Springer, Berlin, pp 147–166. https://doi.org/10.1007/978-3-540-48514-8_9
Griffith GP, Hop H, Vihtakari M et al (2019) Ecological resilience of Arctic marine food-webs to climate change. Nat Clim Chang 9:868–872. https://doi.org/10.1038/s41558-019-0601-y
Hadziavdic K, Lekang K, Lanzen A et al (2014) Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 9(2):e87624. https://doi.org/10.1371/journal.pone.0087624
Hanssen SA, Descamps S, Anker-Nilssen T et al (2023) Sjøfugl i Norge. Resultater fra SEAPOP-programmet. Årsbrosjyre SEAPOP
Harding AMA, Hobson KA, Walkusz W et al (2008) Can stable isotope (δ13C and δ15N) measurements of little auk (Alle alle) adults and chicks be used to track changes in high-Arctic marine foodwebs? Polar Biol 31:725–733. https://doi.org/10.1007/s00300-008-0413-4
Jakubas D, Wojczulanis-Jakubas K, Szeligowska M et al (2022) (2022) Gone with the wind – Wind speed affects prey accessibility for a High Arctic zooplanktivorous seabird, the little auk Alle alle. Sci Tot Env 852:158533. https://doi.org/10.1016/j.scitotenv.2022.158533
Jarman SN, McInnes JC, Faux C et al (2013) Adélie penguin population diet monitoring by analysis of food DNA in scats. PLoS ONE 8:e82227. https://doi.org/10.1371/journal.pone.0082227
Karnovsky NJ, Kwaśniewski S, Węsławski JM et al (2003) Foraging behavior of little auks in a heterogeneous environment. Mar Ecol Prog Ser 253:289–303. https://doi.org/10.3354/meps253289
Keslinka LK, Wojczulanis-Jakubas K, Jakubas D et al (2019) Determinants of the little auk (Alle alle) breeding colony location and size in W and NW coast of Spitsbergen. PLoS ONE 14:e0212668. https://doi.org/10.1371/journal.pone.0212668
Leray M, Yang JY, Meyer CP et al (2013) A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10:34. https://doi.org/10.1186/1742-9994-10-34
Lønne OJ, Gabrielsen GW (1992) Summer diet of seabirds feeding in sea-ice covered waters near Svalbard. Polar Biol 12:685–692. https://doi.org/10.1007/BF00238868
McInnes J (2016) Field collection protocols for DNA dietary analysis of seabird scats. Agreement on the Conservation of Albatrosses and Petrels. http://www.acap.aq/en/resources/acap-conservation-guidelines 2019/01/26
McInnes JC, Alderman R, Deagle BE et al (2016a) Optimised scat collection protocols for DNA metabarcoding in vertebrates. Meth Ecol Evol 8:192–202. https://doi.org/10.1111/2041-210X.12677
McInnes JC, Emmerson L, Southwell C et al (2016b) Simultaneous DNA-based diet analysis of breeding, non-breeding and chick Adélie penguins. Royal Society Open Science 3:150443. https://doi.org/10.1098/rsos.150443
Mehlum F, Gabrielsen GW (1993) The diet of high-arctic seabirds in coastal and ice-covered, pelagic areas near the Svalbard archipelago. Polar Res 12:1–20. https://doi.org/10.3402/polar.v12i1.6698
Meyer Ottesen CA, Hop H, Christiansen JS et al (2011) Early life history of the daubed shanny (Teleostei: Leptoclinus maculatus) in Svalbard waters. Mar Biodivers 41:383–394. https://doi.org/10.1007/s12526-010-0079-3
Meyer Ottesen CA, Hop H, Falk-Petersen S et al (2014) Growth of daubed shanny (Teleostei: Leptoclinus maculatus) in Svalbard waters. Polar Biol 37:809–815. https://doi.org/10.1007/s00300-014-1481-2
Oehm J, Juen A, Nagiller K et al (2011) Molecular scatology: how to improve prey DNA detection success in avian faeces? Mol Ecol Res 11:620–628. https://doi.org/10.1111/j.1755-0998.2011.03001.x
Oehm J, Thalinger B, Eisenkölbl S et al (2017) Diet analysis in piscivorous birds: what can the addition of molecular tools offer? Ecol Evol 7:1984–1995. https://doi.org/10.1002/ece3.2790
Pekkoeva SN, Murzina SA, Nefedova ZA et al (2017) Ecological role of lipids and fatty acids in the early postembryonic development of the daubed shanny, Leptoclinus maculatus (Fries, 1838) from Kongsforden, West Spitsbergen in winter. Russ J Ecol 48(3):240–244. https://doi.org/10.1134/S1067413617030134
Penning E, Verkuil YI, Klunder L et al (2022) Sanderlings feed on a diverse spectrum of prey worldwide but primarily rely on brown shrimp in the Wadden Sea. Ardea 110:187–199. https://doi.org/10.5253/arde.2022.a11
Pompanon F, Deagle BE, Symondson WO et al (2012) Who is eating what: diet assessment using next generation sequencing. Mol Ecol 21:1931–1950. https://doi.org/10.1111/j.1365-294X.2011.05403.x
Sauser C, Angelier F, Blévin P et al (2023) Demographic responses of Arctic seabirds to spring sea-ice variations. Front Ecol Evol. https://doi.org/10.3389/fevo.2023.1107992
Stampe N (2022) Diet variability in black-legged kittiwakes (Rissa tridactyla) in Kongsfjorden in relation to ongoing environmental changes. Master Thesis BIO399, University of Bergen
Steen H, Vogedes D, Broms F (2007) Little auks (Alle alle) breeding in a High Arctic fjord system: bimodal foraging strategies as a response to poor food quality? Polar Res 26:118–125. https://doi.org/10.1111/j.1751-8369.2007.00022.x
Stempniewicz L, Darecki M, Trudnowska EK et al (2013) Visual prey availability and distribution of foraging little auks (Alle alle) in the shelf waters of West Spitsbergen. Polar Biol 36:949–955. https://doi.org/10.1007/s00300-013-1318-4
Stempniewicz L, Weydmann-Zwolicka A, Strzelewicz A et al (2021) Advection of Atlantic water masses influences seabird community foraging in a high-Arctic fjord. Prog Oceanogr 193:102549. https://doi.org/10.1016/j.pocean.2021.102549
Van der Loos LM, Nijland R (2021) Biases in bulk: DNA metabarcoding of marine communities and the methodology involved. Mol Ecol 30:3270–3288. https://doi.org/10.1111/mec.15592
Vihtakari M, Welcker J, Moe B et al (2018) Black-legged kittiwakes as messengers of Atlantification in the Arctic. Sci Rep 8:1178. https://doi.org/10.1038/s41598-017-19118-8
Ware C, Berge J, Sundet JH et al (2014) Climate change, non-indigenous species and shipping: assessing the risk of species introduction to a high-Arctic archipelago. Div Distr 20:10–19. https://doi.org/10.1111/ddi.12117
Węsławski JM, Stempniewicz L, Mehlum F et al (1999) Summer feeding strategy of the little auk (Alle alle) from Bjørnøya, Barents Sea. Polar Biol 21:129–134. https://doi.org/10.1007/PL00013383
Wojczulanis-Jakubas K, Jakubas D, Stempniewicz L (2022) The little auk Alle alle: An ecological indicator of a changing Arctic and a model organism. Polar Biol 45:163–176. https://doi.org/10.1007/s00300-021-02981-7
Wold A, Jæger I, Hop H et al (2011) Arctic seabird food chains explored by fatty acid composition and stable isotopes in Kongsfjorden, Svalbard. Polar Biol 34:1147–1155. https://doi.org/10.1007/s00300-011-0975-4
Acknowledgements
This study was supported by a grant from the Svalbard Environmental Protection Fund (RiS project 1135) and by the SEES2022 expedition of the Netherlands Polar Programme (run by Dutch Research Council NWO). Gijs Breedveld, Michelle van Dijk, Delphin Ruche, Nora Stampe, Joanne Maria Sulich, and Saga Svavarsdottir contributed to field sampling. Maarten Loonen, Frits Steenhuisen, Martine van den Heuvel-Greve, Hans Verdaat, and personnel of AWIPEV and OceanWide Expeditions generously supported sampling logistics. We thank two anonymous reviewers for their constructive comments.
Author information
Authors and Affiliations
Contributions
JL and RN conceived and designed the research. JL acquired funding and collected field samples. XB and RN analyzed samples. All authors analyzed the data. The first draft of the manuscript was written by JL and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Ethical approval
No approval of research ethics committees was required to accomplish the goals of this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
300_2024_3276_MOESM1_ESM.xlsx
Supplementary file1 (XLSX 54 KB) Online Resource 1. DNA metabarcoding results for CO1 and 18S V4-V5 for all faeces samples with sufficient amplification of DNA for taxonomic identification of food items.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Leeuw, J.J., van den Brink, X., Gabrielsen, G.W. et al. DNA metabarcoding reveals high diversity of fish and macrofaunal species in diets of little auks and other Arctic seabird species in Svalbard. Polar Biol (2024). https://doi.org/10.1007/s00300-024-03276-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00300-024-03276-3