Abstract
Biodiversity surveys of Arctic soil ecosystems are limited. Here, we provide a sequence-based inventory of soil fauna from an Arctic tundra ecosystem near the Canadian High Arctic Research Station in Cambridge Bay, Nunavut. Invertebrate communities were extracted at a vegetated and non-vegetated site in three replicates and inventoried using 18S metabarcode sequencing. A total of 361 amplicon sequence variants (ASV) were identified and assigned to the closest matching taxonomic orders, most of which belonged to the Nematoda and Arthropoda. Vegetated soils showed no significantly higher ASV richness relative to non-vegetated soils although they contained a significantly higher diversity of arthropod taxa including insects, mites, and springtails. Most taxa were found only at a single location and even samples from the same site displayed distinct communities, suggesting that belowground species richness in Arctic tundra habitats is highly heterogeneous. Preserving soil biodiversity in a changing Arctic is essential for Inuit communities who rely on intact tundra ecosystems for their health and wellbeing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The importance of belowground biodiversity for terrestrial ecosystem functioning is well-established (Bardgett and van der Putten 2014; Lefcheck et al. 2015), and much recent focus has been put to investigate drivers of biodiversity loss (Duncan et al. 2015). Our understanding of soil community composition and the individual species’ components has increased rapidly following the application of genomic techniques (Geisen et al. 2019), stable isotope work (Ferlian et al. 2015), and network analyses (Derocles et al. 2018; Ramirez et al. 2018). Yet, specific taxonomic knowledge is still very limited in many belowground systems (Tedersoo et al. 2014) and the biotic diversity of soil ecosystems is often overlooked in conservation policy (Guerra et al. 2021).
The association between the loss of soil biodiversity and a resulting loss of soil function is well demonstrated (Wagg et al. 2014). Key functions provided by healthy soil ecosystems include nutrient cycling and carbon sequestration (Koltz et al. 2018) in both natural and agricultural systems (Bender et al. 2016). Unfortunately, such functions are diminished as soil ecosystems become degraded, polluted, or over-exploited—leading to a loss of soil biota (Amundson et al. 2015). Human-caused changes in land use and climate directly influence soil food webs and can impact their resistance and resilience to climate change (de Vries et al. 2012; Griffiths and Philippot 2013). Biodiversity loss further directly threatens the ability of these systems to respond to environmental changes, particularly climate extremes (Isbell et al. 2015).
Arctic ecosystems are particularly vulnerable to global and local human activity as they are experiencing some of the most rapid climate warming (Post et al. 2009). Climate change has resulted in risks to Indigenous country food systems through more unpredictable environmental conditions (Statham et al. 2015), a trend that is predicted to continue (IPCC 2022). Many Arctic regions are also seeing a rapid increase in the exploitation of their terrestrial environments for natural resources (Kumpula et al. 2011) and agriculture (Stephen 2018). The potential effects of these further environmental changes on the biota of previously uncultivated soils could be dramatic. The introduction of non-native species as a result of global warming is predicted to further endanger endemic species’ diversity and lead to shifts in species’ ranges (Nielsen and Wall 2013).
Concurrently, Arctic soil ecosystems have a legacy of habitation by humans, including Inuit (Keith et al. 2007), and provide a critical link in supporting traditional country foods such as caribou and muskox. Socio-economic development of high Arctic regions is predicted to intensify the use of natural resources and lead to a degradation of these environments (Ehrich et al. 2019). Links between changes in micro-invertebrate species’ composition and risks to Indigenous food security have already been demonstrated (Kafle et al. 2020). Potential development in the north thus needs to be guided by an understanding of the diversity of soil invertebrates (free-living and parasitic), especially in high Arctic regions (Bach et al. 2020). Inventories of invertebrate diversity also compliment traditional Indigenous knowledge in Arctic communities, which focus on stewardship of the natural environment (Gadgil et al. 1993).
There is an urgent need to assess the species’ diversity of soil invertebrates in Arctic regions and to identify potential or ongoing risks related to human activities (Pentinsaari et al. 2020). Recent large-scale analysis has shown that nematode densities in sub-Arctic regions are the highest worldwide (van den Hoogen et al. 2019). However, much of this diversity has not been classified taxonomically and knowledge of the ecological roles of many of these groups is very limited. Currently, invertebrate diversity inventories in the Canadian Arctic are only available at some long-term research sites (Rich et al. 2013) and for some highly localized sites (Culjak Mathieu 2020). Unfortunately, this may lead to a biased understanding of patterns of biotic diversity across the whole Arctic (Metcalfe et al. 2018), and may not allow local Indigenous communities to identify and respond to threats to their soil ecosystems. Over the last years, a framework for generating and curating these data is emerging (Ramirez et al. 2015). This has led to a more comprehensive understanding of climate and vegetation effects (Bastida et al. 2020) as well as distribution patterns of soil invertebrates in the Arctic (Phillips et al. 2019; Gillespie et al. 2020).
Sequencing environmental DNA directly from soils can provide an assessment of belowground diversity for Arctic soil invertebrates (Bik et al. 2012) and has been suggested as a tool specific for the monitoring of polar ecosystems and the detection of invasive species (Czechowski et al. 2017). Further, such genomic inventories can provide critical insights into the complexity of these different ecosystems, such as the presence of predatory or parasitic species’ guilds (Creer et al. 2016). The composition of invertebrates in Arctic soils is known to vary depending on soil type (Hansen et al. 2016) and varying habitats respond differently to environmental change (Coulson et al. 1993). Baseline inventories and subsequent monitoring of soil invertebrates is crucial to understanding the broader consequences of environmental change for soil ecosystems, especially in the Arctic (Høye and Culler 2018).
Here, we extracted and sequenced communities of eukaryotes from soil that was collected from vegetated and non-vegetated sites near the Canadian High Arctic Research Station in Iqaluktuuttiaq (Cambridge Bay), Nunavut. This region has been populated by hunters of fish, seal, muskox, and caribou for an estimated 4000 years (Kitikmeot Heritage Society 2012). Soil inventories therefore provide a taxonomic overview of community structure and diversity for an Arctic tundra system that has been managed under Indigenous stewardship for thousands of years. It contributes to building a comprehensive framework of the biodiversity in this community, relevant to larger vertebrate species and aboveground diversity that provides subsistence for the Inuit people of this region.
Methods
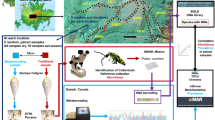
Two sites with differing vegetation cover were identified near the Canadian High Arctic Research Station (CHARS) in Cambridge Bay, Nunavut (vegetated site: 69° 07′ 52.4′′ N 105° 03′ 36.8′′ W, non-vegetated site: 69° 07′ 52.3′′ N 105° 03′ 24.3′′ W, see Fig. 1). Three soil samples were collected from each site in July 2019. Samples at each site were collected within a 50 m radius, selected based on above-ground plant coverage, with Samples V1–V3 obtained from soils with vascular plant coverage and Samples NV4-NV6 obtained from soils from frost-eroded areas with little plant coverage (Fig. 1c, d). The three vegetated samples represented the main plant associations observed on the local tundra system, namely: true sedges (Carex spp.) for Sample V1; a mixture of other sedge grasses and peat moss at Sample V2; and mixed shrub/sedge grass cover for Sample V3. For each sample approximately 250 g of the topsoil was taken from directly underneath any present vegetation (at the soil surface), and collected to a depth of 10 cm. Each sample was placed into a sterile plastic bag, homogenized and transported in insulated containers to CHARS prior to freezing and subsequent transportation frozen (< − 10 °C) to Brigham Young University (BYU) for further analyses.
Once at BYU, a modified sugar-flotation protocol was used to extract micro-invertebrates larger than 40 µm from 100 g of soil from each sample (Freckman and Virginia 1993). DNA was extracted from each sample using a Qiagen DNEasy Blood and Tissue kit (QIAGEN, Germantown, MD), modified by crushing any organisms collected by centrifugation with a sterile pestle before proceeding as per the manufacturer’s instructions. Sequence libraries were constructed using 10 µL of the resulting 100 µL product, which were amplified using standard Illumina sequencing primers and amplicon primer sequences for the 18S small ribosomal subunit marker (18S) as described in Caporaso et al. (2012). The 18S region amplified is the one recommended by the Earth Microbiome Project (Thompson et al. 2017) for identification of micro-eukaryotes commonly found in soils (forward primer 1391f: GTACACACCGCCCGTC; reverse primer EukBr: TGATCCTTCTGCAGGTTCACCTAC), and reference databases for this ribosomal sequence are increasingly found in the literature (Obiol et al. 2020). Sequencing primers and Illumina barcodes were removed before subsequent analysis using cutadapt (v3.2) (Martin 2011). Resulting metabarcoding reads were filtered, trimmed, and clustered to amplicon sequence variants (ASVs) using the DADA2 pipeline (v1.18) (Callahan et al. 2016) in R (v. 4.0.4). Reads were quality filtered and truncated at 140 base pairs for the forward and reverse compliment strands to each pair. DADA2 was used for subsequent alpha and beta diversity analyses and taxonomic assignment to both the SILVA (v 1.32, Quast et al. 2013) and PR2 (v 4, Guillou et al. 2013) databases. Specifically, both the absolute richness (Observed Features) as well as two abundance-controlled metrics (Shannon and Simpson diversity) are used to provide a ranking of the diversity of the individual samples (Lande et al. 2000). Community dissimilarities were investigated using phylogenetically naïve (Bray–Curtis distance) and phylogenetically informed distance metrics, using both abundance weighted and unweighted options (weighted and unweighted UniFrac distance, Lozupone and Knight 2005). Distance metrics were displayed in non-metric multidimensional scaling plots to observe the similarities in community composition between samples (Clarke 1993). Maximum likelihood phylogenetic trees were constructed with the phyloseq package (v. 1.34) (McMurdie and Holmes 2013) in R.
ASVs with poorly resolved taxonomic resolution in the Silva and PR reference databases were further blasted to the NCBI Nucleotide database (Schoch et al. 2020) (May 20th 2021) to be assigned to the taxonomic level for which consensus existed in this database (no other orders showed up as BLAST hits with a score higher than 200). No finer resolution than order-level taxonomy was derived from NCBI BLAST classification; hits to environmental sequences were excluded.
Following preliminary analyses, we found that Nematoda and Arthropoda were the two most commonly sequenced taxa in each sample. Accordingly, the relative frequency of sequences and the diversity of Nematoda and Arthropoda were separately considered across the samples to investigate the similarities in invertebrate community composition between soil types.
Results
Quality control and data retention
Total reads per sample were 39,286, 41,915, 43,318, 66,907, 59,489, and 41,217 for Samples V1–V3 and NV4–NV6, respectively, before trimming, for a total of 292,132 reads. Supplementary Fig. 1 shows data loss during filtering and trimming using the DADA2 pipeline (Callahan et al. 2016). Out of 420 resulting amplicon sequence variants (ASVs) sequences 59 were identified as chimeras and removed, making up 3.83% of total reads. After chimera removal the total of reads across all samples was 176,650, for a data loss of just under 40% (see supplementary Fig. 1 for data loss in each step).
Taxonomic diversity
Based on an initial taxonomic assignment 113 out of 361 total ASVs were identified to the taxonomic Order level in the Silva database, increasing to 147 out of 361 ASVs by combining the Silva and PR databases. Including the NCBI Blast search resolved another 106 ASVs to the Order level, totaling 253 out of 361 ASVs resolved, including all but three of the 50 most common ASVs across all samples. Taxonomic insecurities for the remaining 108 ASVs and the inability to assigned detected taxa to a higher resolution than order level was made difficult by the lack of genomic references for Arctic soil biota. The 18S primers developed for the Earth Microbiome Project are designed to capture a very broad range of eukaryotic diversity but as a result lack the power to resolve species’ taxonomic diversity at a very fine scale (Pawlowski et al. 2012).
The most abundant ASV at the non-vegetated site belonged to the phylum Nematoda, while at the vegetated site Arthropoda made up most of the reads (See supplementary Fig. 2). Platyhelminthes were only found at the vegetated site, and annelid DNA was recovered from Sample V1 only. Across all samples, the other most common phyla after Nematoda and Arthropoda were Rotifera, Platyhelminthes, Ascomycota, Annelida, and Tardigrada. Figure 2 presents the results of taxonomic identification across all samples for these seven most abundant phyla, which make up between 90 and 95% of the total reads for each sample (See Supplementary Fig. 2 for abundance relative to total reads).
Relative proportion of the most common phyla. Relative abundance is calculated as the proportion of reads of each of the seven included Phyla to the total number of reads of only those seven Phyla: Annelida, Arthropoda, Ascomycotes, Nematoda, Platyhelmintha, Rotifera, and Tardigrada. The non-vegetated sites (NV4, NV5, and NV6) showed a higher relative abundance of nematodes than arthropods as compared to the vegetated sites, while Platyhelminthes and annelids were only found in vegetated soils (at all three sites and in the Carex vegetation cover site only, respectively). Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil
Nematodes
Figure 3 shows the distribution of nematode orders across the two sites. Nematode diversity was not significantly higher in vegetated than non-vegetated soils (one-tailed t-test, p < 0.05). The most common ASV in the entire dataset belonged to the Enoplida and was highly abundant in all three non-vegetated soils while very rare in the soils with vegetation cover. While most enoplids are marine in origin, some members of this genus occur as free-living bacterivorous nematodes in soils (Smythe 2015). The third most common ASV belonged to the Araeolaimida and was highly abundant across all six samples. Araeolaimids are also commonly seen as marine free-living nematodes, with some in the order observed living in soils as free-living bacterivores (Yeates 1988). While these two free-living nematode examples occurred in most soil samples, other nematode ASVs were highly restricted to either site or to a single sample, such as the most common nematode ASV (belonging to Enoplida), the sixth (belonging to Mononchida), and eighth (belonging to Tylenchida) most common ASVs all being highly abundant in the non-vegetated site but nearly absent from any of the vegetated soils (see supplementary Table 1).
Diversity of nematode orders and their relative abundance. The much larger proportion of nematode reads in the non-vegetated soil samples is composed mostly of a few very abundant groups in the Araeolaimida, Desmodorida, and Enoplida. Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil
Arthropods
Figure 4 shows the distribution of arthropod orders across the two sites. A higher diversity of arthropod ASVs was found in the vegetated soils than in non-vegetated soils (one-tailed t-test, p < 0.05). Samples NV4–NV6 showed a high similarity in arthropod diversity, with a similar composition of collembolan and sarcoptiform arthropods. However, most collembolan ASVs were unique to one or two samples, except for the most common collembolan ASV which was present in Samples V1, NV4, NV5, and NV6. Trombidiform mites and any insects such as Diptera were only present in the vegetated soils, which were also the only samples to share the presence of the most common platyhelminth, as well as some other platyhelminths occurring only in a single sample. Generally, the vegetated site showed a much higher abundance of arthropod DNA, except for a single collembolan ASV in Sample NV5 that made up around 20% of total sample abundance.
Diversity of arthropod orders and their relative abundance. Unvegetated soil samples’ arthropod communities were limited to very few groups of collembolans and sarcoptiformes, while vegetated soils showed a diversity of insect groups (Diptera, Hemiptera, and Hymenoptera) and a high abundance of sarcoptiform mites. Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil
Inter-site comparison
Seven ASVs were present in all samples, six of which were among the 30 most common ASVs. Of the 361 total ASVs, 247 were unique to a single sample and a further 31 and 18 ASVs were specific to either the vegetated or non-vegetated sampling sites, respectively. The remaining 65 ASVs were not restricted to a single sample or site. However, many were only present in very low abundances in the vegetated soils (as was the case for some of the most common nematode taxa in the non-vegetated soils). The total number of reads that were limited to a single sample or site was 32.17%, indicating that most taxa were relatively rare across samples. Strictly sample-specific taxa made up approximately 16% of total read copies, but more than two-thirds of the number of ASVs identified in this study.
Figure 5 shows the alpha diversity distribution across the six samples, including the observed features measure (A) and Shannon and Simpson diversity metrics which account for the proportional abundance of species (Shannon 1948). The sample with a Carex dominated ground cover (Sample V1) consistently showed high diversity compared to the other communities, while Sample NV6 (non-vegetated) had the fewest observed features and a low biodiversity index.
Alpha diversity at six different sites as measured by the Observed Features metric, Shannon diversity index and Simpson diversity index. Site color is based on soil type (red = vegetated soils, blue = non-vegetated soils). Site V2 and NV66 consistently show lower diversity due to the high abundance of relatively few phyla, with Site NV5 showing the highest overall number of features but a more even distribution of diversity in Site 1. Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil
Patterns of community clustering are shown in Fig. 6 for several measures of beta diversity. Two of the vegetated soils (Samples V2 and V3) clustered together in all NMDS plots, and a general distinction between the non-vegetated (Samples NV4–NV6) and the vegetated (Samples V1–V3) site was observed across all plots. The three vegetated soil samples also show higher within-group differences. The Carex-covered soil community (Sample V1) specifically was less similar to the other two samples from the same site, potentially caused by the effects of vegetation cover on community composition. Both inter-site and intra-site comparisons are broadly consistent between the different distance metrics used and these patterns were also consistent when considering nematode and arthropod diversity separately (Supplementary Figs. 3 and 4, respectively).
Beta diversity community similarities as shown by NMDS. Beta diversity indices are: Bray–Curtis distance (A), Unweighted UniFrac distance (B), Weighted UniFrac distance (C), and Jensen-Shannon distance (D). Samples colored-coded by soil type (red = vegetated soils, blue = non-vegetated soils). Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil
Figure 7 shows the phylogenetic relationship between the identified nematode and arthropod groups and their presence across the two sites. While monophyletic groups of arthropods can be identified as present only in vegetated soils (all Diptera, Hemiptera, and a subset of Collembola) most nematode clades are shown to be present in both soil types.
Phylogenies of all collected nematode and arthropod ASVs and their presence in the different soil types. 139 nematode ASVs and 51 arthropod ASVs are displayed in an unrooted phylogeny to indicate the relationships between the diversity found in the two soil types. Images are of a collembolan and nematode representative of the Kitikmeot region of Nunavut, licensed by the University of Waikato, New Zealand and the Centre for Biodiversity Genomics, Guelph, CA—respectively. Green dots indicate the ASV was found in a vegetated soil sample while orange dots indicate its presence in one of the non-vegetated soils. Size of the dots indicate the number of reads found in that sample. Colored boxes indicate taxonomic order. Gray lines are unassigned, blue lines deviate from the group otherwise indicated by color
Discussion
The composition of the samples we analyzed showed a high degree of heterogeneity. 247 out of 361 total ASVs identified are unique to a single sample. The ecological niches of endemic taxonomic orders can be used as indicators for food web structure (Ferris et al. 2001), although it is important to caution against over-interpreting relative abundance data obtained from metabarcoding (see Schenk et al. 2019). Nematodes such as Rhabditida and Tylenchida are shown here to be prevalent in non-vegetated soil samples and are likely to be free-living bacterivores (Qing and Bert 2019), while Dorylaimida are seen more frequently in the three samples from the vegetated site where they comprise free-living or plant and animal parasitic groups (Lee 2002). Most Mononchida are predators and observed in non-vegetated soils (Ahmad and Jairajpuri 2010), occupying a higher level in the food chain in these ecosystems. Few Trombidiformes were found at the non-vegetated site while making up roughly one third and one fourth of ASV reads in the vegetated Samples V2 and V3, respectively, suggesting that only vegetated soils host enough prey items for these predatory mites (Seniczak et al. 2020). Sarcoptiform mites were instead found at both the vegetated and non-vegetated site and are mainly expected to have a mycophagous and saprophagous ecology in the Arctic (Young et al. 2012).
The composition of arthropods in these soils show distinct communities exist between the vegetated and non-vegetated sites, with only three out of the 25 total types of Collembola shared between the two soil types, and three out of 12 total mite taxa. Many of the arthropod ASVs were only found in a single sample, indicating that some ASVs were potentially associated with specific plant communities, similar to patterns observed for bacterial and fungal diversity elsewhere in the Arctic (Bölter 2003). Both phylogenetic trees indicate a high degree of heterogeneity of nematode and arthropod taxa in these soils, particularly among the rarer observed ASVs in this study. Arthropod species with such patchy distributions are suggested as key indicator species of changing Arctic ecosystems as they are likely to be highly vulnerable to climate changes and invasive species (Hodkinson et al. 2013; Gillespie et al. 2020).
These ecosystems are warming rapidly and resulting declines in biodiversity and biomass have been demonstrated aboveground (CAFF 2013)—with further declines predicted (Niittynen et al. 2018). However, the belowground components of these ecosystems are generally less well known. The diversity of belowground systems is a driving force behind the above-ground diversity of plants (Frouz 2018) and is therefore of great importance in supporting country food such as muskox and caribou. Arctic microinvertebrates have also been shown to directly affect this food supply through the transmission of diseases such as brucellosis in wild muskox (Tomaselli et al. 2019), and arthropods such as biting flies can cause behavioral changes in large mammals (Witter et al. 2012). Soil microbiota thus play key roles in Arctic foodwebs supporting the health and wellbeing of Indigenous communities (Pedersen et al. 2020). Understanding the dispersal mechanisms (Coulson et al. 2003) and responses of below-ground fauna to climate and vegetation changes (Joly et al. 2009) can provide Indigenous communities with crucial knowledge for responding to changing environmental conditions.
Our study provides an inventory of belowground invertebrate diversity from the Canadian central Arctic. Many of the ASVs we identified in this study were found only in a single sample and point to a high degree of heterogeneity on the small spatial scale at which we investigated these belowground invertebrate communities. While some ecological information can be inferred from metabarcoding (Makiola et al. 2020), the incomplete nature of publicly available genetic repositories and biases in geographic and taxonomic representation can reduce confidence in individual species identification and their function in local habitats. Similar and ongoing studies for above-ground invertebrate communities (Pentinsaari et al. 2020) have also highlighted similar issues with existing morphological and molecular taxonomic databases. Our study and ongoing genomic and taxonomic characterization of the eukaryotic soil diversity in the Arctic are first steps in redressing this knowledge gap.
Data availability
Data used for this study are available at https://www.ncbi.nlm.nih.gov/bioproject/817555.
Code availability
Code used for data analysis is available as a supplementary file (Sup File 1).
References
Ahmad W, Jairajpuri MS (2010) Mononchida—the predatory soil nematodes. In: Hunt DJ, Perry RN (eds) Nematology monographs and perspectives. Brill Academic Publishing, Leiden
Amundson R, Berhe AA, Hopmans JW et al (2015) Soil and human security in the 21st century. Science 348:1261071. https://doi.org/10.1126/science.1261071
Bach EM, Ramirez KS, Fraser TD, Wall DH (2020) Soil Biodiversity integrates solutions for a sustainable future. Sustainability 12:2662. https://doi.org/10.3390/su12072662
Bardgett RD, van der Putten WH (2014) Belowground biodiversity and ecosystem functioning. Nature 515:505–511. https://doi.org/10.1038/nature13855
Bastida F, Eldridge DJ, Abades S et al (2020) Climatic vulnerabilities and ecological preferences of soil invertebrates across biomes. Mol Ecol 29:752–761. https://doi.org/10.1111/mec.15299
Bender SF, Wagg C, van der Heijden MGA (2016) An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31:440–452. https://doi.org/10.1016/j.tree.2016.02.016
Bik HM, Porazinska DL, Creer S et al (2012) Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol Evol 27:233–243. https://doi.org/10.1016/j.tree.2011.11.010
Bölter M (2003) Microbiological communities and properties of arctic soils: results of the tundra northwest expedition 1999 (Nunavut and Northwest Territories, Canada). Polarforschung 73:103–110
CAFF (2013) Arctic biodiversity assessment. The Conservation of Arctic Flora and Fauna, Akureyri
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Caporaso JG, Lauber CL, Walters WA et al (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. https://doi.org/10.1038/ismej.2012.8
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. https://doi.org/10.1111/j.1442-9993.1993.tb00438.x
Coulson S, Hodkinson ID, Strathdee A et al (1993) Simulated climate change: the interaction between vegetation type and microhabitat temperatures at Ny Ålesund, Svalbard. Polar Biol 13:67–70. https://doi.org/10.1007/BF00236585
Coulson SJ, Hodkinson ID, Webb NR (2003) Aerial dispersal of invertebrates over a high-Arctic glacier foreland: Midtre Lovénbreen, Svalbard. Polar Biol 26:530–537. https://doi.org/10.1007/s00300-003-0516-x
Creer S, Deiner K, Frey S et al (2016) The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol Evol 7:1008–1018. https://doi.org/10.1111/2041-210X.12574
Culjak Mathieu V (2020) Habitat heterogeneity drives plant-pollinator network diversity on the tundra of Victoria Island, Nunavut. McGill University, Canada
Czechowski P, Clarke LJ, Cooper A, Stevens MI (2017) A primer to metabarcoding surveys of Antarctic terrestrial biodiversity. Antarct Sci 29:3–15. https://doi.org/10.1017/S0954102016000389
de Vries FT, Liiri ME, Bjørnlund L et al (2012) Land use alters the resistance and resilience of soil food webs to drought. Nat Clim Change 2:276–280. https://doi.org/10.1038/nclimate1368
Derocles SAP, Bohan DA, Dumbrell AJ et al (2018) Chapter One—Biomonitoring for the 21st century: integrating next-generation sequencing into ecological network analysis. In: Bohan DA, Dumbrell AJ, Woodward G, Jackson M (eds) Advances in ecological research. Academic Press, pp 1–62
Duncan C, Thompson JR, Pettorelli N (2015) The quest for a mechanistic understanding of biodiversity–ecosystem services relationships. Proc Royal Soc Biol Sci 282:20151348. https://doi.org/10.1098/rspb.2015.1348
Ehrich D, Thuestad AE, Tømmervik H et al (2019) Local land use associated with socio-economic development in six arctic regions. Ambio 48:649–660. https://doi.org/10.1007/s13280-018-1095-y
Ferlian O, Klarner B, Langeneckert AE, Scheu S (2015) Trophic niche differentiation and utilisation of food resources in collembolans based on complementary analyses of fatty acids and stable isotopes. Soil Biol Biochem 82:28–35. https://doi.org/10.1016/j.soilbio.2014.12.012
Ferris H, Bongers T, de Goede RGM (2001) A framework for soil food web diagnostics: extension of the nematode faunal analysis concept. Appl Soil Ecol 18:13–29. https://doi.org/10.1016/S0929-1393(01)00152-4
Freckman DW, Virginia RA (1993) Extraction of nematodes from dry valley antarctic soils. Polar Biol 13:483–487
Frouz J (2018) Effects of soil macro- and mesofauna on litter decomposition and soil organic matter stabilization. Geoderma 332:161–172. https://doi.org/10.1016/j.geoderma.2017.08.039
Gadgil M, Berkes F, Folke C (1993) Indigenous knowledge for biodiversity conservation. Ambio 22:151–156
Geisen S, Briones MJI, Gan H et al (2019) A methodological framework to embrace soil biodiversity. Soil Biol Biochem 136:107536. https://doi.org/10.1016/j.soilbio.2019.107536
Gillespie MAK, Alfredsson M, Barrio IC et al (2020) Status and trends of terrestrial arthropod abundance and diversity in the North Atlantic region of the Arctic. Ambio 49:718–731. https://doi.org/10.1007/s13280-019-01162-5
Griffiths BS, Philippot L (2013) Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev 37:112–129. https://doi.org/10.1111/j.1574-6976.2012.00343.x
Guerra CA, Bardgett RD, Caon L et al (2021) Tracking, targeting, and conserving soil biodiversity. Science 371:239–241. https://doi.org/10.1126/science.abd7926
Guillou L, Bachar D, Audic S et al (2013) The Protist Ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res 41:597–604. https://doi.org/10.1093/nar/gks1160
Hansen RR, Hansen OLP, Bowden JJ et al (2016) Meter scale variation in shrub dominance and soil moisture structure Arctic arthropod communities. PeerJ. https://doi.org/10.7717/peerj.2224
Hodkinson ID, Babenko A, Behan-Pelletier VM et al (2013) Terrestrial and freshwater invertebrates. In: Meltofte H, Josefson AB, Payer D (eds) Arctic biodiversity assessment. Status and trends in Arctic biodiversity. Conservation of Arctic Flora and Fauna, Akureyri, pp 194–223
Høye TT, Culler LE (2018) Tundra arthropods provide key insights into ecological responses to environmental change. Polar Biol 41:1523–1529. https://doi.org/10.1007/s00300-018-2370-x
IPCC (2022) Climate change 2022: impacts, adaptation, and vulnerability. Contribution of working group ii to the sixth assessment report of the intergovernmental panel on climate change. Cambridge University Press
Isbell F, Craven D, Connolly J et al (2015) Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 526:574–577. https://doi.org/10.1038/nature15374
Joly K, Jandt RR, Klein DR (2009) Decrease of lichens in Arctic ecosystems: the role of wildfire, caribou, reindeer, competition and climate in north-western Alaska. Polar Res 28:433–442. https://doi.org/10.1111/j.1751-8369.2009.00113.x
Kafle P, Peller P, Massolo A et al (2020) Range expansion of muskox lungworms track rapid arctic warming: implications for geographic colonization under climate forcing. Sci Rep 10:17323. https://doi.org/10.1038/s41598-020-74358-5
Keith D, Friesen M, Kitikmeot Heritage Society (2007) Iqaluktuurmiutat: life at Iqaluktuuq. Artisan Press Ltd., Yellowknife, NT
Kitikmeot Heritage Society (2012). https://web.archive.org/web/20120110092547/http://www.kitikmeotheritage.ca/research/iqaluktk/predorset.htm. Accessed 7 Jun 2022
Koltz AM, Asmus A, Gough L et al (2018) The detritus-based microbial-invertebrate food web contributes disproportionately to carbon and nitrogen cycling in the Arctic. Polar Biol 41:1531–1545. https://doi.org/10.1007/s00300-017-2201-5
Kumpula T, Pajunen A, Kaarlejärvi E et al (2011) Land use and land cover change in Arctic Russia: ecological and social implications of industrial development. Glob Environ Chang 21:550–562. https://doi.org/10.1016/j.gloenvcha.2010.12.010
Lande R, DeVries PJ, Walla TR (2000) When species accumulation curves intersect: implications for ranking diversity using small samples. Oikos 89:601–605. https://doi.org/10.1034/j.1600-0706.2000.890320.x
Lee DL (2002) The biology of nematodes. Taylor and Francis, London
Lefcheck JS, Byrnes JEK, Isbell F et al (2015) Biodiversity enhances ecosystem multifunctionality across trophic levels and habitats. Nat Commun 6:1–7. https://doi.org/10.1038/ncomms7936
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. https://doi.org/10.1128/AEM.71.12.8228-8235.2005
Makiola A, Compson ZG, Baird DJ et al (2020) Key questions for next-generation biomonitoring. Front Environ Sci. https://doi.org/10.3389/fenvs.2019.00197
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Metcalfe DB, Hermans TDG, Ahlstrand J et al (2018) Patchy field sampling biases understanding of climate change impacts across the Arctic. Nat Ecol Evol 2:1443–1448. https://doi.org/10.1038/s41559-018-0612-5
Nielsen UN, Wall DH (2013) The future of soil invertebrate communities in polar regions: different climate change responses in the Arctic and Antarctic? Ecol Lett 16:409–419. https://doi.org/10.1111/ele.12058
Niittynen P, Heikkinen RK, Luoto M (2018) Snow cover is a neglected driver of Arctic biodiversity loss. Nat Clim Change 8:997–1001. https://doi.org/10.1038/s41558-018-0311-x
Obiol A, Giner CR, Sánchez P et al (2020) A metagenomic assessment of microbial eukaryotic diversity in the global ocean. Mol Ecol Resour 20:718–731. https://doi.org/10.1111/1755-0998.13147
Pawlowski J, Audic S, Adl S et al (2012) CBOL protist working group: Barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLOS Biol 10:e1001419. https://doi.org/10.1371/journal.pbio.1001419
Pedersen C, Otokiak M, Koonoo I et al (2020) ScIQ: an invitation and recommendations to combine science and Inuit Qaujimajatuqangit for meaningful engagement of Inuit communities in research. Arctic Sci 6:326–339. https://doi.org/10.1139/as-2020-0015
Pentinsaari M, Blagoev GA, Hogg ID et al (2020) A DNA barcoding survey of an arctic arthropod community: implications for future monitoring. InSects 11:46. https://doi.org/10.3390/insects11010046
Phillips HRP, Guerra CA, Bartz MLC et al (2019) Global distribution of earthworm diversity. Science 366:480–485. https://doi.org/10.1126/science.aax4851
Post E, Forchhammer MC, Bret-Harte MS et al (2009) Ecological dynamics across the arctic associated with recent climate change. Science 325:1355–1358. https://doi.org/10.1126/science.1173113
Qing X, Bert W (2019) Family Tylenchidae (Nematoda): an overview and perspectives. Org Divers Evol 19:391–408. https://doi.org/10.1007/s13127-019-00404-4
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Ramirez K, Döring M, Eisenhauer N et al (2015) Toward a global platform for linking soil biodiversity data. Front Ecol Evol 3:91
Ramirez KS, Geisen S, Morriën E et al (2018) Network analyses can advance above-belowground ecology. Trends Plant Sci 23:759–768. https://doi.org/10.1016/j.tplants.2018.06.009
Rich ME, Gough L, Boelman NT (2013) Arctic arthropod assemblages in habitats of differing shrub dominance. Ecography 36:994–1003. https://doi.org/10.1111/j.1600-0587.2012.00078.x
Schenk J, Geisen S, Kleinboelting N, Traunspurger W (2019) Metabarcoding data allow for reliable biomass estimates in the most abundant animals on earth. Metabarcoding Metagenomics. https://doi.org/10.3897/mbmg.3.46704
Schoch CL, Ciufo S, Domrachev M et al (2020) NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database 2020:2020. https://doi.org/10.1093/database/baaa062
Seniczak A, Seniczak S, Schwarzfeld MD et al (2020) Diversity and distribution of mites (Acari: Ixodida, Mesostigmata, Trombidiformes, Sarcoptiformes) in the Svalbard Archipelago. Diversity 12:323. https://doi.org/10.3390/d12090323
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x
Smythe AB (2015) Evolution of feeding structures in the marine nematode order enoplida. Integr Comp Biol 55:228–240. https://doi.org/10.1093/icb/icv043
Statham S, Ford J, Berrang-Ford L et al (2015) Anomalous climatic conditions during winter 2010–2011 and vulnerability of the traditional Inuit food system in Iqaluit, Nunavut. Polar Rec 51:301–317. https://doi.org/10.1017/S0032247414000151
Stephen K (2018) Societal impacts of a rapidly changing Arctic. Curr Clim Change Rep 4:223–237. https://doi.org/10.1007/s40641-018-0106-1
Tedersoo L, Bahram M, Põlme S et al (2014) Global diversity and geography of soil fungi. Science 346:1256688. https://doi.org/10.1126/science.1256688
Thompson LR, Sanders JG, McDonald D et al (2017) A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. https://doi.org/10.1038/nature24621
Tomaselli M, Elkin B, Kutz S et al (2019) A transdisciplinary approach to Brucella in muskoxen of the Western Canadian Arctic 1989–2016. EcoHealth 16:488–501. https://doi.org/10.1007/s10393-019-01433-3
van den Hoogen J, Geisen S, Routh D et al (2019) Soil nematode abundance and functional group composition at a global scale. Nature 572:194–198. https://doi.org/10.1038/s41586-019-1418-6
Wagg C, Bender SF, Widmer F, van der Heijden MGA (2014) Soil biodiversity and soil community composition determine ecosystem multifunctionality. PNAS 111:5266–5270. https://doi.org/10.1073/pnas.1320054111
Witter LA, Johnson CJ, Croft B et al (2012) Behavioural trade-offs in response to external stimuli: time allocation of an Arctic ungulate during varying intensities of harassment by parasitic flies. J Anim Ecol 81:284–295. https://doi.org/10.1111/j.1365-2656.2011.01905.x
Yeates GW (1988) Distribution of Plectidae (Nematoda: Araeolaimida) in pasture soils. N Z J Zool 15:191–202. https://doi.org/10.1080/03014223.1988.10422614
Young MR, Behan-Pelletier VM, Hebert PDN (2012) Revealing the hyperdiverse mite fauna of subarctic Canada through DNA barcoding. PLoS ONE 7:e48755. https://doi.org/10.1371/journal.pone.0048755
Acknowledgements
Financial and logistic support for this research was provided by Polar Knowledge Canada. Collembolan pictures were provided by Barry O’Brien.
Author information
Authors and Affiliations
Contributions
IH, DW and BA conceived research objectives. JJ and BA designed research approach. BV and IH collected samples. JJ processed samples and analyzed data. JJ wrote the first draft of the manuscript. All authors contributed to the interpretation of data and subsequent drafts.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
300_2023_3131_MOESM1_ESM.png
Supplementary file1 (PNG 75 kb)—Data loss visualization in the DADA2 pipeline. Samples showed roughly equal proportions of data lost during filtering, denoising and chimera removal steps. Truncating and filtering low-quality reads was responsible for a loss of data of up to 40% while subsequent steps resulted in little data loss.
300_2023_3131_MOESM3_ESM.xlsx
Supplementary file3 (XLSX 59 kb)—Amplicon sequence variant (ASV) table including counts. ASV names and sequences are reported along with the combined taxonomic assignment from the Silva and PR databases and Genbank blasts. Counts of each ASV across the samples are also reported, as are the BLAST scores used to fill in taxonomic gaps.
300_2023_3131_MOESM4_ESM.png
Supplementary file4 (PNG 18 kb)—Number of reads of all classes within the most abundant phyla across samples relative to the total number of reads in each sample. The top four and bottom four facets have separate legends. Total diversity of phyla was greater than the eight phyla reported on in this study, but most of these phyla had a relative abundance of less than 2%. The number of sequences not assigned to a taxonomic order was very small, except at Site 1 where it reached almost 5%. Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil.
300_2023_3131_MOESM5_ESM.png
Supplementary file5 (PNG 25 kb)—Beta diversity community similarities as shown by non-metric multidimensional scaling (NMDS) of the nematode diversity. Four different beta diversity indexes are used: Bray-Curtis distance (A), Unweighted UniFrac distance (B), Weighted UniFrac distance (C), and Jensen-Shannon distance (D). Samples are colored based on soil type (red = vegetated soils, blue = non-vegetated soils). All non-vegetated soil samples show more similarities to each other than they do to the vegetated soils. The three vegetated soil samples also show higher similarity, with the Carex covered soils less similar to the other two sites. Results are thus highly similar to the overall Beta diversity results shown in Figure 3. Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil.
300_2023_3131_MOESM6_ESM.png
Supplementary file6 (PNG 28 kb)—Beta diversity community similarities as shown by non-metric multidimensional scaling (NMDS) of the arthropod diversity. Four different beta diversity indexes are used: Bray-Curtis distance (A), Unweighted UniFrac distance (B), Weighted UniFrac distance (C), and Jensen-Shannon distance (D). Samples are colored based on soil type (red = vegetated soils, blue = non-vegetated soils). All non-vegetated soil samples show more similarities to each other than they do to the vegetated soils. The three vegetated soil samples also show higher similarity, with the Carex covered soils less similar to the other two sites. Results are thus highly similar to the overall Beta diversity results shown in Figure 3. Sample vegetation: V1) True sedge grasses (Carex spp.), V2) Sedge grass and peat moss, V3) Mixed sedge/shrub cover, V4:V6) Non-vegetated soil.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jorna, J., Vandenbrink, B., Hogg, I.D. et al. Metabarcoding inventory of an arctic tundra soil ecosystem reveals highly heterogeneous communities at a small scale. Polar Biol 46, 461–471 (2023). https://doi.org/10.1007/s00300-023-03131-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-023-03131-x