Abstract
Understanding the diversity and functioning of Arctic sea ice ecosystems is vital to evaluate and predict the impact of current and future climate change. In the microscopic communities inhabiting the brine channels inside sea ice, nematodes often dominate numerically and act as bacterivores and herbivores. Despite nematodes great abundances and known ecological roles, molecular tools have not been applied to investigate their species diversity in sea ice. In an attempt to begin establishing a molecular baseline for species diversity of sea ice nematodes, we Sanger sequenced 74 specimens from four locations around Svalbard (European Arctic), using the 18S rRNA barcode. Currently available nucleotide reference databases are both underpopulated with representative marine nematode taxa and contain a substantial number of misidentified organisms. Together, these limitations inhibited the ability to identify marine specimens collected in this study with certainty. Nevertheless, our molecular data indicate the presence of two genera in sea ice on Svalbard—Theristus and Halomonhystera. While it is possible that the latter represents a novel ice nematode species, future studies, including morphometric analysis, are needed to verify our molecular findings. We leverage the assignment of molecular information to robustly identify nematodes and provide the first insight into the diversity of sea ice nematodes in the European Arctic. We advocate for an intensified cooperation between molecular and morphological taxonomists to expedite the establishment of baseline surveys that are required to predict biological consequences of the diminishing sea ice habitat in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nematodes are among the most abundant and species-rich metazoans on earth, comprising taxonomic and functionally diverse communities in terrestrial, freshwater and marine habitats (Heip et al. 1985; Yeates 2007; Moens et al. 2013). Free-living marine nematodes occupy various ecological niches by acting as predators, by influencing bacteria activity, stimulating decompositional processes and by serving as prey themselves (Heip et al. 1985; Moens et al. 2013). Nematodes are commonly the most abundant meiofauna group in aquatic systems (Platt and Warwick 1980; Semprucci 2013), including the unique Arctic coastal sea ice habitat (Bluhm et al. 2018).
Despite their broad distribution and functionality, only 30,000 species have been described globally (Hugot et al. 2001), which is assumed to present only 3% of the estimated 1 million species that are thought to exist (Creer et al. 2010). Much of this undiscovered diversity is thought to be hidden in the marine environment from which only around 6000 species have been described thus far (Bezerra et al. 2020). For sea ice, it is even less with only five described species so far: Theristus melnikovi from the Central Arctic Ocean, Laptev Sea, and Fram Strait (Tchesunov 1986; Riemann and Ngando 1997), Cryonema crassum (Tchesunov and Riemann 1995; Tchesunov and Portnova 2005) and Cryonema tenue (Tchesunov and Riemann 1995) from the White Sea, off the coast of Siberia, and the Fram Strait, and Hieminema obliquorum (Tchesunov and Portnova 2005) from the White Sea. In Antarctic sea ice, only one individual registration of Halomonhystera glaciei exists (Blome and Riemann 1999). Nematodes inhabit the brine channels that form inside of sea ice when salt is rejected from seawater. Here, they can feed on the readily available bacteria and ice algae while being safe from predators. They may eventually reach adulthood and reproduce inside the sea ice (e.g. Bluhm et al. 2018).
Cryptic speciation, small size and limited prominent external morphological features challenge the identification of nematodes to species level (Lee et al. 2017). In an effort to circumvent the challenges associated with morphological identification and begin resolving uncharacterized evolutionary relationships among closely related clades within Nematoda, molecular phylogenies and thresholds of pairwise analysis of DNA sequences are used as a diagnostic tool (e.g. Bik et al. 2013). Specifically, the use of mitochondrial DNA and nuclear loci that encode the small ribosomal subunits (e.g. 18S rRNA) has revealed higher genetic diversity than expected, even over small spatial scales (Derycke et al. 2010; Tchesunov et al. 2015; Fonseca et al. 2017). Still, the absence of molecular data corresponding to morphological descriptions of Arctic metazoans hinders high-throughput approaches to inventory diversity.
In this study, our main objective was therefore to do the first screening of the biodiversity and distribution of sea ice nematodes in Svalbard by analysing the 18S small ribosomal subunit loci of isolated specimens. Svalbard is an archipelago located between 74° and 81°N. The largest island is called Spitsbergen, which is influenced to the west by a branch of the warm, saline Gulf Stream (namely the West Spitsbergen Current) and cold currents to the east. Out of all Arctic regions, Svalbard has experienced some of the most severe changes with warmer winter temperatures, loss and failure in sea ice formation (Søreide et al. 2021). This will have yet unknown implications for the Arctic coastal (sea ice) ecosystem. However, these changes are difficult to assess accurately, as sea ice studies in Svalbard are generally limited to physical aspects and ice algae, neglecting the metazoan component. The few existing in-ice fauna studies indicate that nematodes are the most numerous land-fast ice metazoans in Svalbard (e.g. Andreasen et al. 2019; Pitusi et al. 2019), but so far no species identification of nematodes has been conducted from the sea ice habitat in Svalbard (Bluhm et al. 2018). Thus, information on ice nematode species diversity and distribution is lacking from Svalbard. Knowledge about species diversity is needed to accurately evaluate ecological changes that might be occurring due to changing sea ice conditions. In this study, ice cores from land-fast sea ice were collected and analysed during 2017–2019 to provide the first insight into the molecular diversity of sympagic (= ‘ice-associated’) nematodes from the west and east coast of Svalbard, by using nematode-specific 18S rRNA primers.
Materials and methods
Study area
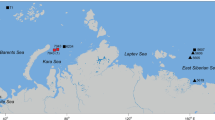
Samples were collected from four sea ice covered fjords and bays located in western and eastern Svalbard (Fig. 1): Van Mijenfjorden, Inglefieldbukta, Wahlenbergfjorden and Palanderbukta. Sampling occurred throughout spring 2017 and 2018 (Table 1).
Van Mijenfjorden (77.7°N, 15.5°E) is a relatively shallow fjord (up to 120 m-deep) that is 70-km-long and 10-km-wide and located along the southwestern coast of Spitsbergen. It is a partially enclosed fjord, which efficiently regulates the exchange of warm and saline Atlantic water that follows along the shelf via the West Spitsbergen Current (WSC) (Høyland et al. 2009). This partial separation from the WSC, coupled with the strong freshwater input, plays a role in the relatively reliable perennial formation of sea ice. Comparatively, Inglefieldbukta (77.9°N, 18.3°E) is a 2.5-km-wide and long bay situated on the southeastern coast of Spitsbergen, in connection with Storfjorden. The hydrography of the bay is influenced by glacial runoff to the west and by cold Arctic water to the east. Wahlenbergfjorden (79.7°N, 20.5°E) is a 50-km-long fjord situated on Nordaustlandet, in north-eastern Svalbard. To the south, the 35-km-long Palanderbukta (79.5°N, 20.4°E) branches in a northwest–southeast direction. Both locations are surrounded by glaciers and connected to the 400-m-deep Hinlopen Strait, through which both Atlantic and Arctic water flows.
In Van Mijenfjorden and Inglefieldbukta, land-fast ice was sampled by coring ridge-free areas in triplicates (∅ = 9 cm, Kovacs Ice Coring Equipment, USA). Triplicates were sampled within 0.5 to 1 m of each other. After retrieval, cores were sectioned into 0–1, 1–2, 2–3 and 3–10-cm from the bottom up and then continually in 10-cm sections to the top of the core. To avoid osmotic shock of the nematodes during melting, 100 mL of 0.7-um filtered seawater (GF/F, Whatman) was added per 1-cm ice (Garrison and Buck 1986; Spindler and Dieckmann 1986). Ice cores were kept separate, in different bags, and thawed in darkness at 4 °C at the University Centre in Svalbard (UNIS, Longyearbyen). To investigate the similarity of nematode species composition in the benthic and sympagic realm, Van Veen grab (0.1-m2) samples were collected in Van Mijenfjorden. From each of the grab’s four doors, duplicates of the upper 3 cm of the sediment were collected using a plastic syringe (∅ = 2.6-cm). Firstly, the sample was passed through a 500-µm sieve followed by a 63-µm sieve. The duplicates were stored in 96% ethanol and 4% formalin to enable both molecular and community analysis. Ethanol samples were washed in GF/F filtered seawater back at UNIS and 10 nematodes per sample site were picked randomly for sequencing; formalin samples were not analysed in this study.
In Wahlenbergfjorden, qualitative sea ice pieces (volume not quantified) were collected to which GF/F filtered seawater was added before melting, in the dark, on board the R/V Helmer Hansen. In Palanderbukta, three sea ice pieces (~ 0.1 m3) were cut from along the melting sea ice edge (approximately 20-cm thick and highly porous). GF/F filtered seawater was added to the samples and then melted, in the dark, and processed on board the M/S Spitsbergen.
After melting, ice core samples were concentrated over a 20-μm sieve and transferred to a petri dish. A Leica MZ16 stereomicroscope (Wetzler, Germany), with magnification 0.71–11.5 x, was used to pick and measure live nematodes and suspected nematode eggs. Picked specimens and potential nematode eggs were stored in individual Eppendorf tubes in 96% ethanol until DNA extraction. A Sony microscope camera was used to photograph individual nematodes and eggs. Photos were taken for reference and visual documentation of basic morphological features (such as egg shape and colour). Length measurements, with the microscope eye piece, were made to gather information about size distribution between stations (in the same fjord) and locations (west versus east Svalbard). Albeit, size is not the most distinguishable body characteristics needed to determine genera or species morphologically, it was the only information alongside reproduction mode that we could easily gather without expertise knowledge. By reproduction mode, we refer to any visible signs of sexual maturation of ice nematodes (e.g. development of reproductive organs, appearance of eggs and juveniles) (Schierenberg and Sommer 2013).
DNA barcoding
Ethanol-preserved nematodes and eggs (n = 164) were rinsed in MilliQ water prior to DNA extraction; 30 nematodes were lost during this washing step due to individuals sticking to the inside of pipettes or disintegrating. DNA was extracted from 134 whole specimens using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), per manufacturer’s instructions. HotSHOT extraction (Truett el al. 2000) was applied to 30 of the 134 samples, in order to increase DNA extraction success after experiencing failure with the DNeasy kit. After extraction, DNA was PCR amplified using nematode-specific 18S rRNA primers: MN18F (forward) (5′-CGCGAATRGCTCATTACAACAGC-3′) (IDT, USA) and Nem_18S_R (reverse) (3′-GGGCGGTATCTGATCGCC-5′) (Bhadury et al. 2006). These primers target a 925 base pair locus within the 18S rRNA region. PCR was conducted with 2.5 μL 10 × DreamTaq Buffer (Thermo Scientific, USA), 2 μL dNTPS (2.5 mM), 0.25 μL forward and reverse primers (10 μM), 0.2 μL DreamTaq Polymerase (Thermo Scientific, USA), 17.8 μL MilliQ water and 2 μL DNA to have a total volume of 25 μL. Thermocycling was conducted with an initial denaturation at 95°C for 5 min, 37 cycles of 955′-CGCGAATRGCTCATTACAACAGC-3′) C for 1 min, 545′-CGCGAATRGCTCATTACAACAGC-3′) C for 1 min and 725′-CGCGAATRGCTCATTACAACAGC—3′) C for 2 min, with a final extension period at 725′-CGCGAATRGCTCATTACAACAGC—3′) C for 5 min, with a Eppendorf Mastercycler EP Gradient S Thermal Cycler (Eppendorf, Germany). Amplicons were purified using HigherPurity solid-phase reversible immobilization (SPRI) cleaning (Canvax, Córdoba, Spain). During the cleaning process, the amplified DNA of 40 nematodes was lost. Subsequently, amplicons of 94 specimens were sequenced bi-directionally to increase the confidence of the base call along the length of the amplicon. Sanger sequencing was conducted at Eurofins Genomics (GATC Biotech LightRun, Germany). Sequences were analysed in FinchTV V 1.4 (http://www.geospiza.com) and explored for the presence of ambiguous base calls and double peaks. Bi-directionally sequenced DNA were aligned and end-trimmed in Geneious V 11.0.4 (Kearse et al. 2012), to form a high-quality consensus sequence; 74 sequences were of good read quality. These sequences were exported and compared to deposited sequences in NCBI/GenBank using BLAST.
The same procedure was applied to the benthic samples (n = 32), from which 18 good quality sequences were obtained. One of these was included in the phylogenetic tree to determine the placement of the benthic to the sympagic samples.
Phylogenetic tree
High-quality consensus DNA sequences (n = 75) were imported into MEGA7 (Kumar et al. 2016) and uniformly oriented. This multiple sequence file was expanded with 18S rRNA sequences that represent all taxonomically characterized nematode clades. This sequence file was further expanded with closely allied (> 90% identity) environmental sequence data, retrieved from NCBI by querying GenBank with the already derived nematode sequences, using the BLASTn algorithm. In total, our 18S rRNA phylogenetic tree analysed the relationship of our environmental sequences relative to two different taxonomic classes (Enoplea and Chromadorea) and five different orders (Monhysterida, Araeolaimida, Rhabditida, Desmodorida, Chromadorida) that circumscribed 69 genera. Diplolaimella dievengatensis was chosen as an outgroup, following Tchesunov et al. (2015). In total, phylogenies were assessed with a total of 544 sequences. All sequences were then aligned using MAFFT 7 (Katoh et al. 2017) with default parameters. This multiple sequence alignment (MSA) was then vetted with trimAl (-gt 0.3 -st 0.001 -cons 30, Capella-Gutierre et al. 2009). The high-quality MSA was then phylogenetically analysed in RAxML using the GTRCATI hill climbing algorithm and subsequently bootstrapped with 1000 pseudo-replications with the T-Rex server (Boc et al. 2012). The resulting phylogeny was then modified with FigTree v1.4.4 (Rambaut et al. 2018). The alignment and tree used in this analysis were deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S26176).
Results
Sampling events within surveyed fjords and bays revealed the presence of nematodes in all ice cores collected at locations with a water depth > 4 m. According to BLAST queries, 70 specimens (including the suspected nematode eggs) had the closest molecular identity to Halomonhystera spp. (92–93% identity), while 4 specimens were most genetically similar to Theristus spp. (94–96% identity) (see Online Resource 1). For the benthic samples, all nematodes identified molecularly are known to be associated solely with the benthos and not sea ice. Morphologically they appeared different from the ice specimens (pers. obser.). BLAST results indicated the presence of 8 nematode genera in sediment samples—Anticoma, Ascolaimus, Axonolaimus, Calomicrolaimus, Cephalanticoma, Microlaimus, Sabatieria, and Sphaerolaimus.
Throughout spring, one particular morphotype of egg cluster was observed after the appearance of sexually mature nematodes, in early April, which were confirmed to be nematode eggs based on observations of hatching and DNA sequencing. Eggs occurred in clusters (often with more than 20 eggs) that were contained within a ‘sack’. The shell of the eggs was transparent with the zygote filling nearly all of the egg shell (photos in Online Resource 2). Larvae developed from the zygote in late spring until hatching in mid to late April. Length measurements of nematodes showed that ice nematodes from Inglefieldbukta had a smaller size range than specimens from Van Mijenfjorden (Table 2).
To estimate evolutionary relationships among our nematode isolates, we tested phylogenetic hypotheses using the 18S ribosomal subunit gene locus. In general, many publicly available nematode sequences phylogenetically clustered into their, respectively, assigned clades, especially at higher taxonomic ranks (Fig. 2). Although many taxonomic orders were well supported, most with bootstrap support > 80%, many taxa in the same order formed separate well-supported clades, including the Desmodorida that branch sister to a larger clade containing many sequences within the Desmodorida (100% bootstrap support). In addition to split taxonomic orders, there were numerous single atypical sequences that grouped among otherwise well-supported monophyletic clades (AY692344). One Anticoma among Enoplus sequences (LK054717) and one Valvaelaimus among Bathylaimus species, a non-nematode apicomplexan branching sister to Monhystrella parvella. Moreover, there was a clade comprised six different genera that spanned four taxonomic orders and two classes grouping within the Rhabditida. When sequences from this clad were culled, aligned and comparatively explored, there were 10 SNPs out of 3522 base pairs analysed. Although the results of our de novo phylogeny suggested some misidentified nematode taxa, Svalbard nematodes grouped within the Chromadorea (100% bootstrap support), primarily in Halomonhystera (100% bootstrap support). Four isolates (two from Wahlenbergfjorden and two from Van Mijenfjorden) grouped in the genus Theristus (100% bootstrap support), with one additional sequence of a benthic nematode from Inglefieldbukta branching sister to one of the Araeolaimida.
Phylogenetic tree of Nematoda (18S rRNA) inferred with maximum likelihood methods in RAxML. Bootstrap values are generated after 1000 pseudo-replications. The placement of Svalbard sequences is denoted with arrows. For Svalbard nematode sequences sampling locations (VMF = Van Mijenfjorden, IB = Inglefieldbukta, PAL = Palanderbukta and WB = Wahlenbergfjorden) and number of individuals (n) are noted
Discussion
This study presents the first insight into the genetic diversity of coastal sympagic nematodes in the Arctic and foremost in Svalbard. Our results based on the 18S rRNA barcode strongly indicate the presence of at least two genera inhabiting the land-fast ice in western and eastern Svalbard—Theristus and Halomonhystera. One of the genera (Theristus) is well known and commonly described from (Arctic) benthic and often sea ice samples (e.g. Bluhm et al. 2018; Portnova et al. 2019). The second genus (Halomonhystera sp. n.) found is a novel discovery to land-fast ice and could represent a new sympagic nematode species. Thus far, this genus has only been found in the Arctic at the Håkon Mosby Mud Volcanoes in the Barents Sea (Tchesunov et al. 2015). Molecular tools enabled the identification of nematode eggs making it possible to confirm one of the morphotypes of eggs present in ice samples. This morphotype was targeted specifically after nematode larvae were observed developing inside the egg shell and after observations of hatching prior to the ice algal spring bloom. Benthic sequences were not included in the phylogenetic tree (apart from one), because none of the genera overlapped with the ones detected in the sea ice samples. All benthic sequences matched with genera that are commonly found in sediment samples, including Spitsbergen (Gerlach 1965; Urban-Malinga et al. 2005). Additionally, while screening through the sediment samples it appeared that the benthic nematodes had a different morphology than the sympagic specimens. They appeared to have a more distinct tail shape and be wider than the individuals found in the land-fast ice samples. Despite the obvious differences in their morphology, they were picked to confirm molecularly that they were not associated with the ice.
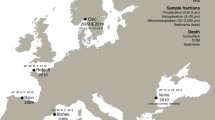
To date, morphological descriptions of four sympagic nematodes exist for the Arctic (Tchesunov 1986; Tchesunov and Riemann 1995; Tchesunov and Portnova 2005). However, these sympagic nematodes have not been assigned a molecular barcode yet, making it difficult to confirm whether our findings represent a new species (Halomonhystera sp. n.), individuals of the previously described ice nematode species or both. Nevertheless, four of our sequences strongly indicate the presence of the genus Theristus in coastal sea ice in Wahlenbergfjorden and Van Mijenfjorden, adding to the pan-Arctic distribution of this genus (Fig. 3). The Theristus Svalbard sequences phylogenetically branch sister to four Theristus acer sequences, which can be frequently observed in sediment samples in the Arctic including Svalbard (Urban-Malinga et al. 2005). Furthermore, our sequences have 95% identity with T. acer, underscoring that our isolates are closely related, yet molecularly distinct from T. acer. Therefore, we hypothesize that our sequences represent the first molecular record for the sympagic nematode: Theristus melnikovi. T. melnikovi is known from perennial sea ice over the Arctic Ocean (Tchesunov 1986) and seasonal sea ice from the Laptev Sea (Tchesunov and Riemann 1995).
Illustrative map of published records of known ice-associated nematode genera (Theristus melnikovi, Theristus sp., Cryonema crassum, Cryonema tenue, Cryonema sp., Hieminema obliquorum) in the Arctic. Species observed in sea ice are highlighted in orange and from the benthos in brown (based on Tchesunov 1986; Tchesunov and Riemann 1995; Riemann and Ngando 1997; Portnova and Tchesunov 2005; Derycke et al. 2007; Tchesunov et al. 2015)
Nematodes from Van Mijenfjorden, Inglefieldbukta and Palanderbukta were most genetically similar to reference sequences within the genus Halomonhystera. Halomonhystera is known from a wide range of habitats globally and is usually associated with the benthos and macroalgal detritus (e.g. Derycke et al. 2007; Tchesunov et al. 2015). The functional plasticity of this genus, which has made it successful at colonizing a wide range of marine habitats (e.g. Derycke et al. 2007; Leduc 2014; Tchesunov et al. 2015), is well documented. Within Halomonhystera, 21 species (Bezerra et al. 2020) have been accepted of which Halomonhystera disjuncta has been studied most intensely with molecular tools (e.g. Derycke et al. 2007; Van Campenhout et al. 2014). The ice nematode sequences from Svalbard form their own clade, indicating that they cannot be considered an integral part of the intertidal H. disjuncta species complex. Subsequently, our ice nematodes could potentially represent a new species of Halomonhystera that resides within seasonal land-fast ice. Despite no individuals of Halomonhystera (nor Theristus) detected in sediment samples, from Van Mijenfjorden, their absence cannot be confirmed due to small sample sizes analysed combined with a potentially too coarse mesh size (63 and 500 µm). Based on our own observations, the nematodes present in the ice have an oviparous reproduction mode, which has also been recorded for other species of Halomonhystera (see Tchesunov et al. 2015 for an overview). Sympagic nematodes from Van Mijenfjorden and Inglefieldbukta were similar in size with slightly larger specimens found in the western location, potentially due to environmental factors, such as food availability. No in-depth taxonomic information, such as body setae or form of spicules, was collected to enable morphology-based identification due to lack of expertise knowledge and necessary equipment. Subsequently, Dr Alexei Tchesunov (Lomonosov Moscow State University) and Dr Daria Portnova (P. P. Shirshov Institute of Oceanology) were consulted and a few selected photographs of ice nematodes were shared. They pointed out the high morphological similarity to Cryonema or Hieminema, which are both well described from coastal sea ice in the Russian Arctic (Tchesunov and Riemann 1995; Tchesunov and Portnova 2005) and closely related to Halomonhystera within the Geomonhysterini tribe (Fonseca and Decraemer 2017). However, dissections are needed to confirm these observations. In future studies, a close collaboration between morphological and molecular taxonomists are needed to determine whether this unique habitat harbours more than two species, as determined molecularly.
Nematodes lack many prominent morphological features that allow rapid diagnostic assessment of diversity. Through our analysis of publicly available nematode sequences, we found evidence that many nematodes have been assigned erroneous taxonomic names, underscoring the need for collaboration between molecular and trained morphological taxonomists. Moreover, we identified many clades that were split, yet supported with high bootstrap values, suggesting that many nematode clades might be unnatural. Polyphyletic clades have been previously reported (Meldal et al. 2007; Park et al. 2011), supporting our findings of molecularly polyphyletic clades.
This study provided a first insight into the diversity of ice nematodes colonizing coastal sea ice around Svalbard. The sequences and reproductive information from this study represent the first steps towards building a sympagic fauna database, which will facilitate future work on sea ice meiofauna biodiversity and their dependence on sea ice. However, several knowledge gaps remain to be filled and more combined morphological and molecular approaches are critical to accurately identify the nematode species that inhabit coastal sea ice. When nematode species can be identified accurately using molecular tools, we can more efficiently study their life cycle and dependency on land-fast ice, known unknowns that are vital in order to assess their ecological response to current and future changing sea ice conditions in the Arctic.
References
Andreasen MH (2019) Community composition, population structure and phylogeny of coastal sympagic meiofauna in eastern Svalbard. Master thesis, Bergen: UiB Universitet i Bergen. https://hdl.handle.net/1956/20217. Accessed 01 Oct 2020
Bezerra TN, Decraemer W, Eisendle-Flöckner U, Hodda M, Holovachov O, Leduc D, Miljutin D, Mokievsky V, Peña Santiago, R, Sharma J, Smol N, Tchesunov A, Venekey V, Zhao Z, Vanreusel A (2020) Nemys: World database of Nematodes, Nematoda. World Register of Marine Species. http://www.marinespecies.org/aphia.php?p=taxdetails&id=799%20on%202020-10-1. Accessed 1 Oct 2020.
Bhadury P, Austen MC, Bilton DT, Lambshead PJ, Rogers AD, Smerdon GR (2006) Development and evaluation of a DNA-barcoding approach for the rapid identification of nematodes. Mar Ecol Prog Ser 320:1–9. https://doi.org/10.3354/meps320001
Bik HM, Fournier D, Sung W, Bergeron RD, Thomas WK (2013) Intra-genomic variation in the ribosomal repeats of nematodes. PLoS ONE 8(10):e78230. https://doi.org/10.1371/journal.pone.0078230
Blome D, Riemann F (1999) Antarctic sea ice nematodes, with description of Geomonhystera glaciei sp. n. (Monhysteridae). Mitt Hambg Zool Mus Inst 96:15–20
Bluhm BA, Hop H, Vihtakari M, Gradinger R, Iken K, Melnikov IA, Søreide JE (2018) Sea ice meiofauna distribution on local to pan-Arctic scales. Ecol Evol 8:2350–2364. https://doi.org/10.1002/ece3.3797
Boc A, Diallo AB, Makarenkov V (2012) T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res 40:W573-579
Bütschli O (1874) Zur Kenntnis der freilebenden Nematoden, insbesondere der des Kieler Hafens. Abhandlungen hrsg. v. d. Senckenb. Naturf. Ges. Winter, Füssen, Germany
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Creer S, Fonseca VG, Porazinska DL, Giblin-Davis RM, Sung W, Power DM, Packer M, Carvalho GR, Blaxter ML, Lambshead PJ, Thomas WK (2010) Ultrasequencing of the meiofaunal biosphere: practice, pitfalls and promises. Mol Ecol 19:4–20. https://doi.org/10.1111/j.1365-294X.2009.04473.x
Derycke S, Backeljau T, Vlaeminck C, Vierstraete A, Vanfleteren J, Vincx M, Moens T (2007) Spatiotemporal analysis of population genetic structure in Geomonhystera disjuncta (Nematoda, Monhysteridae) reveals high levels of molecular diversity. Mar Biol 151:1799–1812. https://doi.org/10.1007/s00227-007-0609-0
Derycke S, De Ley P, Tandingan De Ley I, Holovachov O, Rigaux A, Moens T (2010) Linking DNA sequences to morphology: cryptic diversity and population genetic structure in the marine nematode Thoracostoma trachygaster (Nematoda, Leptosomatidae). Zool Scr 39:276–289. https://doi.org/10.1111/j.1463-6409.2009.00420.xFonseca
Fonseca G, Decraemer W (2008) State of the art of the free-living marine Monhysteridae (Nematoda). JMBA 88:1371–1390. https://doi.org/10.1017/S0025315408001719
Fonseca VG, Sinniger F, Gaspar JM, Quince C, Creer S, Power DM, Peck LS, Clark MS (2017) Revealing higher than expected meiofaunal diversity in Antarctic sediments: a metabarcoding approach. Sci Rep 7:6094. https://doi.org/10.1038/s41598-017-06687-x
Garrison DL, Buck KR (1986) Organism losses during ice melting: a serious bias in sea ice community studies. Polar Biol 6:237–239. https://doi.org/10.1007/BF00443401
Geospiza Research Team (2006) FinchTV 1.4. Geospiza, Inc., Seattle, WA, USA. http://www.geospiza.com. Accessed Dec 2017
Gerlach SA (1965) Freilebende Meeresnematoden aus der Gezeitenzone von Spitzbergen. Veröff Inst Meeresforsch Bremerhav 9:109–172
Heip C, Vincx M, Vranken G (1985) The ecology of marine nematodes. Aberdeen University Press, Aberdeen
Høyland KV (2009) Ice thickness, growth and salinity in Van Mijenfjorden Svalbard, Norway. Polar Res 28(3):339–352. https://doi.org/10.1111/j.1751-8369.2009.00133.x
Hugot JP, Baujard P, Morand S (2001) Biodiversity in helminths and Nematodes as a field of study: an overview. Nematology 3:199–208. https://doi.org/10.1163/156854101750413270
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice, and visualization. Brief Bioinform 20:1160–1166. https://doi.org/10.1093/bib/bbx108
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lambshead PJD, Boucher G (2003) Marine Nematode deep-sea biodiversity—hyperdiverse or hype? J Biogeogr 30:475–485. https://doi.org/10.1046/j.1365-2699.2003.00843.x
Leduc D (2014) Free-living nematodes of the genus Halomonhystera (Monhysteridae) from the Southwest Pacific region and Ross Sea. N. Z J Zool 41:46–57. https://doi.org/10.1080/03014223.2013.827581
Lee MR, Canales-Aguirre CB, Nunez D, Pérez K, Hernández CE, Brante A (2017) The identification of sympatric cryptic free-living nematode species in the Antarctic intertidal. PLoS ONE 12:e0186140. https://doi.org/10.1371/journal.pone.0186140
Meldal BH, Debenham NJ, De Ley P, De Ley IT, Vanfleteren JR, Vierstraete AR, Bert W, Borgonie G, Moens T, Tyler PA, Austen MC, Blaxter ML, Rogers AD, Lambshead PJD (2007) An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol 42:622–636. https://doi.org/10.1016/j.ympev.2006.08.025
Moens T, Braeckman U, Derycke S, Fonseca G, Gallucci F, Ingels J, Leduc D, Vanaverbeke J, Van Colen C, Vanreusel A, Vincx M (2013) Ecology of free-living marine nematodes. In: Schmidt-Rhaesa A (ed) Handbook of zoology: Gastrotricha, Cycloneuralia and Gnathifera, Nematoda, vol 2. De Gruyter, Berlin, pp 109–152. https://doi.org/10.1515/9783110274257.109
Norwegian Polar Institute (2020) Geodata Norwegian Polar Institute. https://geodata.npolar.no/. Accessed Dec 2020
Park JK, Sultana T, Lee SH, Kang S, Kim HK, Min GS, Eom KS, Nadler SA (2011) Monophyly of clade III nematodes is not supported by phylogenetic analysis of complete mitochondrial genome sequences. BMC Genom 12:392. https://doi.org/10.1186/1471-2164-12-392
Pitusi V (2019) Seasonal abundance and activity of sympagic meiofauna in Van Mijenfjorden, Svalbard. Master thesis, Tromsø: UiT Norges arktiske universitet. https://hdl.handle.net/10037/15941
Platt HM, Warwick RM (1980) The significant of free-living nematodes to the littoral ecosystem. In: Price JH, Irvine DEG, Farnham WF (eds) The shore environment ecosystems, vol 2. Academic Press, Cambridge, pp 729–759
Portnova D, Fedyaeva MA, Udalov AA, Tchesunov AV (2019) Community structure of nematodes in the laptev sea shelf with notes on the lives of ice nematodes. Reg Stud Mar Sci 31:100757. https://doi.org/10.1016/j.rsma.2019.100757
Rambaut A, Suchard MA, Xie D, Drummond AJ (2018) FigTree v1.4.4. Available at: https://github.com/rambaut/figtree/releases. Accessed 1 Dec 2019
Riemann F, Sime-Ngando T (1997) Note on sea-ice nematodes (Monhysteroidea) from resolute passage, canadian high Arctic. Polar Biol 18:70–75. https://doi.org/10.1007/s003000050160
Schierenberg E, Sommer RJ (2013) Reproduction and development in nematodes. In: Schmidt-Rhaesa A (ed) Handbook of zoology: Gastrotricha, Cycloneuralia and Gnathifera, Nematoda. Vol.2 De Gruyter, Berlin, pp 61-108
Semprucci F (2013) Marine nematodes from the shallow subtidal coast of the Adriatic sea: species list and distribution. IJBC 1:1–9. https://doi.org/10.1155/2013/187659
Søreide JE, Pitusi V, Vader A et al (2021) Environmental status of Svalbard coastal waters: coastscapes and focal ecosystem components (SvalCoast). In: Moreno-Ibáñez et al (eds) SESS report 2020, Svalbard Integrated Arctic Earth Observing System, Longyearbyen, pp 142–175. https://doi.org/10.5281/zenodo.4293849. Accessed 15 Jan 2021
Spindler M, Dieckmann GS (1986) Distribution and abundance of the planktic foraminifer Neogloboquadrina pachyderma in sea ice of the Weddell Sea (Antarctica). Polar Biol 5:185–191. https://doi.org/10.1007/BF00441699
Tchesunov AV (1986) A new free-living nematode connected with sea Arctic ice. Zool Zh 65:1782–1787
Tchesunov AV, Portnova DA (2005) Free-living nematodes in seasonal coastal ice of the White Sea. Description of Hieminema obliquorum gen. et sp. n. (Nematoda, Monhysteroidea). Zool Zh 84:899–914
Tchesunov AV, Riemann F (1995) Arctic sea ice nematodes (Monhysteroidea), with descriptions of Cryonema crassum gen. n., sp. n. and C. tenue sp. n. Nematologica 41:35–50. https://doi.org/10.1163/003925995X00035
Tchesunov AV, Portnova DA, Van Campenhout J (2015) Description of two free-living nematode species of Halomonhystera disjuncta complex (Nematoda: Monhysterida) from two peculiar habitats in the sea. Helgol Mar Res 69:57–85. https://doi.org/10.1007/s10152-014-0416-1
Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29:52–54. https://doi.org/10.2144/00291bm09
Urban-Malinga B, Wiktor J, Jabłońska A, Moens T (2005) Intertidal meiofauna of a high-latitude glacial Arctic fjord (Kongsfjorden, Svalbard) with emphasis on the structure of free-living nematode communities. Polar Biol 28:940–950. https://doi.org/10.1007/s00300-005-0022-4
Van Campenhout J, Derycke S, Tchesunov AV, Portnova D, Vanreusel A (2014) The dominant Håkon Mosby mud volcano nematode is genetically differentiated from its shallow–water relatives and shows genetic structure within the mud volcano. J Zool Syst Evol Res 52:203–216
Yeates GW (2007) Abundance, diversity, and resilience of nematode assemblages in forest soils. Can J Forest Res 37:216–225. https://doi.org/10.1139/x06-172
Acknowledgements
The authors would like to thank the UNIS logistics department, AB-330 participants of 2018, Malin Daase and Rupert Krapp for their support in the field. They thank the Norwegian Polar Institute for lending us their small field station in Inglefieldbukta. They appreciate the help from Stuart Thompson in the laboratory. They wish to express their gratitude to Tove Gabrielsen and Marie Davey for advice on processing of molecular data, and Alexei Tchesunov and Daria Portnova for advising us on the morphology of three of our nematodes. They would like to thank the reviewers, namely, Vadmir Mokievsky and Daria Portnova, for their constructive feedback which helped improve this manuscript. This research was supported by the Fram Centre (Tromsø), Arctic Ocean Flagship (project FADE; RiS-ID 11311), the 2017–2018 Belmont Forum and BiodivERsA joint call for research proposals, under the BiodivScen ERA-Net COFUND programme (project ACCES), the Norwegian Research Council, project nr 243702 (FAABulous) and Arctic Field Grants with Research in Svalbard project nr 10641, 10907 and 10931and the UiT/Tromsø Research Foundation for project ArcticSIZE (project Number 01vm/h15).
Funding
Open access funding provided by The University Centre in Svalbard.
Author information
Authors and Affiliations
Contributions
VP, MHA, MM and JES contributed to the study concept and design. Funding for the study was acquired by VP and MHA (fieldwork), and JES (laboratory work). Sample collection was performed by JES and VP in 2017 and 2018, and by MHA in 2018. Sample processing and analysis was conducted by VP and MHA; MM was a support in the molecular laboratory. Data processing and analysis were performed by VP, MHA and BTH (who was responsible for the creation of a robust phylogenetic tree). The first draft of the manuscript was written by VP and MHA, and all the authors commented on the previous versions of the manuscript before reading and approving the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pitusi, V., Søreide, J.E., Hassett, B.T. et al. The occurrence of Nematoda in coastal sea ice on Svalbard (European Arctic) determined with the 18S small subunit rRNA gene. Polar Biol 44, 1153–1162 (2021). https://doi.org/10.1007/s00300-021-02863-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-021-02863-y