Abstract
Key message
A wheat protein phosphatase PP2C-a10, which interacted with TaDOG1L1 and TaDOG1L4, promoted seed germination and decreased drought tolerance of transgenic Arabidopsis.
Abstract
Seed dormancy and germination are critical to plant fitness. DELAY OF GERMINATION 1 (DOG1) is a quantitative trait locus for dormancy in Arabidopsis thaliana. Some interactions between DOG1 and the type 2C protein phosphatases (PP2Cs) have been reported in Arabidopsis. However, the research on molecular functions and regulations of DOG1Ls and group A PP2Cs in wheat (Triticum aestivum. L), an important crop plant, is rare. In this study, the whole TaDOG1L family was identified. Expression analysis revealed that TaDOG1L2, TaDOG1L4 and TaDOG1L-N2 specially expressed in wheat grains, while others displayed distinct expression patterns. Yeast two-hybrid analysis of TaDOG1Ls and group A TaPP2Cs revealed interaction patterns differed from those in Arabidopsis, and TaDOG1L1 and TaDOG1L4 interacted with TaPP2C-a10. The qRT-PCR analysis showed that TaPP2C-a10 exhibited the highest transcript level in wheat grains. Further investigation showed that ectopic expression of TaPP2C-a10 in Arabidopsis promoted seed germination and decreased sensitivity to ABA during germination stage. Additionally, TaPP2C-a10 transgenic Arabidopsis exhibited decreased tolerance to drought stress. Finally, the phylogenetic analysis indicated that TaPP2C-a10 gene was conserved in angiosperm during evolutionary process. Overall, our results reveal the role of TaPP2C-a10 in seed germination and abiotic stress response, as well as the functional diversity of TaDOG1L family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seed dormancy is critical to plants by preventing germination under unfavorable conditions. Insufficient dormancy can lead to pre-harvest sprouting which significantly decreases the grain yield and quality in agricultural production, however too deep dormancy delays germination, thus decreasing the growth time at the appropriate season (Gubler et al. 2005; Finch-Savage and Leubner 2006).
The induction and release of dormancy are mainly controlled by two plant hormones: abscisic acid (ABA) and gibberellins (GA) (Yan and Chen 2017; Vishal and Kumar 2018). ABA participates in the processes of plant growth, development and responses to various abiotic stresses, while GA is involved in various stages of plant growth and development. ABA and GA have antagonistic effects on seed dormancy and germination, i.e., endogenous ABA content is gradually decreased from dormancy to germination phase in the seed, whereas the GA level is increased during this process (Golldack et al. 2013; Shu et al. 2016). In Arabidopsis, ABA-deficient mutants accelerate germination (Frey et al. 2011), while ABA catabolism mutants display deeper dormancy phenotype (Matakiadis et al. 2009).
In addition to ABA synthesis- and catabolism-related factors, proteins in ABA signaling pathway also affect seed dormancy and germination. ABA insensitive 3 (ABI3), ABI4, and ABI5 which are the transcriptional regulators of ABA signaling, are involved in seed dormancy and germination in Arabidopsis (Söderman et al. 2000; Finkelstein et al. 2002). MOTHER OF FT AND TFL1 (MFT), a phosphatidyl ethanolamine-binding protein (PEBP), plays a role in both ABA and GA signaling pathways to regulate seed germination in Arabidopsis (Xi et al. 2010). Group A protein phosphatase 2Cs (PP2Cs) which are ABA co-receptors, negatively regulate ABA signaling pathway. ABI1 and ABI2 are the group A PP2C proteins in Arabidopsis, their single mutants exhibited reduced sensitivity to exogenous ABA at seed germination stage (Leung et al. 1997; Gosti et al. 1999). However, their double mutant displays increased response to ABA comparing to the single mutants, suggesting that ABI1 and ABI2 form a negative feedback loop in regulating ABA signaling pathway (Merlot et al. 2001). Another two group A PP2Cs in Arabidopsis, ABA hypersensitive germination 1 (AHG1) and AHG3/AtPP2CA, play distinct and overlapping roles in seed germination (Yoshida et al. 2006; Nishimura et al. 2007). HON, a group A PP2C specifically expresses in seeds, inhibits ABA signaling directly while activating GA signaling indirectly in imbibed seeds, thus regulating seed dormancy homeostatically (Kim et al. 2013). The reduced dormancy 5 (RDO5), an ungrouped PP2C, suppresses Arabidopsis PUMILIO 9 (APUM9) transcript levels to regulate seed dormancy (Xiang et al. 2014).
Apart from phytohormone related proteins, other key regulators of seed dormancy have been characterized. DELAY OF GERMINATION 1 (DOG1) is identified as quantitative trait locus for dormancy in Arabidopsis (Alonso-Blanco et al. 2003). DOG1 belongs to a novel gene family which is plant-specific; other four members DOG1-like 1–4 (DOGL1–4) have been identified. DOG1 specifically expresses in the seed, and displays sequence diversity in the coding regions of different Arabidopsis varieties (Bentsink et al. 2006). DOG1 can regulate seed dormancy in an ABA-independent way (Nakabayashi et al. 2012). Further study has demonstrated that DOG1 acts as a timer for seed germination in a temperature-dependent manner (Graeber et al. 2014). Besides, DOG1 also affects seed development via genetic interaction with ABI3 (Dekkers et al. 2016), flowering time via microRNA pathway (Huo et al. 2016). It was assumed that DOG1 functioned in parallel to ABA signaling as both ABA and DOG1 have effects on seed dormancy (Nakabayashi et al. 2012). Recent studies found that DOG1 directly interacted with AHG1 and AHG3, which were members of group A PP2Cs, to control seed dormancy (Née et al. 2017; Nishimura et al. 2018).
Seed dormancy is an important agronomic trait for cereals, especially for wheat (T. aestivum. L) which is the most widely cultivated cereal around the world. Wheat dormancy and pre-harvest sprouting can be affected by TaDOG1Ls. Four DOG1-like genes in wheat have been identified, including TaDOG1L1, TaDOG1L2, TaDOG1L4 and TaDOG1L5-1 (Ashikawa et al. 2013). Ectopic expression of TaDOG1L1 increased seed dormancy in Arabidopsis (Ashikawa et al. 2010). Overexpression and RNA interference of TaDOG1L4 in wheat confirmed the role of TaDOG1L4 in seed dormancy and germination (Ashikawa et al. 2014). The fully annotated reference genome of hexaploid wheat has been completed; this greatly facilitates the identification of gene families in wheat genome. In this study, genome-wide identification and expression analyses of TaDOG1L genes were conducted. Several group A TaPP2Cs were isolated in our previous study (Yu et al. 2019). To examine the regulation of TaDOG1Ls by group A TaPP2Cs in wheat, yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays were performed and TaPP2C-a10 was found to interact with TaDOG1L4 in nuclei. Subsequently, the role of TaPP2C-a10 and possible regulatory mechanism in seed dormancy and germination were investigated in Arabidopsis. In addition to seed dormancy, TaPP2C-a10 also functioned in drought stress response. These findings enrich the functional studies of TaDOG1Ls and group A PP2Cs in wheat.
Materials and methods
Identification and phylogenetic analysis of TaDOG1L in wheat genome
To identity all the TaDOG1L proteins, DOG1 domain (PF14144) in Pfam 32.0 database (https://pfam.xfam.org) (El-Gebali et al. 2018) was used to search against the wheat annotated reference genome database (IWGSC RefSeq v1.0) by the hmmsearch program of the HMMER software 3.2.1 (https://hmmer.org/download.html) (Wheeler and Eddy 2013). The obtained proteins were further screened for conserved domains by InterProScan (https://www.ebi.ac.uk/interpro/download/) (Mitchell et al. 2018) in STANDALONE mode and NCBI CD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al. 2014). Protein sequences containing DOG1 domains were used to perform multiple sequence alignment by ClustalX 2.1 (Larkin et al. 2007), and then the phylogenetic tree was constructed by MEGA 6.0 based on the neighbor-joining (NJ) method with 1000 bootstrap replicates (Tamura et al. 2011). To analyze the exon–intron structures of TaDOG1Ls, the coding sequences and genome sequences were downloaded from Ensembl Plants database (https://plants.ensembl.org/index.html) (Howe et al. 2019). Gene Structure Display Server was then used to determine and visualize gene structures (https://gsds.cbi.pku.edu.ch) (Hu et al. 2014). To identify the AHG-like proteins in various species, amino sequences of Arabidopsis AHG1 and AHG3 were used to BLASTp against the annotated reference genome databases of A. thaliana (TAIR10), Brassica napus (AST_PRJEB5043_v1), Glycine max (Glycine_max_v2.1), Medicago truncatula (MedtrA17_4.0), Capsicum annuum (ASM51225v2), Nicotiana attenuata (NIATTr2), Brachypodium distachyon (Brachypodium_distachyon_v3.0), Oryza sativa Japonica Group (IRGSP-1.0), Aegilops tauschii (Aet_v4.0), Hordeum vulgare subsp. Vulgare (IBSC_v2), T. aestivum, Sorghum bicolor (Sorghum_bicolor_NCBIv3), Zea mays (B73_RefGen_v4), Amborella trichopoda (AMTR1.0), Selaginella moellendorffii (v1.0), Physcomitrella patens (Phypa_V3). Proteins with highest score and similarity were selected. Multiple sequence alignment was done by ClustalX 2.1, poorly aligned regions were removed, and then the polygenic tree was constructed by IQ-TREE web server (https://iqtree.cibiv.univie.ac.at/) (Trifinopoulos et al. 2016) using maximum-likehood (ML) method with 1000 bootstrap replicates. Subsequently, the phylogenetic trees were annotated and colored by Evolview webserver (https://www.evolgenius.info/evolview/) (Subramanian et al. 2019). Conserved motifs were discovered by MEME tool (https://meme-suite.org/tools/meme) (Bailey et al. 2006). Approximately 2 kb upstream genome sequence of gene was obtained to analyze the cis-regulatory elements by PlantCARE search tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002).
Expression analysis of TaDOG1L family in silico
To perform the expression analysis of TaDOG1L genes in different tissues at different wheat developmental stages, pubic RNA-seq data of the project choulet_URGI (Ramirez-Gonzalez et al. 2018) and ERP004505 (Pfeifer et al. 2014) were downloaded from the Wheat Expression Browser powered by expVIP (https://www.wheat-expression.com/) (Borrill et al. 2016). The transcripts per million (TPM) values were log 2 transformed to create heatmap by pheatmap package of R project (https://www.r-project.org/).
The plasmid construction of TaDOG1Ls and TaPP2C-a10
Primers were designed to isolate TaDOG1L genes from mixed cDNA templates of wheat by PCR, and A homoeolog of TaDOG1L1, B homoeolog of TaDOG1L2 and A homoeolog of TaDOG1L4 were cloned. For yeast two-hybrid assay, the coding regions of TaDOG1L1, 2 and 4 were amplified and inserted into vector pGBKT7 via DNA recombination. The pGADT7-TaPP2Cs plasmids were obtained from our lab (Yu et al. 2019). For subcellular localization assay, the coding sequences of TaDOG1L1, 4 and TaPP2C-a10 were cloned into vector pBI121 containing eGFP reporter gene. For BiFC assay, the coding sequences of TaDOG1Ls and TaPP2C-a10 were cloned and inserted into SpYCE and SpYNE, respectively. For Arabidopsis transformation, the coding region of TaPP2C-a10 was inserted into vector pSN1301 containing GUS reporter gene. TaPP2C-a10 and GUS gene were driven by individual CaMV 35S promoter. All the primers used for cloning and plasmid construction are listed in Supplementary Table S1.
Yeast two-hybrid assay
Yeast two-hybrid assay was performed by the Matchmaker GAL4 system (Clontech, USA). The pGADT7-TaPP2Cs, pGBKT7-TaDOG1Ls and the control vectors were co-transformed into yeast strain AH109. Interaction of SV40T and p53 or lamin-C was used as positive or negative control, respectively. The transformants were first selected by growing on double-dropout medium (SD/–Trp–Leu, DDO), then appraised by transferring to triple-dropout medium (SD/–His–Trp–Leu, TDO) and quadruple-dropout medium (SD/–Ade–His–Trp–Leu, QDO).
Subcellular localization and BiFC assays
Vectors pBI121-TaDOG1Ls, pBI121-TaPP2C-a10, SpYCE-TaDOG1Ls and SpYNE-TaPP2C-a10 were transformed into Agrobacterium tumefaciens strain EHA105, respectively. The positive transformants were then cultured and injected into young leaves of 4-week-old tobacco (Nicotiana tabacum) by Agrobacterium-mediated infiltration. For BiFC assay, leaves were co-infiltrated with mixtures of an equal amount of SpYCE/SpYNE culture. After 48 h, GFP or YFP signals of the epidermal cell from the infiltrated leaves were checked by fluorescence microscopy (OLYMPUS LX71, Japan). Cell nuclei were stained by the 4′,6-diamidino-2-phenylindole (DAPI) dye.
Plant materials and growth conditions
The wheat cultivar Chinese Spring was grown in a greenhouse (16 h light/8 h dark cycle at 22 ℃). Roots, stems, leaves from seedlings at three leaf stage, roots, stems, leaves, flag leaves, pistils, stamens from mature plants at flowering stage, and grains at different days post anthesis (dpa) were collected to perform tissue-specific expression analysis. The Arabidopsis wild-type Columbia was used in this study. Arabidopsis seeds were first sterilized and sown on 1/2 Murashige and Skoog (MS) medium, and then transferred to greenhouse. The whole plants of 7- or 10-day-old seedlings were collected for ABA- or drought-responsive gene expression analysis. All samples were stored at – 80 ℃ before extraction.
Total RNA extraction and qRT-PCR analysis
Total RNA from tissue sample was isolated by Plant Total RNA extraction Kit (Zomanbio, China), subsequently examined by agarose gel electrophoresis. Then, the first-strand cDNA was created by FastKing RT Kit (Tiangen, China). The qRT-PCR analysis was conducted using the AceQ qPCR SYBR Green Master Mix (Vazyme, China) on a real-time PCR instrument (CFX96; Bio-Rad, USA). The reaction procedure was as follow: 95 ℃ for 5 min, 40 cycles of 95 ℃ for 10 s, 58 ℃ for 20 s, 72 ℃ for 20 s. The 2−ΔΔCT method was used for qRT-PCR analysis (Livak and Schmittgen 2001). The wheat actin and Arabidopsis actin genes were used as reference genes. All primers used for qRT-PCR are listed in Supplementary Table S1.
Generation of transgenic Arabidopsis plants
The pSN1301-TaPP2C-a10 plasmid was transformed into strain EHA105. Positive transformants were cultured to infiltrate Arabidopsis through the floral-dip method (Clough and Bent 1998). Transgenic seeds were selected by growing on 1/2 MS medium (pH 5.8) with 20 mg/L hygromycin B. In addition, GUS staining and PCR detection were used to confirm the transgenic lines. Homozygous lines were used for further analysis. The abundance of the TaPP2C-a10 transcript in transgenic Arabidopsis was assessed by RT-PCR.
Germination and root growth assays of Arabidopsis
For germination assay, about 50 seeds were sown on 1/2 MS plates with various concentrations of ABA. Germination (radicles emergence) and post-germination growth (expanded green cotyledons) were counted daily for 7 days. For the root growth assay, 5-day-old seedlings from hormone-free 1/2 MS were transferred to 1/2 MS plates with various concentrations of ABA. The primary root length was measured after 7 days. To analyze the expression levels of ABA-responsive genes in transgenic Arabidopsis and the wild type, 7-day-old seedlings from 1/2 MS plates without ABA were collected for qRT-PCR assay.
Drought treatment and water loss assay of Arabidopsis plants
For drought treatment, seedlings were grown in soil for 4–5 weeks, then were deprived of water for 7 days before the bolting stage. For water loss assay, rosette leaves were collected from 4-week-old plants, and then immediately weighed at each time point in the lab environment (23–25 ℃). For dry treatment of 10-day-old seedlings, whole plants were exposed in the air of the lab for 1 h. The untreated seedlings were taken as controls to perform qRT-PCR analysis of drought-responsive genes.
Results
Genome-wide identification of TaDOG1L family
Common wheat is allohexaploid with three homoeologous subgenomes (A, B, and D), therefore, each gene in common wheat should potentially have three homoeologs. Previous study has identified four DOG1-like genes (12 homoeologs) in wheat based on the amino acid sequence similarity with AtDOG1 (Ashikawa et al. 2013). In this study, hmmsearch program was applied to search DOG1L genes using the latest wheat reference database. After genome-wide searching, 54 proteins were found to have the DOG1 domains (Fig. 1, Table S2). However, there were 39 proteins also containing the typical bZIP domains. Further phylogenetic analysis indicated that these 39 proteins belonged to the clade D bZIP transcription factors including three previous identified proteins of TaDOG1L5-1 homoeologs. Therefore, a total of six TaDOG1L genes (15 homoeologs) were identified including three genes TaDOG1L1, TaDOG1L2 and TaDOG1L4 which had been identified before (Fig. 1). Another new TaDOG1L genes were renamed as TaDOG1L-N1, -N2 and -N3 (Fig. 2a, Table S2). TaDOG1L1, TaDOG1L2, TaDOG1L4 and TaDOG1L-N3 genes having three homoeologs, TaDOG1L-N2 lacking D homoeolog, and TaDOG1L-N1 only having A homoeolog. These genes were located on wheat chromosomes 1, 2, 3 and 6, respectively. In Arabidopsis, except for DOG1, four DOG1-like genes (AtDOGL1-4) were identified (Bentsink et al. 2006), so wheat contained one more DOG1-like member than Arabidopsis. Unlike AtDOG1 which had many transcripts (Bentsink et al. 2006; Cyrek et al. 2016), each TaDOG1L had only one transcript. Analyses of the exon–intron structures of TaDOG1Ls revealed that most genes had one exon (Fig. 2b), while AtDOG1 had three exons.
Phylogenetic analysis of all wheat proteins containing DOG domains. Amino acid sequences of 54 wheat proteins were used to construct the phylogenetic tree using the NJ method by ClustalX 2.1 and MEGA 6.0 with 1000 bootstrap replicates. Conserved domains and gene expression pattern are indicated with different shapes
Gene structures and expression patterns of TaDOG1L gene family. a The phylogenetic tree of TaDOG1L family. The scale bar indicates the average number of amino acid substitutions per site. b Exon–intron structures of TaDOG1L genes. c Expression profiles of TaDOG1L genes at different developmental stages and tissues. dpa days post anthesis. d Expression profiles of TaDOG1L genes in different cell types at three developmental stages of wheat endosperm. Cell types include whole endosperm (WE), starchy endosperm (SE), aleurone layer (AL), aleurone layer and starchy endosperm (ALCE), transfer cells (TCs). The color scale below represents the relative gene expression level, and red or blue indicates relative higher or lower expression level (c, d) (color figure online)
Tissue-specific expression analysis of TaDOG1L family
Pubic RNA-seq data of wheat variety Chinese Spring were acquired to analyze the tissue-specific expression patterns of TaDOG1Ls in 15 tissues (stem, spike, root, leaf, grain tissues at three developmental stages) under non-stress condition (Fig. 2c). TaDOG1L2, TaDOG1L4 and TaDOG1L-N2 exhibited grain-specific expression especially in grains at 15 and 30 dpa stages, excepting that TaDOG1L4-A also expressed in spike at flag leaf stage. TaDOG1L1 had broad range of expression levels in stems, spikes, roots and grains, and preferentially expressed in roots at all developmental stages. TaDOG1L-N3 displayed non-specific expression in stems, spikes and grains. Overall, all the TaDOG1Ls expressed in grains except for two homoeologs without detectable expression value. Furthermore, the expression patterns of TaDOG1Ls in different cell types at three different developmental stages of endosperms were evaluated (Fig. 2d). As a result, most TaDOG1Ls displayed higher transcript levels in 20 dpa endosperms, mainly in the aleurone layer (AL), merely in starchy endosperm (SE). TaDOG1L1-A and TaDOG1L-N3-A showed higher transcript abundance in transfer cells (TCs) than AL comparing to their B and D homoeologs, indicating the expression divergence within homoeologs.
Interactions between TaDOG1Ls and group A TaPP2Cs
Three TaDOG1Ls (TaDOG1L1, 2 and 4) were cloned to perform interaction analysis between TaDOG1Ls and group A TaPP2Cs by yeast two-hybrid assay (Fig. 3). TaDOG1L1 had strong interaction with TaPP2C-a10, and weak interactions with TaPP2C-a5 and -a9. TaDOG1L4 also interacted with TaPP2C-a10. However, no interaction was found between TaDOG1L2 and the eight group A TaPP2Cs. Phylogenetic analysis of all tested TaPP2Cs and group A AtPP2Cs revealed that TaPP2C-a1, -a2, -a3 and -a4 belonged to the ABI subfamily, while TaPP2C-a5, -a8, -a9 and -a10 belonged to the AHG1 subfamily (Fig. 4a). Besides, TaPP2C-a10 was closer to AHG1 than the other three proteins. To discover motifs within group A TaPP2Cs and AtPP2Cs, MEME motif search tool was applied. As a result, ten motifs were identified (Fig. 4b, Table S3). After comparing with the motifs in AtPP2Cs and TaPP2Cs identified by previous studies (Xue et al. 2008; Yu et al. 2019), seven motifs were found to exist in all PP2Cs including group A PP2Cs, and motifs 7, 8 and 10 were specifically found in group A PP2Cs. Moreover, motifs 8 and 10 only existed in ABI subfamily (Fig. 4c).
Phylogenetic analysis and motif distribution pattern of group A TaPP2Cs and AtPP2Cs. a Phylogenetic analysis of group A TaPP2Cs and AtPP2Cs. The phylogenetic tree was constructed using the NJ method with ClustalX 2.1 and MEGA 6.0 with 1000 bootstrap replicates. The scale bar represents 0.1 amino acid substitutions per site. b Motif distribution pattern of group A TaPP2Cs and AtPP2Cs. Ten motifs were discovered by MEME searching tool. The scale bar below corresponds to the length of amino acid sequence. c Sequence logos of motif 7, 8 and 10
To further confirm the interactions of TaDOG1L1, TaDOG1L4 with TaPP2C-a10 in vivo, BiFC assay was performed using young tobacco leaves. Before BiFC assay, subcellular localizations of TaDOG1L1, TaDOG1L4 and TaPP2C-a10 were determined by transient expressions of their corresponding coding sequences fused with GFP. The locations of nuclei were confirmed by DAPI staining. As shown in Fig. 5a, TaDOG1L1 and TaDOG1L4 exhibited distinct subcellular localization patterns. For TaDOG1L1-GFP fusion protein, green fluorescence signals were detected in both cytoplasm and nuclei, while TaDOG1L4-GFP fusion protein accumulated only in nuclei. Moreover, TaPP2C-a10-GFP fusion protein also displayed nuclear localization. In BiFC assay, fluorescence signals were observed in nuclei when co-expressing TaPP2C-a10-YNE and TaDOG1L4-YCE, while no fluorescence was detected in the vector control (Fig. 5b). However, we did not observe YFP signal when co-expressing TaPP2C-a10-YNE and TaDOG1L1-YCE. Taken together, TaPP2C-a10 interacts with TaDOG1L4 in the nuclei.
The subcellular localization and interactions of TaDOG1Ls and TaPP2C-a10. a The subcellular localization of TaDOG1L1, TaDOG1L4 and TaPP2C-a10 in tobacco leaves. The pBI121-GFP vector was transformed as control. b BiFC analysis of TaDOG1L4 and TaPP2C-a10. Leaves were co-infiltrated with plasmids expressing TaPP2C-a10 fused with YNE and DOG1L4 fused with YCE. The GFP and YFP signals were observed after 48–72 h. Scale bars 100 μm
TaPP2C-a10 exhibits the highest transcript level in wheat grains
The qRT-PCR analysis was applied to examine the expression patterns of TaDOG1L1, TaDOG1L4 and TaPP2C-a10 in different wheat tissues. TaDOG1L1 mainly expressed in roots, stems and grains, particularly in 20 dpa grains (Fig. 6a). This finding partially corresponded to the RNA-seq data of TaDOG1L1 which displayed higher transcript abundance in roots than stems and grains. TaDOG1L4 specifically expressed in grains, and had significant increase (more than 20-fold) at late grain development stage (Fig. 6b). TaPP2C-a10 expressed largely in grains and less in young leaves. The transcript level of TaPP2C-a10 reached the highest (nearly 60-fold) at 20 dpa grains, and decreased with time after that (Fig. 6c). Although expression patterns of TaDOG1L1, TaDOG1L4 and TaPP2C-a10 were distinct from each other in these tissues, they crossed and overlapped in grains, especially at late development stage.
Quantitative RT-PCR analysis of TaDOG1L1, TaDOG1L4 and TaPP2C-a10 genes in various tissues at different developmental stages. Expressions of TaDOG1L1 (a), TaDOG1L4 (b) and TaPP2C-a10 (c) in root, stem, leaf tissues from seedlings at three leaf stage, root, stem leaf, flag leaf, stamen and pistil tissues from mature plants at flowering stage and grains at different dpa stages were analyzed. Error bars represent the standard deviation of three independent replicates
TaPP2C-a10 promotes seed germination in Arabidopsis
Yeast two-hybrid assay showed that TaPP2C-a10 interacted with TaDOG1L1 and TaDOG1L4 which were involved in seed dormancy and germination (Ashikawa et al. 2010, 2014), and qRT-PCR analysis showed that TaPP2C-a10 preferentially expressed in grains. These results indicated that TaPP2C-a10 might affect seed dormancy and germination by interacting with DOG1Ls. To investigate whether TaPP2C-a10 has such roles, we subsequently introduced TaPP2C-a10 into Arabidopsis by Agrobacterium mediated floral-dip method for functional characterization. GUS staining was applied to detect the transgenic lines (Fig. S1a). Six independent transgenic Arabidopsis lines were obtained, and expression levels of TaPP2C-a10 in these lines were verified by RT-PCR (Fig. S1b). Apart from to transgenic line 6, lines 1–5 all had relative high TaPP2C-a10 expression levels. Therefore, homozygous lines 2, 3 and 4 (L2, L3 and L4) were randomly chosen to perform germination assay. Transgenic Arabidopsis of pSN1301 empty vector was used for negative control.
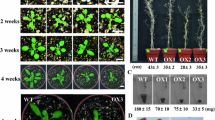
To analyze the effect of seed stratification on the germination of TaPP2C-a10 transgenic Arabidopsis in the absence of exogenous ABA, germination and post-germination growth efficiencies after stratification for 0 and 4 days were calculated. Without stratification, TaPP2C-a10 transgenic line L4 germinated and grew slightly faster than the wild type (Fig. 7a, b). No apparent difference was observed between L4 and the wild type after stratification treatment. As our previous study showed that TaPP2C-a10 significantly responded to ABA stress, to elucidate the role of TaPP2C-a10 in the ABA response of transgenic Arabidopsis, germination and post-germination growth efficiencies under various concentrations of ABA after stratification were examined. Under low concentration of ABA treatment, no significant difference was found among TaPP2C-a10 transgenic lines, VC and WT (Fig. 7c, g). However, at higher ABA content (1.0 and 1.5 μM) medium, the germination efficiencies of TaPP2C-a10 transgenic lines were less inhibited by ABA comparing to VC and WT lines (Fig. 7d, e). Additionally, the post-germination growth efficiencies of TaPP2C-a10 transgenic lines retained 70–90% with high ABA content, exhibiting significant difference (**p < 0.01) from VC and WT which had dramatically reduced post-germination growth efficiencies (less than 20% at 1.0 μM ABA) (Fig. 7f, g). These observations indicate that TaPP2C-a10 promotes seed germination in Arabidopsis and decreases sensitivity to ABA during germination.
TaPP2C-a10 decreases ABA sensitivity of transgenic Arabidopsis during germination. Germination (a) and post-germination (b) efficiencies of TaPP2C-a10 transgenic seeds (closed symbols) and wild-type (WT) seeds (open symbols) after stratification for 0 day (circles) or 4 days (triangles). Germination (c–e) and post-germination (f) efficiencies of seeds from TaPP2C-a10 transgenic lines, vector control (VC) and WT plants in the presence of 0.5 (c), 1.0 (d), 1.5 (e) μM ABA after stratification for 4 days. Error bars indicate the standard deviation of three independent experiments (a–f). g Phenotypes of TaPP2C-a10 transgenic lines, VC and WT plants. Seeds were grown on 1/2 MS plates containing various concentrations of ABA for 7 days
TaPP2C-a10 regulates seed germination and root growth though ABA signaling
Besides seed germination, ABA also affects plant growth including primary root growth (Hong et al. 2013). To validate whether constitutive expression of TaPP2C-a10 gene in Arabidopsis could affect root growth, 5-day-old seedlings of same size and consistent growth status were transferred to medium containing various concentrations of ABA and the primary root lengths were measured after seven days. In the absence of ABA, primary root lengths of TaPP2C-a10 transgenic lines were almost the same as the controls (Fig. 8a, b). With high concentrations of ABA (no less than 10 μM), primary root elongations of VC and WT were visibly inhibited by ABA, while those of TaPP2C-a10 transgenic lines seemed to be unaffected, even under treatment with 40 μM ABA, suggesting that TaPP2C-a10 also decreases ABA sensitivity of root during plant development.
TaPP2C-a10 regulates seed germination and primary root growth though ABA signaling. a Primary root in TaPP2C-a10 transgenic lines is insensitive to ABA. Five-day-old seedlings from hormone-free medium were transferred to 1/2 MS with various concentrations of ABA. Primary root length was measured after 7 days. b Statistical analysis of primary root lengths of TaPP2C-a10 transgenic lines, VC and WT in a. c Expression analysis of ABA-responsive genes in TaPP2C-a10 transgenic lines and WT. Whole plants of 7-day-old seedlings were used for analysis. Error bars indicate the standard deviation of three independent replicates (b, c). The asterisks indicate significant differences compared with the wild type (*P < 0.05, **P < 0.01; Tukey test)
To further examine whether ABA signaling in TaPP2C-a10 transgenic Arabidopsis was affected comparing to the wild type, transcript levels of ABA-responsive genes, such as SnRK2s (Nakashima et al. 2009), ABI3 (Giraudat et al. 1992), ABI4 (Finkelstein et al. 1998), ABI5 (Finkelstein and Lynch 2000), Em1 and Em6 (Carles et al. 2002) were confirmed by qRT-PCR using 7-day-old seedlings (Fig. 8c). The result showed that the expression levels of all tested ABA-responsive genes were reduced at different degrees in TaPP2C-a10 transgenic Arabidopsis. Additionally, the expression of Em1 was significantly suppressed in all TaPP2C-a10 transgenic lines (**P < 0.01). These findings agreed with the ABA-insensitive phenotypes of TaPP2C-a10 transgenic lines. As these ABA-responsive genes were involved in regulating seed germination and plant growth, we could deduce that TaPP2C-a10 takes part in ABA signaling pathway to regulate seed germination and root growth.
TaPP2C-a10 decreases drought tolerance in Arabidopsis
As the expression level of TaPP2C-a10 was greatly increased after PEG treatment (Yu et al. 2019), the effect of drought stress on TaPP2C-a10 transgenic Arabidopsis was checked. For drought treatment assay, adult plants grown in soil for 4–5 weeks were withheld water. After 7 days, TaPP2C-a10 transgenic plant withered and died, while WT plants seemed much healthier (Fig. 9a). Furthermore, detached leaves of TaPP2C-a10 transgenic plants exhibited higher relative water loss rate than WT (Fig. 9b). These results suggest that TaPP2C-a10 decreases drought tolerance ability of transgenic Arabidopsis.
TaPP2C-a10 negatively regulates drought tolerance of transgenic Arabidopsis. a Phenotypes of TaPP2C-a10 transgenic line and WT plants under drought conditions. Four-week-old plants grown on soil were withheld water for 7 days. b Relative water loss of detached rosette leaves from TaPP2C-a10 transgenic lines (circles) and WT plants (triangles). Error bars indicate the standard deviation of three replicates. c Expression analysis of drought-responsive genes in TaPP2C-a10 transgenic lines and WT plants. Ten-day-old plants with or without drought treatment were used for analysis. Error bars indicate the standard deviation of three independent replicates. The asterisks indicate significant differences compared with the wild type (*P < 0.05, **P < 0.01; Tukey test)
To further confirm the role of TaPP2C-a10 in drought stress response, the transcript levels of drought-responsive genes in 10-day-old plants before and after drought treatment were analyzed by qRT-PCR. Under normal conditions, the expression levels of SnRK2.2, SnRK2.6, ABI5, ABF3, ABF4 and RD29B in transgenic Arabidopsis were significantly reduced than those in WT (Fig. 9c). After drought treatment, this difference was still evident between TaPP2C-a10 transgenic lines and WT, indicating that expression of TaPP2C-a10 affects the drought response of transgenic Arabidopsis.
TaPP2C-a10 gene is conserved during evolutionary process
Phylogenetic analysis revealed that TaPP2C-a10 belonged to AHG1 subfamily (Fig. 4), and TaPP2C-a10 functioned like AHG1 and AHG3 by regulating seed dormancy and germination with DOG1Ls. To further survey the evolutionary relationships between these AHG-like proteins, 31 AHG-like proteins including TaPP2C-a10 were identified by BLAST tool (Table 1). These proteins were from different families and genera in eudicotyledon, monocotyledon, amborellales, lycopodiopsida and bryopsida. The phylogenetic tree was constructed by ML method with 1000 bootstrap replicates (Fig. 10a). The AHG-like proteins in angiosperms happened to fall into two groups: AHG1- and AHG3-like groups. From evolution perspective, the evolution pattern of AHG1/AHG3-like gene accorded with the evolutionary process of angiosperms. This indicates that AHG1/AHG3-like gene including TaPP2C-a10 is conserved during evolutionary process. Eleven conserved motifs were discovered within these AHG-like proteins (Fig. 10b, Table S3), and motifs 1–7 existed in the entire PP2C family. Apart from these seven motifs, motif 8 was only found in group A PP2Cs, motif 10 was specifically possessed by the AHG-like proteins, and the rest (motifs 9 and 11) merely existed in AHG3-like group. Moreover, motif 10 was rich in arginine which is a kind of alkaline amino acid (Fig. 10c).
Phylogenetic analysis and motif distribution pattern of AHG-like proteins. a Phylogenetic analysis of AHG-like proteins. The phylogenetic tree was constructed using the ML method by IQ-TREE with 1000 bootstrap replicates. The red and green branches represent eudicotyledon and monocotyledon species, respectively. The scale bar indicates 0.1 amino acid substitutions per site. b Motif distribution pattern of AHG-like proteins in angiosperm. Eleven motifs were discovered by MEME searching tool. The scale bar below corresponds to the number of amino acid residues. c Sequence logo of motif 10 (color figure online)
Discussion
Functional diversity of TaDOG1L family
Seed dormancy and germination are important traits for crops. DOG1 is critical to the induction of seed dormancy. In the present study, six TaDOG1L genes (15 homoeologs) were identified by genome-wide searching (Fig. 1). Actually, another 39 proteins also contained the DOG1 domains, but technically belonged to the clade D bZIP transcript factors according to the description of DOG1 family (Bentsink et al. 2006). Expression patterns of TaDOG1Ls in different tissues showed almost all TaDOG1Ls had expressions in grains. Further expression analyses of TaDOG1Ls in different cell types of endosperms showed that most genes expressed in the aleurone layer. As aleurone cells are essential for grain germination (Pfeifer et al. 2014), TaDOG1Ls potentially participate in seed germination. In fact, TaDOG1L1 and TaDOG1L4 have been verified to regulate seed dormancy and germination (Ashikawa et al. 2010, 2014). Expression analyses of TaDOG1Ls also revealed that three genes including TaDOG1L2 and 4 specifically expressed in grains, while others genes like TaDOG1L1 had non-specific and broad expression in wheat except for TaDOG1L-N1 which had no detectable value in all tissues (Fig. 2). The diverse expression patterns of TaDOG1L genes suggest functional difference between them.
Additionally, TaDOG1L1, TaDOG1L2 and TaDOG1L4 displayed different interactions with group A TaPP2Cs. While TaDOG1L1 interacted with TaPP2C-a5, -a9 and -a10, TaDOG1L2 had no interactions with any TaPP2Cs. Meanwhile, TaDOG1L4 only interacted with TaPP2C-a10 (Fig. 3). Transient expression of TaDOG1Ls fused with GFP showed that TaDOG1L1 was observed in cytoplasm and nuclei, whereas TaDOG1L4 was only detected in nuclei (Fig. 4). These results also suggest the functional diversity of TaDOG1L family. In Arabidopsis, unlike AtDOG1 mutant, AtDOGL1, AtDOGL2, and AtDOGL3 mutants germinated normally (Bentsink et al. 2006). Therefore, DOG1L family members play roles in more than regulating seed dormancy and germination. Two studies have demonstrated that DOG1L family is involved in regulating flowering time (Huo et al. 2016) and drought tolerance (Zhang et al. 2019). Besides, two-hybrid assay showed that TaDOG1L1 had interactions with TaPP2C-a5, -a9 and -a10 which were all from the AHG1 subfamily, but not ABI1 subfamily (Fig. 4); in other words, TaDOG1L1 only interacted with TaPP2Cs from AHG1 subfamily. This is partly in accordance with the result in Arabidopsis that AtDOG1 had interactions with all AHG1 subfamily members rather than ABI1 subfamily members from group A PP2Cs (Nishimura et al. 2018). However, there is apparent difference that TaDOG1L1 could not interact with all AHG1 subfamily members, suggesting different interaction patterns of DOG1 and group A PP2Cs in wheat and Arabidopsis. Additionally, to confirm the relationship between TaDOG1Ls and TaPP2C-a10, other protein–protein interaction methods need to be considered in the further study.
TaPP2C-a10 can regulate seed germination and plant growth though ABA signaling
Two-hybrid and BiFC assays exhibited that TaPP2C-a10 could interact with TaDOG1L4 (Figs. 3, 5b). Phylogenetic analysis revealed that TaPP2C-a10 was highly homologous to Arabidopsis AHG1 (Fig. 4), which interacts with N-terminus of AtDOG1 to control seed dormancy and germination (Née et al. 2017; Nishimura et al. 2018). Meanwhile, qRT-PCR analysis displayed that TaPP2C-a10 abundantly expressed in grains (Fig. 6c). Thus, TaPP2C-a10 might function in seed dormancy and germination. Further investigation showed that TaPP2C-a10 transgenic Arabidopsis germinated faster than WT, especially in the presence of high content ABA (Fig. 7). The primary roots of transgenic seedlings also elongated longer than those of the controls on the mediums with various concentrations of ABA (Fig. 8a, b). Therefore, TaPP2C-a10 indeed plays roles in seed germination and plant growth.
Expression analysis displayed that transcript levels of some ABA-responsive genes were dramatically decreased in TaPP2C-a10 transgenic Arabidopsis (Fig. 8c); while the AtDOG1 expression level in freshly harvested seeds of TaPP2C-a10 transgenic Arabidopsis had no significant difference from those of WT plants (Fig. S2). These ABA-responsive genes included subclass III SnRK2s (SnRk2.2, SnRk2.3 and SnRk2.6), ABI3, ABI4, ABI5, Em1 and Em6 (Nakashima and Yamaguchi-Shinozaki 2013). Previous study showed that DOG1 genetically interacted with ABI3, and affected the expression of downstream ABI5 (Dekkers et al. 2016). Another study confirmed synergistic action between ABI4 and ABI5 in regulating gene expression (Reeves et al. 2011). As ABI3, ABI4 and ABI5 play essential roles in seed development, and their mutants decreased sensitivity to ABA inhibition of germination (Giraudat et al. 1992; Finkelstein et al. 1998; Finkelstein and Lynch 2000), we deduce that the accumulation of TaPP2C-a10 protein in transgenic Arabidopsis suppresses the DOG1-ABI3-ABI5 pathway, thus decreasing sensitivity to ABA during germination. Moreover, ABI5 directly regulated expressions of Em1 and Em6 by binding to their promoters (Bensmihen et al. 2002; Carles et al. 2002), this can explain the lower expression levels of Em1 and Em6 in TaPP2C-a10 transgenic lines. On the other hand, subclass III SnRK2s are positive regulators of ABA signaling pathway, which phosphorylate ABFs and ABI5 to activate ABRE-driven gene expression (Umezawa et al. 2010; Fujita et al. 2013). Group A PP2Cs can dephosphorylate and inactivate subclass III SnRK2s. Additionally, SnRk2.2, SnRk2.3 and SnRk2.6 were shown to control seed dormancy and development through ABA signaling (Fujii et al. 2007; Nakashima et al. 2009). Thus, TaPP2C-a10 protein might also regulate seed germination by affecting SnRK2-ABI5 pathway. These findings suggest the cross-talk and signal integration between DOG1 and ABA signaling pathways, furthermore, group A PP2Cs, such as TaPP2C-a10 and AHG1, act like cross points therein.
Interestingly, analysis of upstream regulatory sequences of TaDOG1L1 and TaDOG1L4 revealed that there were some ABRE-motifs existing in their promoters (Table S4). Previous study reported that DOG1 antisense strongly responded to ABA stress (Yatusevich et al. 2017). We further analyzed the response of TaDO1L1 to ABA stress, and the expression of TaDO1L1 was significantly decreased after ABA treatment (Fig. S3). To figure out whether TaDOG1Ls have roles in ABA signaling, deep researches are required. Additionally, our previous study showed that TaPP2C-a10 was significantly up-regulated after GA treatment, and further investigation on the promoter of TaPP2C-a10 revealed the presence of GA-responsive element (Yu et al. 2019). A previous report has revealed that a group A PP2Cs was involved in both ABA and GA signaling pathways (Kim et al. 2013). Therefore, TaPP2C-a10 might also participate in GA signaling pathway to regulate seed dormancy and germination, still this assumption needs experimental verification. Nevertheless, the reason why expression levels of subclass III SnRK2s were reduced in TaPP2C-a10 transgenic lines remains unclear, as little is known about regulatory mechanism of SnRK2s at expression level.
TaPP2C-a10 is also involved in drought stress response
Group A PP2Cs were shown to repress the intrinsic desiccation tolerance of the vegetative tissue (Komatsu et al. 2013). TaPP2C-a10 transgenic plants exhibited weaker tolerance to drought stress comparing to WT, and its detached leaves had relative higher water loss rate (Fig. 9a, b). Meanwhile, TaPP2C-a10 transcript level was notably induced by PEG treatment (Yu et al. 2019). Therefore, TaPP2C-a10 negatively regulates drought stress response. A recent study also showed that group A PP2C genes ZmPP2C-A2 and ZmPP2C-A6 negatively regulated drought responses in maize (Zea mays) (He et al. 2019). Our further analysis showed that expression levels of several drought stress-responsive genes were lower in TaPP2C-a10 transgenic lines than the controls under normal condition, and were still lower than WT after drought treatment (Fig. 9c). Previous studies showed that subclass III SnRK2s responded to multiple stresses including drought stress in both ABA-independent and -dependent pathways (Fujita et al. 2009; Nakashima et al. 2009). SnRK2 kinases can activate the ABF transcription factors to regulate gene expression in ABA-dependent manner, thus responding to osmotic stress (Fujita et al. 2013; Yoshida et al. 2014). The reduced expression levels of SnRk2.2, SnRK2.6, ABF3, ABF4 and ABI5 suggest that the SnRK2-ABF pathway was suppressed in TaPP2C-a10 transgenic Arabidopsis. Additionally, TaPP2C-a10 was found to interact with subclass III SnRK2s (Yu et al. 2019). Consequently, TaPP2C-a10 probably suppresses SnRK2-ABF pathway to affect plant drought stress response. Besides, the DOG1L family was shown to be involved in drought stress tolerance, even though the molecular mechanism was unclear. NtabDOG1L positively regulated drought stress tolerance in N. tabacum (Zhang et al. 2019). Inactivation of DOG1 by its antisense transcript (asDOG1) resulted in enhanced drought sensitivity (Yatusevich et al. 2017). Thus, there is a possibility that TaPP2C-a10 regulates DOG1Ls to respond to drought stress.
AHG-like gene is conserved in angiosperm
The evolutionary process of AHG-like gene in angiosperm suggests conserved structures and functions of AHG1/AHG3-like proteins (Fig. 10a). This was verified by the functional study of TaPP2C-a10. Moreover, the motif distribution pattern supports their evolutionary relationship (Fig. 10b). Motif 8 identified in Fig. 10 is similar to motif 7 identified in Fig. 4, this confirms their group A-specific characteristic. Besides, motif 10, which is arginine-rich, is peculiar to AHG-like protein and located at the N-terminus of peptide chains (Fig. 10c), suggesting that motif 10 might be required for functions of AHG-like proteins.
In conclusion, our study revealed functional diversity of the DOG1L proteins in wheat and physical interactions between TaDOG1L4 and TaPP2C-a10. Further investigation showed that TaPP2C-a10 regulated seed germination and plant growth though ABA signaling. Additionally, TaPP2C-a10 also negatively regulated drought stress response. The phylogenetic analysis of AHG-like proteins suggests that TaPP2C-a10 gene is conserved during evolutionary process. These results provide valuable information for the functional studies of TaDOG1Ls and AHG-like proteins, and additional insights into the roles of group A PP2Cs.
References
Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164:711–729
Ashikawa I, Abe F, Nakamura S (2010) Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci 179:536–542
Ashikawa I, Abe F, Nakamura S (2013) DOG1-like genes in cereals: investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci 208:1–9
Ashikawa I, Mori M, Nakamura S, Abe F (2014) A transgenic approach to controlling wheat seed dormancy level by using Triticeae DOG1-like genes. Transgenic Res 23:621–629
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14:1391–1403
Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103:17042–17047
Borrill P, Ramirez-Gonzalez R, Uauy C (2016) expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol 170:2172–2186
Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel K, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30:373–383
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cyrek M, Fedak H, Ciesielski A et al (2016) Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol 170:947–955
Dekkers BJW, He H, Hanson J et al (2016) The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J 85:451–465
El-Gebali S, Mistry J, Bateman A et al (2018) The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432
Finch-Savage WE, Leubner G (2006) Seed dormancy and control of germination. New Phytol 171:501–523
Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10:1043–1054
Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl):S15–S45
Frey A, Effroy D, Lefebvre V et al (2011) Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J 70:501–512
Fujii H, Verslues P, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19:485–494
Fujita Y, Nakashima K, Yoshida T et al (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50:2123–2132
Fujita Y, Yoshida T, Yamaguchi-Shinozaki K (2013) Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant 147:15–27
Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4:1251–1261
Golldack D, Li C, Mohan H, Probst N (2013) Gibberellins and abscisic acid signal crosstalk: living and developing under unfavorable conditions. Plant Cell Rep 32:1007–1016
Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11:1897–1910
Graeber K, Linkies A, Steinbrecher T et al (2014) DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc Natl Acad Sci USA 111:E3571–E3580
Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8:183–187
He Z, Wu J, Sun X, Dai M (2019) The maize clade A PP2C phosphatases play critical roles in multiple abiotic stress responses. Int J Mol Sci 20:3573
Hong JH, Seah SW, Xu J (2013) The root of ABA action in environmental stress response. Plant Cell Rep 32:971–983
Howe KL, Contreras-Moreira B, De Silva N et al (2019) Ensembl genomes 2020—enabling non-vertebrate genomic research. Nucleic Acids Res 48:D689–D695
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Huo H, Wei S, Bradford KJ (2016) DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc Natl Acad Sci USA 113:E2199–E2206
Kim W, Lee Y, Park J, Lee N, Choi G (2013) HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol 54:555–572
Komatsu K, Suzuki N, Kuwamura M et al (2013) Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat Commun 4:2219
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9:759–771
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 25:402–408
Marchler-Bauer A, Derbyshire MK, Gonzales NR et al (2014) CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226
Matakiadis T, Alboresi A, Jikumaru Y et al (2009) The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol 149:949–960
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25:295–303
Mitchell AL, Attwood TK, Babbitt PC et al (2018) InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360
Nakabayashi K, Bartsch M, Xiang Y, Miatton E, Pellengahr S, Yano R, Seo M, Soppe WJJ (2012) The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24:2826–2838
Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32:959–970
Nakashima K, Fujita Y, Kanamori N, Katagiri T et al (2009) Three arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50:1345–1363
Née G, Kramer K, Nakabayashi K, Yuan B, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ (2017) DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat Commun 8:72
Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination 1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50:935–949
Nishimura N, Tsuchiya W, Moresco JJ et al (2018) Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat Commun 9:2132–2132
Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, Mayer KFX, Olsen OA (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345:1250091
Ramirez-Gonzalez RH, Borrill P, Lang D et al (2018) The transcriptional landscape of polyploid wheat. Science 361:eaar6089
Reeves WM, Lynch TJ, Mobin R, Finkelstein RR (2011) Direct targets of the transcription factors ABA-insensitive (ABI) 4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol Biol 75:347–363
Shu K, Liu X, Xie Q, He Z (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9:34–45
Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive 4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124:1752–1765
Subramanian B, Gao S, Lercher MJ, Hu S, Chen WH (2019) Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res 47:270–275
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51:1821–1839
Vishal B, Kumar PP (2018) Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front Plant Sci 9:838
Wheeler TJ, Eddy SR (2013) nhmmer: DNA homology search with profile HMMs. Bioinformatics 29:2487–2489
Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22:1733–1748
Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJJ (2014) REDUCED DORMANCY 5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26:4362
Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y (2008) Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom 9:550
Yan A, Chen Z (2017) The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep 36:689–703
Yatusevich R, Fedak H, Ciesielski A, Dziasek K, Kulik A, Dobrowolska G, Swiezewski S (2017) Antisense transcription represses Arabidopsis seed dormancy QTL DOG1 to regulate drought tolerance. EMBO Rep 18:e201744862
Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination 3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140:115–126
Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2014) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic-acid signaling in response to osmotic stress. Plant Cell Environ 38:35–49
Yu X, Han J, Wang E, Xiao J, Hu R, Yang G, He G (2019) Genome-wide identification and homoeologous expression analysis of PP2C genes in wheat (Triticum aestivum L.). Front Genet 10:561
Zhang X, Wei X, Wang M, Zhu X, Zhao Y, Wei F, Xia Z (2019) Overexpression of NtabDOG1L promotes plant growth and enhances drought tolerance in Nicotiana tabacum. Plant Sci 287:110186
Acknowledgements
The research was supported by the National Genetically Modified New Varieties of Major Projects of China (2016ZX08010004-004), the National Natural Science Foundation of China (nos. 31771418 and 31570261), and Key Project of Hubei Province (2017AHB041).
Author information
Authors and Affiliations
Contributions
GH and GY conceived the study; XY and JH performed the experiments and data analysis; LL and QZ assisted to the management of the materials; XY wrote the draft manuscript; GY and GH revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Communicated by Zheng-Yi Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, X., Han, J., Li, L. et al. Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis. Plant Cell Rep 39, 635–651 (2020). https://doi.org/10.1007/s00299-020-02520-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02520-4