Abstract
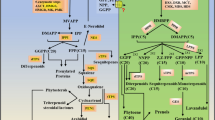
The 12-oxo-phytodienoic acid reductases (OPRs) belong to the old yellow enzyme family of flavoenzymes and form multiple subfamilies in angiosperm plants. In our previous study, a comparative genomic analysis showed that five OPR subfamilies (subs. I–V) occur in monocots, and two subfamilies (subs. I and II) in dicots. Here, a comparative study of five OsOPR genes, representing five subfamilies (I–V) in rice, was performed to provide insights into OPR biochemical properties and physiological importance. Comparative analysis of the three-dimensional structure by homology modeling indicated all five OsOPR proteins contained a highly conserved backbone with (α/β)8-barrels, while two middle variable regions (MVR i and ii) were also detected and defined. Analysis of enzymatic characteristics revealed that all five OsOPR fusion proteins exhibit distinct substrate specificity. Different catalytic activity was observed using racemic OPDA and trans-2-hexen-1-al as substrates, suggesting OsOPR family genes participate in two main branches of the octadecanoid pathway, including the allene oxide synthase and hydroperoxide lyase pathways which regulate various developmental processes and/or defense responses. The transcript profiles of five OsOPR genes exhibited strong tissue-specific and inducible expression patterns under abiotic stress, hormones and plant wounding treatments. Furthermore, the transcriptions of OsOPR04-1 (OsOPR11) and OsOPR08-1 (OsOPR7), representing subs. I and II, respectively, were observed in all six selected tissues and with all above-stress treatments. This suggests that these two subfamilies play an important role during different developmental stages and in response to stresses; while the expressions of OsOPR06-1 (OsOPR6), OsOPR01-1 (OsOPR10) and OsOPR02-1 (OsOPR8), representing subs. III, IV and V respectively, were strongly up-regulated with abscisic acid (ABA) and indoleacetic acid (IAA) treatments in roots, suggesting these three subfamilies play an important role in responding to hormones especially ABA and IAA signals in roots.







Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Agrawal GK, Jwa NS, Shibato J, Han O, Iwahashi H, Rakwal R (2003) Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochem Biophys Res Commun 310:1073–1082
Agrawal GK, Tamogami S, Han O, Iwahashi H, Rakwal R (2004) Rice octadecanoid pathway. Biochem Biophys Res Commun 317:1–15
Baker D, Sali A (2001) Protein structure prediction and structural genomics. Science 294:93–96
Biesgen C, Weiler EW (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208:155–165
Breithaupt C, Kurzbauer R, Schaller F, Stintzi A, Schaller A, Huber R, Macheroux P, Clausen T (2009) Structural basis of substrate specificity of plant 12-oxophytodienoate reductases. J Mol Biol 392:1266–1277
Brown BJ, Deng Z, Karplus PA, Massey V (1998) On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194. J Biol Chem 273:32753–32762
Capener CE, Shrivastava IH, Ranatunga KM, Forrest LR, Smith GR, Sansom MS (2000) Homology modeling and molecular dynamics simulation studies of an inward rectifier potassium channel. Biophys J 78:2929–2942
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chehab EW, Kaspi R, Savchenko T, Rowe H, Negre-Zakharov F, Kliebenstein D, Dehesh K (2008) Distinct roles of jasmonates and aldehydes in plant-defense responses. PloS one 3:e1904
Feussner I, Wasternack C (2002) The lipoxygenase pathway. An Rev Plant Biol 53:275–297
Fox KM, Karplus PA (1994) Old yellow enzyme at 2 A resolution: overall structure, ligand binding, and comparison with related flavoproteins. Structure 2:1089–1105
Fox KM, Karplus PA (1999) The flavin environment in old yellow enzyme. An evaluation of insights from spectroscopic and artificial flavin studies. J Biol Chem 274:9357–9362
Gfeller A, Liechti R, Farmer EE (2006) Arabidopsis jasmonate signaling pathway. Sci STKE 2006:cm1
Goodsell DS, Morris GM, Olson AJ (1996) Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit 9:1–5
Gopal S, Schroeder M, Pieper U, Sczyrba A, Aytekin-Kurban G, Bekiranov S, Fajardo JE, Eswar N, Sanchez R, Sali A, Gaasterland T (2001) Homology-based annotation yields 1, 042 new candidate genes in the Drosophila melanogaster genome. Nat Genet 27:337–340
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–S173
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33:W557–W559
Karplus PA, Fox KM, Massey V (1995) Flavoprotein structure and mechanism. 8. Structure-function relations for old yellow enzyme. FASEB J 9:1518–1526
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486
Li W, Liu B, Yu L, Feng D, Wang H, Wang J (2009) Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol Biol 9:90
Liechti R, Farmer EE (2006) Jasmonate biochemical pathway. Sci STKE 2006:cm3
Liechti R, Gfeller A, Farmer EE (2006) Jasmonate signaling pathway. Sci STKE 2006:cm2
Matsui K (2006) Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr Opin Plant Biol 9:274–280
Matsui H, Nakamura G, Ishiga Y, Toshima H, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2004) Structure and expression of 12-oxophytodienoate reductase (subgroup I) genes in pea, and characterization of the oxidoreductase activities of their recombinant products. Mol Genet Genomics 271:1–10
Miura R, Yamaichi K, Tagawa K, Miyake Y (1987) On the structure of old yellow enzyme studied by specific limited proteolysis. J Biochem 102:1311–1320
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Neuhoff V, Stamm R, Eibl H (1985) Clear background and highly sensitive protein staining with Coomassie blue dyes in poly-acrylamide gels: a systematic analysis. Electrophoresis 6:427–448
Ogawa H, Toyoshima C (2002) Homology modeling of the cation binding sites of Na+K+-ATPase. Proc Natl Acad Sci USA 99:15977–15982
Raine AR, Scrutton NS, Mathews FS (1994) On the evolution of alternate core packing in eightfold beta/alpha-barrels. Protein Sci 3:1889–1892
Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12:1041–1061
Schaller F (2001) Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot 52:11–23
Schaller F, Weiler EW (1997) Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana. Structural and functional relationship to yeast old yellow enzyme. J Biol Chem 272:28066–28072
Schaller F, Hennig P, Weiler EW (1998) 12-Oxophytodienoate-10,11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiol 118:1345–1351
Schaller F, Biesgen C, Mussig C, Altmann T, Weiler EW (2000) 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210:979–984
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Sobajima H, Takeda M, Sugimori M, Kobashi N, Kiribuchi K, Cho EM, Akimoto C, Yamaguchi T, Minami E, Shibuya N, Schaller F, Weiler EW, Yoshihara T, Nishida H, Nojiri H, Omori T, Nishiyama M, Yamane H (2003) Cloning and characterization of a jasmonic acid-responsive gene encoding 12-oxophytodienoic acid reductase in suspension-cultured rice cells. Planta 216:692–698
Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97:10625–10630
Stintzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98:12837–12842
Strassner J, Furholz A, Macheroux P, Amrhein N, Schaller A (1999) A homolog of old yellow enzyme in tomato. Spectral properties and substrate specificity of the recombinant protein. J Biol Chem 274:35067–35073
Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32:585–601
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577
Tani T, Sobajima H, Okada K, Chujo T, Arimura S, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H (2008) Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227:517–526
Warburg O, Christian W (1933) Uber das gelbe oxydationsferment. Biochem Z 263:228–229
Zhang J, Simmons C, Yalpani N, Crane V, Wilkinson H, Kolomiets M (2005) Genomic analysis of the 12-oxo-phytodienoic acid reductase gene family of Zea mays. Plant Mol Biol 59:323–343
Zhou J, Goldsbrough PB (1995) Structure, organization and expression of the metallothionein gene family in Arabidopsis. Mol Gen Genet 248:318–328
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (No. 30800600 and No. 30970237), the Natural Science Foundation of Guangdong Province, P R China (No. 8151027501000016) and the Fundamental Research Funds for the Central Universities (No. 10lgpy34).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Jordan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, W., Zhou, F., Liu, B. et al. Comparative characterization, expression pattern and function analysis of the 12-oxo-phytodienoic acid reductase gene family in rice. Plant Cell Rep 30, 981–995 (2011). https://doi.org/10.1007/s00299-011-1002-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1002-5